Gene Page: CD63
Summary ?
| GeneID | 967 |
| Symbol | CD63 |
| Synonyms | LAMP-3|ME491|MLA1|OMA81H|TSPAN30 |
| Description | CD63 molecule |
| Reference | MIM:155740|HGNC:HGNC:1692|Ensembl:ENSG00000135404|HPRD:01121|Vega:OTTHUMG00000170454 |
| Gene type | protein-coding |
| Map location | 12q13.2 |
| Pascal p-value | 0.636 |
| Sherlock p-value | 0.288 |
| Fetal beta | -0.745 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21601059 | 12 | 56122983 | CD63 | 3.09E-4 | -0.186 | 0.04 | DMG:Wockner_2014 |
| cg19987288 | 12 | 56122958 | CD63 | 4.436E-4 | -0.607 | 0.045 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9313483 | chr5 | 169448229 | CD63 | 967 | 0.16 | trans |
Section II. Transcriptome annotation
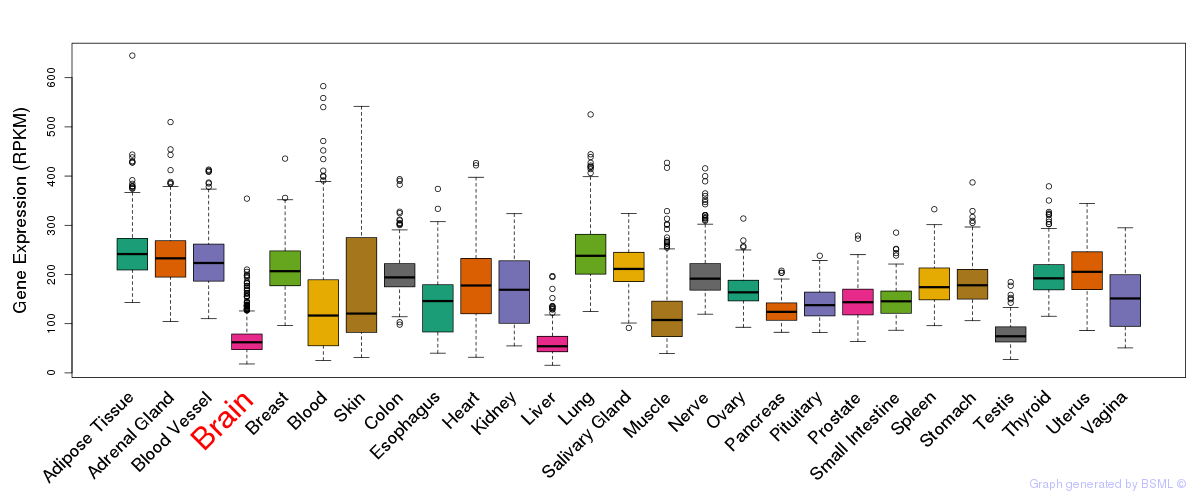
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-DPA1 | 0.93 | 0.94 |
| HLA-DRA | 0.88 | 0.90 |
| HLA-DRB1 | 0.84 | 0.88 |
| HLA-DQA1 | 0.80 | 0.85 |
| HLA-DRB5 | 0.79 | 0.83 |
| HLA-DPB1 | 0.79 | 0.86 |
| HLA-DMB | 0.78 | 0.81 |
| EVI2B | 0.78 | 0.85 |
| HLA-DMA | 0.77 | 0.78 |
| MAP7 | 0.75 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1949 | -0.54 | -0.66 |
| TUBB2B | -0.54 | -0.72 |
| NKIRAS2 | -0.53 | -0.63 |
| ZNF311 | -0.52 | -0.65 |
| ZNF821 | -0.52 | -0.63 |
| SH3BP2 | -0.52 | -0.70 |
| ZNF300 | -0.52 | -0.64 |
| AC004017.1 | -0.52 | -0.64 |
| ZNF551 | -0.51 | -0.64 |
| TIGD1 | -0.51 | -0.63 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AP4M1 | MU-4 | MU-ARP2 | adaptor-related protein complex 4, mu 1 subunit | - | HPRD,BioGRID | 10436028 |
| CD81 | S5.7 | TAPA1 | TSPAN28 | CD81 molecule | - | HPRD | 12036870 |
| CD81 | S5.7 | TAPA1 | TSPAN28 | CD81 molecule | in vivo | BioGRID | 9006891 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | - | HPRD,BioGRID | 9759843 |
| CD9 | 5H9 | BA2 | BTCC-1 | DRAP-27 | GIG2 | MIC3 | MRP-1 | P24 | TSPAN29 | CD9 molecule | - | HPRD,BioGRID | 8630057 |11204565 |12175627 |
| HLA-DMB | D6S221E | RING7 | major histocompatibility complex, class II, DM beta | - | HPRD,BioGRID | 9759843 |
| HLA-DOB | DOB | major histocompatibility complex, class II, DO beta | Affinity Capture-Western | BioGRID | 9759843 |
| HLA-DRA | HLA-DRA1 | major histocompatibility complex, class II, DR alpha | - | HPRD,BioGRID | 9759843 |
| ITGA6 | CD49f | DKFZp686J01244 | FLJ18737 | ITGA6B | VLA-6 | integrin, alpha 6 | - | HPRD,BioGRID | 7629079 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | Affinity Capture-Western | BioGRID | 12175627 |
| KIT | C-Kit | CD117 | PBT | SCFR | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | - | HPRD,BioGRID | 12036870 |
| PTGFRN | CD315 | CD9P-1 | EWI-F | FLJ11001 | FPRP | KIAA1436 | SMAP-6 | prostaglandin F2 receptor negative regulator | - | HPRD,BioGRID | 11278880 |
| SRF | MCM1 | serum response factor (c-fos serum response element-binding transcription factor) | Two-hybrid | BioGRID | 9271374 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG LYSOSOME | 121 | 83 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA UP | 205 | 140 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| JOHANSSON GLIOMAGENESIS BY PDGFB UP | 58 | 44 | All SZGR 2.0 genes in this pathway |
| BORLAK LIVER CANCER EGF UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA UP | 61 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER ACOX1 UP | 64 | 40 | All SZGR 2.0 genes in this pathway |
| BYSTROEM CORRELATED WITH IL5 UP | 51 | 30 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC E2F1 UP | 56 | 34 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER CIPROFIBRATE UP | 60 | 42 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER DENA UP | 60 | 40 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER E2F1 UP | 62 | 35 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA DN | 52 | 33 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| LEIN ASTROCYTE MARKERS | 42 | 35 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| BASSO HAIRY CELL LEUKEMIA DN | 80 | 66 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS EMT UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS NEURAL UP | 18 | 13 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| KAPOSI LIVER CANCER MET UP | 18 | 11 | All SZGR 2.0 genes in this pathway |
| FONTAINE THYROID TUMOR UNCERTAIN MALIGNANCY UP | 36 | 23 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA UP | 66 | 38 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| ACOSTA PROLIFERATION INDEPENDENT MYC TARGETS DN | 116 | 74 | All SZGR 2.0 genes in this pathway |