Gene Page: CREB5
Summary ?
| GeneID | 9586 |
| Symbol | CREB5 |
| Synonyms | CRE-BPA|CREB-5 |
| Description | cAMP responsive element binding protein 5 |
| Reference | HGNC:HGNC:16844|Ensembl:ENSG00000146592|HPRD:09896|Vega:OTTHUMG00000097081 |
| Gene type | protein-coding |
| Map location | 7p15.1 |
| Pascal p-value | 7.609E-4 |
| Sherlock p-value | 0.372 |
| DEG p-value | DEG:Zhao_2015:p=2.53e-04:q=0.0872 |
| Fetal beta | 0.386 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6480275 | chr10 | 53237440 | CREB5 | 9586 | 0.12 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
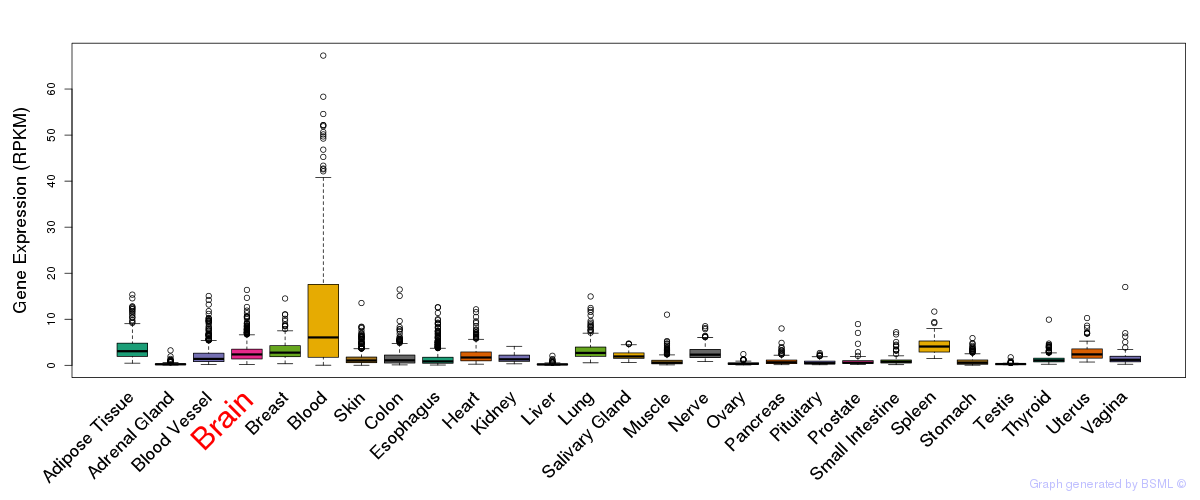
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CTCF | 0.98 | 0.98 |
| SIN3A | 0.97 | 0.98 |
| SF3B3 | 0.97 | 0.97 |
| XPO5 | 0.97 | 0.95 |
| PHF20 | 0.96 | 0.97 |
| CNOT1 | 0.96 | 0.97 |
| KDM4A | 0.96 | 0.97 |
| TNPO3 | 0.96 | 0.96 |
| PRPF8 | 0.96 | 0.98 |
| MSL2 | 0.96 | 0.97 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.77 | -0.91 |
| FXYD1 | -0.76 | -0.91 |
| MT-CO2 | -0.76 | -0.91 |
| IFI27 | -0.75 | -0.89 |
| AF347015.27 | -0.74 | -0.87 |
| AF347015.33 | -0.74 | -0.87 |
| S100B | -0.74 | -0.85 |
| C5orf53 | -0.74 | -0.77 |
| AIFM3 | -0.74 | -0.79 |
| TSC22D4 | -0.73 | -0.81 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| KEGG PROSTATE CANCER | 89 | 75 | All SZGR 2.0 genes in this pathway |
| ST DIFFERENTIATION PATHWAY IN PC12 CELLS | 45 | 35 | All SZGR 2.0 genes in this pathway |
| ST ERK1 ERK2 MAPK PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| ST G ALPHA S PATHWAY | 16 | 11 | All SZGR 2.0 genes in this pathway |
| ST P38 MAPK PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| ST GRANULE CELL SURVIVAL PATHWAY | 27 | 23 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL DN | 118 | 79 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS UP | 214 | 134 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA DN | 77 | 52 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD DN | 84 | 63 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| WEI MIR34A TARGETS | 148 | 97 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 2 | 114 | 76 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |
| GHANDHI BYSTANDER IRRADIATION UP | 86 | 54 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |