Gene Page: BAG3
Summary ?
| GeneID | 9531 |
| Symbol | BAG3 |
| Synonyms | BAG-3|BIS|CAIR-1|MFM6 |
| Description | BCL2 associated athanogene 3 |
| Reference | MIM:603883|HGNC:HGNC:939|Ensembl:ENSG00000151929|HPRD:04860|Vega:OTTHUMG00000019155 |
| Gene type | protein-coding |
| Map location | 10q25.2-q26.2 |
| Pascal p-value | 0.319 |
| Sherlock p-value | 0.433 |
| Fetal beta | -1.837 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10520238 | chr4 | 173525310 | BAG3 | 9531 | 0.06 | trans | ||
| snp_a-2084518 | 0 | BAG3 | 9531 | 0.06 | trans | |||
| rs7706288 | chr5 | 141864248 | BAG3 | 9531 | 0.1 | trans | ||
| rs2700772 | chr8 | 5837584 | BAG3 | 9531 | 0.11 | trans | ||
| rs2700770 | chr8 | 5837808 | BAG3 | 9531 | 0.11 | trans | ||
| rs10842557 | 0 | BAG3 | 9531 | 0.02 | trans | |||
| rs10492401 | chr13 | 33489671 | BAG3 | 9531 | 0.16 | trans | ||
| rs17103412 | chr14 | 66725774 | BAG3 | 9531 | 0.1 | trans | ||
| rs7215675 | chr17 | 10905819 | BAG3 | 9531 | 0.16 | trans | ||
| rs16944595 | chr17 | 11242834 | BAG3 | 9531 | 0 | trans | ||
| rs16944600 | chr17 | 11245098 | BAG3 | 9531 | 0.06 | trans | ||
| rs16944601 | chr17 | 11245219 | BAG3 | 9531 | 0.03 | trans | ||
| rs2073206 | chr20 | 948279 | BAG3 | 9531 | 0.01 | trans |
Section II. Transcriptome annotation
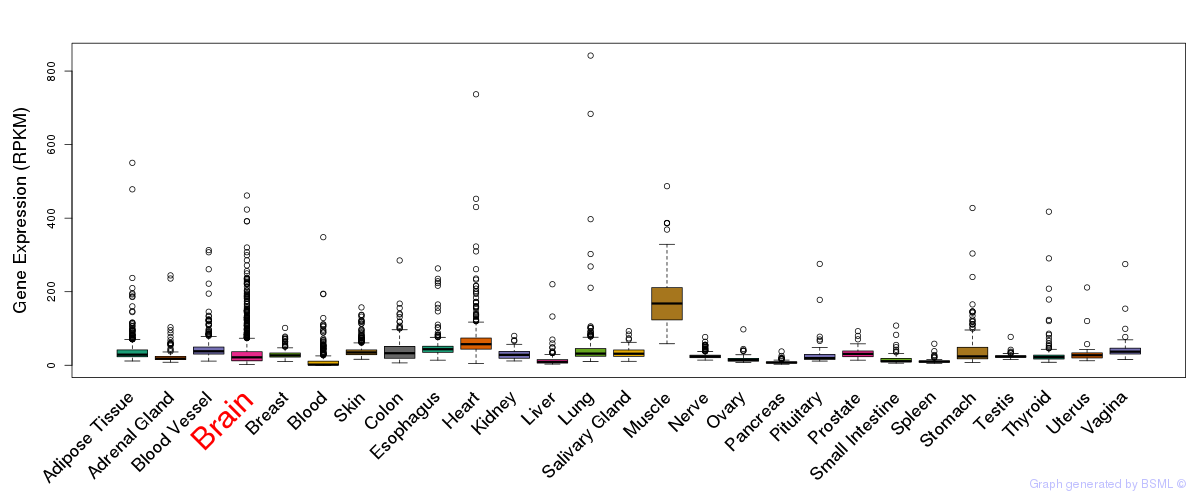
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| QRICH1 | 0.95 | 0.97 |
| KIAA1967 | 0.95 | 0.96 |
| WDR5 | 0.95 | 0.95 |
| DHX8 | 0.95 | 0.96 |
| SART3 | 0.95 | 0.95 |
| KIAA0174 | 0.95 | 0.94 |
| EFTUD2 | 0.94 | 0.95 |
| EDC3 | 0.94 | 0.95 |
| USP10 | 0.94 | 0.94 |
| C20orf11 | 0.94 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.84 | -0.90 |
| MT-CO2 | -0.83 | -0.91 |
| AF347015.27 | -0.80 | -0.88 |
| AF347015.33 | -0.80 | -0.86 |
| AF347015.8 | -0.80 | -0.90 |
| MT-CYB | -0.80 | -0.87 |
| FXYD1 | -0.78 | -0.87 |
| AF347015.21 | -0.77 | -0.92 |
| AF347015.15 | -0.77 | -0.87 |
| HIGD1B | -0.77 | -0.86 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| NOJIMA SFRP2 TARGETS UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS BLUE UP | 136 | 80 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| YORDY RECIPROCAL REGULATION BY ETS1 AND SP100 DN | 87 | 48 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP | 110 | 71 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| GOLDRATH IMMUNE MEMORY | 65 | 42 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP | 78 | 56 | All SZGR 2.0 genes in this pathway |
| GERY CEBP TARGETS | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 24HR | 35 | 22 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 2HR | 51 | 36 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 1 | 33 | 24 | All SZGR 2.0 genes in this pathway |
| TRACEY RESISTANCE TO IFNA2 DN | 32 | 23 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION DN | 105 | 67 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |