Gene Page: FEZ1
Summary ?
| GeneID | 9638 |
| Symbol | FEZ1 |
| Synonyms | UNC-76 |
| Description | fasciculation and elongation protein zeta 1 |
| Reference | MIM:604825|HGNC:HGNC:3659|Ensembl:ENSG00000149557|HPRD:09212|Vega:OTTHUMG00000165886 |
| Gene type | protein-coding |
| Map location | 11q24.2 |
| Pascal p-value | 0.009 |
| Fetal beta | -1.407 |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 31 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1594142 | chr2 | 57263761 | FEZ1 | 9638 | 0.17 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
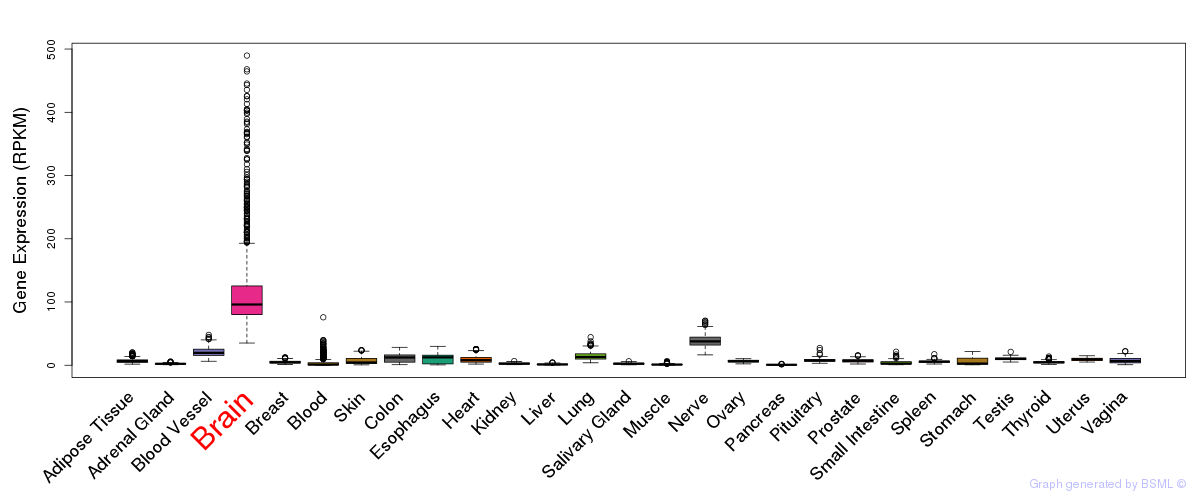
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| COIL | 0.97 | 0.97 |
| CAND1 | 0.97 | 0.97 |
| KLHL7 | 0.97 | 0.97 |
| XRCC5 | 0.97 | 0.97 |
| POLR2B | 0.97 | 0.97 |
| ACTR3 | 0.97 | 0.97 |
| MARCH7 | 0.97 | 0.97 |
| ABCE1 | 0.96 | 0.96 |
| CDC5L | 0.96 | 0.97 |
| CCT6A | 0.96 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.73 | -0.90 |
| FXYD1 | -0.73 | -0.90 |
| AF347015.31 | -0.73 | -0.89 |
| AF347015.33 | -0.72 | -0.88 |
| HLA-F | -0.71 | -0.81 |
| AF347015.27 | -0.71 | -0.87 |
| MT-CYB | -0.71 | -0.87 |
| AIFM3 | -0.70 | -0.80 |
| AF347015.8 | -0.70 | -0.88 |
| IFI27 | -0.70 | -0.86 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 14690447 |15383276 |16169070 | |
| GO:0043015 | gamma-tubulin binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | TAS | axon (GO term level: 13) | 9096408 |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9096408 |
| GO:0007155 | cell adhesion | TAS | 9096408 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005813 | centrosome | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTG1 | ACT | ACTG | DFNA20 | DFNA26 | actin, gamma 1 | Affinity Capture-Western | BioGRID | 12874605 |
| C13orf34 | BORA | FLJ22624 | chromosome 13 open reading frame 34 | Two-hybrid | BioGRID | 16169070 |
| CCL7 | FIC | MARC | MCP-3 | MCP3 | MGC138463 | MGC138465 | NC28 | SCYA6 | SCYA7 | chemokine (C-C motif) ligand 7 | Two-hybrid | BioGRID | 16169070 |
| CIB1 | CIB | KIP | KIP1 | SIP2-28 | calcium and integrin binding 1 (calmyrin) | - | HPRD | 11856312 |
| COL9A2 | DJ39G22.4 | EDM2 | MED | collagen, type IX, alpha 2 | Two-hybrid | BioGRID | 16169070 |
| CSTF2 | CstF-64 | cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa | Two-hybrid | BioGRID | 16169070 |
| DIS3L2 | FAM6A | FLJ36974 | MGC42174 | DIS3 mitotic control homolog (S. cerevisiae)-like 2 | Two-hybrid | BioGRID | 16169070 |
| DISC1 | C1orf136 | FLJ13381 | FLJ21640 | FLJ25311 | FLJ41105 | KIAA0457 | SCZD9 | disrupted in schizophrenia 1 | - | HPRD,BioGRID | 12874605 |
| GTF2F1 | BTF4 | RAP74 | TF2F1 | TFIIF | general transcription factor IIF, polypeptide 1, 74kDa | Two-hybrid | BioGRID | 16169070 |
| HEATR2 | FLJ20397 | FLJ25564 | FLJ31671 | FLJ39381 | HEAT repeat containing 2 | Two-hybrid | BioGRID | 16169070 |
| HTATSF1 | TAT-SF1 | dJ196E23.2 | HIV-1 Tat specific factor 1 | Two-hybrid | BioGRID | 16169070 |
| Magmas | CGI-136 | mitochondria-associated protein involved in granulocyte-macrophage colony-stimulating factor signal transduction | Two-hybrid | BioGRID | 16169070 |
| NBR1 | 1A1-3B | KIAA0049 | M17S2 | MIG19 | neighbor of BRCA1 gene 1 | - | HPRD,BioGRID | 11856312 |
| NDUFB9 | B22 | DKFZp566O173 | FLJ22885 | LYRM3 | UQOR22 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa | Two-hybrid | BioGRID | 16169070 |
| NEK1 | DKFZp686D06121 | DKFZp686K12169 | KIAA1901 | MGC138800 | NY-REN-55 | NIMA (never in mitosis gene a)-related kinase 1 | NEK1 interacts with FEZ-1. | BIND | 14690447 |
| OLFML3 | HNOEL-iso | OLF44 | olfactomedin-like 3 | Two-hybrid | BioGRID | 16169070 |
| P4HB | DSI | ERBA2L | GIT | PDI | PDIA1 | PHDB | PO4DB | PO4HB | PROHB | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), beta polypeptide | Two-hybrid | BioGRID | 16169070 |
| PDCD7 | ES18 | HES18 | MGC22015 | programmed cell death 7 | Two-hybrid | BioGRID | 16169070 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | - | HPRD,BioGRID | 9971736 |
| PTH | PTH1 | parathyroid hormone | Two-hybrid | BioGRID | 16169070 |
| PTPRS | PTPSIGMA | protein tyrosine phosphatase, receptor type, S | Two-hybrid | BioGRID | 16169070 |
| SMEK2 | FLFL2 | FLJ31474 | KIAA1387 | PP4R3B | PSY2 | smk1 | SMEK homolog 2, suppressor of mek1 (Dictyostelium) | Two-hybrid | BioGRID | 16169070 |
| STAR | STARD1 | steroidogenic acute regulatory protein | Two-hybrid | BioGRID | 16169070 |
| TAC3 | NKB | NKNB | PRO1155 | ZNEUROK1 | tachykinin 3 | Two-hybrid | BioGRID | 16169070 |
| TOMM20 | KIAA0016 | MAS20 | MGC117367 | MOM19 | TOM20 | translocase of outer mitochondrial membrane 20 homolog (yeast) | Two-hybrid | BioGRID | 16169070 |
| TTR | HsT2651 | PALB | TBPA | transthyretin | Two-hybrid | BioGRID | 16169070 |
| TXNDC9 | APACD | thioredoxin domain containing 9 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 UP | 146 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| SHIRAISHI PLZF TARGETS UP | 10 | 5 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS | 179 | 105 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 UP | 118 | 66 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS UP | 58 | 40 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH MLL FUSIONS | 78 | 45 | All SZGR 2.0 genes in this pathway |
| DER IFN GAMMA RESPONSE DN | 11 | 9 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL UP | 54 | 29 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| ROSS LEUKEMIA WITH MLL FUSIONS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS DN | 74 | 50 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL DN | 29 | 11 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 16 | 79 | 47 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 DN | 39 | 21 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 UP | 121 | 70 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |