Gene Page: SDC3
Summary ?
| GeneID | 9672 |
| Symbol | SDC3 |
| Synonyms | SDCN|SYND3 |
| Description | syndecan 3 |
| Reference | MIM:186357|HGNC:HGNC:10660|HPRD:01719| |
| Gene type | protein-coding |
| Map location | 1p35.2 |
| Pascal p-value | 0.079 |
| Fetal beta | -0.495 |
| Support | Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
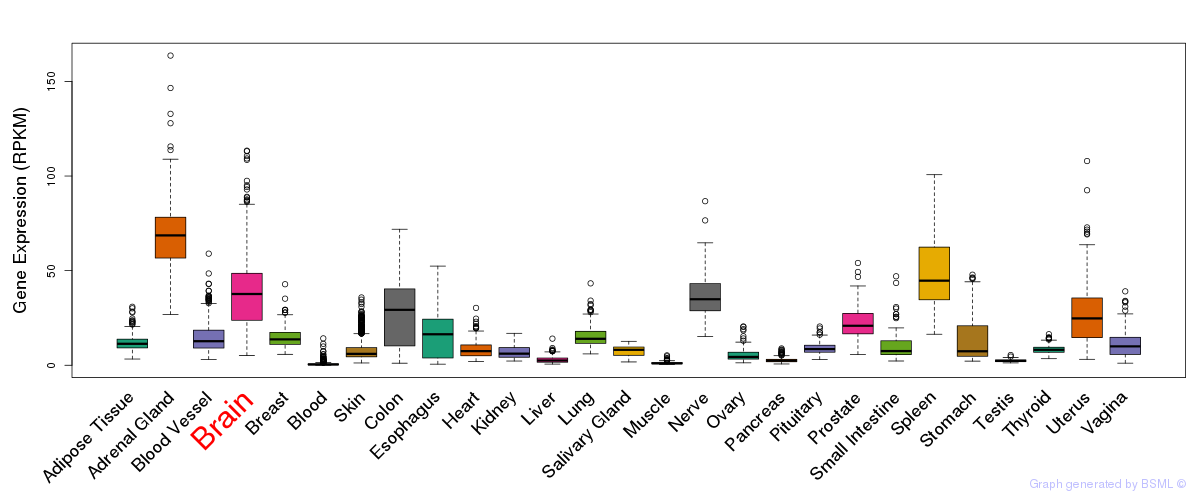
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ADNP2 | 0.95 | 0.96 |
| RRP1B | 0.94 | 0.93 |
| BMPR1A | 0.93 | 0.91 |
| C20orf11 | 0.93 | 0.92 |
| MEX3C | 0.93 | 0.94 |
| DCP1A | 0.93 | 0.93 |
| ZXDA | 0.93 | 0.94 |
| RBM15 | 0.93 | 0.93 |
| AMMECR1L | 0.93 | 0.93 |
| ZNF778 | 0.93 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.68 | -0.84 |
| MT-CO2 | -0.68 | -0.85 |
| FXYD1 | -0.67 | -0.82 |
| AF347015.27 | -0.67 | -0.81 |
| AF347015.33 | -0.66 | -0.80 |
| C5orf53 | -0.66 | -0.72 |
| AF347015.8 | -0.65 | -0.83 |
| S100B | -0.65 | -0.77 |
| IFI27 | -0.65 | -0.77 |
| MT-CYB | -0.65 | -0.81 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | - | HPRD | 10460248 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD,BioGRID | 9553134 |
| CTTN | EMS1 | FLJ34459 | cortactin | - | HPRD,BioGRID | 9553134 |
| CUX1 | CASP | CDP | CDP/Cut | CDP1 | COY1 | CUTL1 | CUX | Clox | Cux/CDP | GOLIM6 | Nbla10317 | p100 | p110 | p200 | p75 | cut-like homeobox 1 | - | HPRD | 8662884 |
| FGF2 | BFGF | FGFB | HBGF-2 | fibroblast growth factor 2 (basic) | - | HPRD,BioGRID | 8344959 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 9553134 |
| MDK | FLJ27379 | MK | NEGF2 | midkine (neurite growth-promoting factor 2) | - | HPRD | 9089390 |
| PTN | HARP | HBGF8 | HBNF | NEGF1 | pleiotrophin | - | HPRD | 8175719 |
| SDCBP | MDA-9 | ST1 | SYCL | TACIP18 | syndecan binding protein (syntenin) | Syndecan-3 interacts with ST-1. This interaction was modelled on a demonstrated interaction between human syndecan-3 and rat ST-1. | BIND | 11152476 |
| TUBA1A | B-ALPHA-1 | FLJ25113 | LIS3 | TUBA3 | tubulin, alpha 1a | - | HPRD,BioGRID | 9553134 |
| TUBB | M40 | MGC117247 | MGC16435 | OK/SW-cl.56 | TUBB1 | TUBB5 | tubulin, beta | Reconstituted Complex | BioGRID | 9553134 |
| TUBB2A | TUBB | TUBB2 | dJ40E16.7 | tubulin, beta 2A | - | HPRD,BioGRID | 9553134 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ECM RECEPTOR INTERACTION | 84 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 3 PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME HS GAG DEGRADATION | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | 49 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME HS GAG BIOSYNTHESIS | 31 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | 52 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOSAMINOGLYCAN METABOLISM | 111 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | 25 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| JOHANSSON GLIOMAGENESIS BY PDGFB UP | 58 | 44 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 UP | 256 | 159 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1B TARGETS | 92 | 68 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| BASSO HAIRY CELL LEUKEMIA DN | 80 | 66 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| WIEDERSCHAIN TARGETS OF BMI1 AND PCGF2 | 57 | 39 | All SZGR 2.0 genes in this pathway |
| BOSCO INTERFERON INDUCED ANTIVIRAL MODULE | 78 | 48 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
| NABA ECM AFFILIATED | 171 | 89 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |