Gene Page: ENTPD6
Summary ?
| GeneID | 955 |
| Symbol | ENTPD6 |
| Synonyms | CD39L2|IL-6SAG|IL6ST2|NTPDase-6|dJ738P15.3 |
| Description | ectonucleoside triphosphate diphosphohydrolase 6 (putative) |
| Reference | MIM:603160|HGNC:HGNC:3368|Ensembl:ENSG00000197586|HPRD:16013|Vega:OTTHUMG00000032116 |
| Gene type | protein-coding |
| Map location | 20p11.21 |
| Pascal p-value | 0.051 |
| Sherlock p-value | 0.994 |
| DMG | 2 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00926318 | 20 | 25177340 | ENTPD6 | 4.662E-4 | 0.474 | 0.046 | DMG:Wockner_2014 |
| cg04974899 | 20 | 25177027 | ENTPD6 | 4.32E-8 | -0.013 | 1.19E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
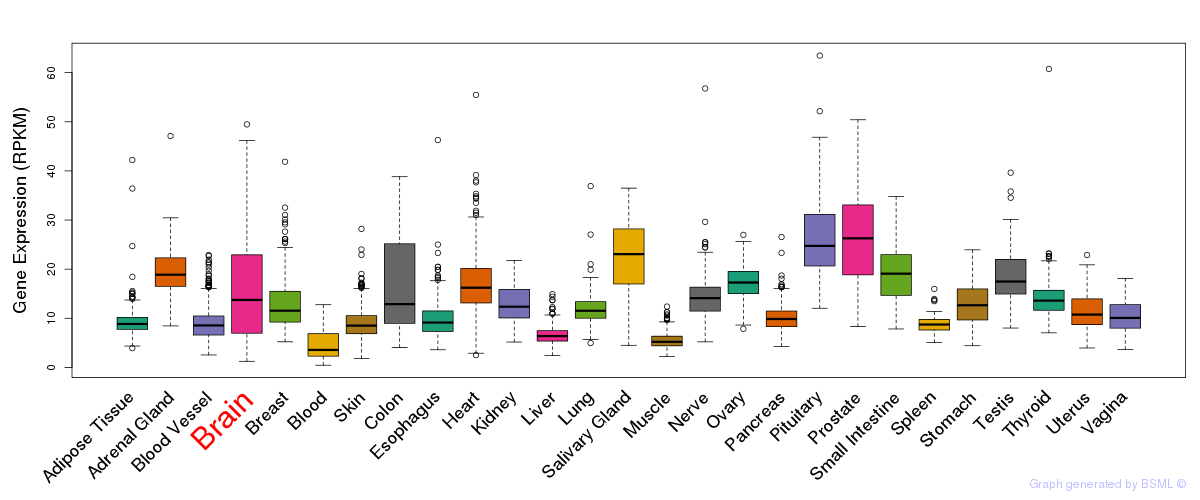
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SFRP4 | 0.82 | 0.73 |
| AC015908.2 | 0.76 | 0.59 |
| STK33 | 0.75 | 0.70 |
| WDR49 | 0.74 | 0.59 |
| AK7 | 0.72 | 0.54 |
| SULT1C4 | 0.71 | 0.65 |
| CCDC135 | 0.71 | 0.36 |
| DNAH9 | 0.71 | 0.57 |
| ANXA1 | 0.71 | 0.55 |
| NEK5 | 0.71 | 0.46 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IER5L | -0.38 | -0.49 |
| MPPED1 | -0.35 | -0.52 |
| MEF2C | -0.35 | -0.48 |
| KLHL1 | -0.34 | -0.45 |
| SATB2 | -0.33 | -0.50 |
| GPR22 | -0.33 | -0.50 |
| SLC26A4 | -0.32 | -0.47 |
| GPR21 | -0.32 | -0.45 |
| SLA | -0.32 | -0.40 |
| TIAM2 | -0.32 | -0.46 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG PYRIMIDINE METABOLISM | 98 | 53 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER UP | 96 | 57 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS DN | 66 | 39 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 4 | 48 | 32 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE UP | 43 | 34 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 VIA ERCC6 UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 1 DN | 48 | 30 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP C | 92 | 60 | All SZGR 2.0 genes in this pathway |