Gene Page: ZNF536
Summary ?
| GeneID | 9745 |
| Symbol | ZNF536 |
| Synonyms | - |
| Description | zinc finger protein 536 |
| Reference | HGNC:HGNC:29025|Ensembl:ENSG00000198597|HPRD:15825|Vega:OTTHUMG00000182112 |
| Gene type | protein-coding |
| Map location | 19q12 |
| Pascal p-value | 1.388E-6 |
| Sherlock p-value | 0.56 |
| TADA p-value | 0.025 |
| Fetal beta | -1.472 |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs2053079 | chr19 | 30987423 | AG | 3.789E-9 | intronic | ZNF536 |
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ZNF536 | chr19 | 30935608 | G | T | NM_014717 | p.380R>L | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs184583 | chr19 | 30371589 | ZNF536 | 9745 | 0.12 | cis | ||
| rs9676954 | chr19 | 31008053 | ZNF536 | 9745 | 0.18 | cis | ||
| rs8112930 | chr19 | 31036268 | ZNF536 | 9745 | 0.09 | cis | ||
| rs2322319 | chr3 | 3637199 | ZNF536 | 9745 | 0.04 | trans | ||
| rs11157436 | chr14 | 22636873 | ZNF536 | 9745 | 0.2 | trans | ||
| rs17137051 | chr16 | 4521748 | ZNF536 | 9745 | 0.13 | trans |
Section II. Transcriptome annotation
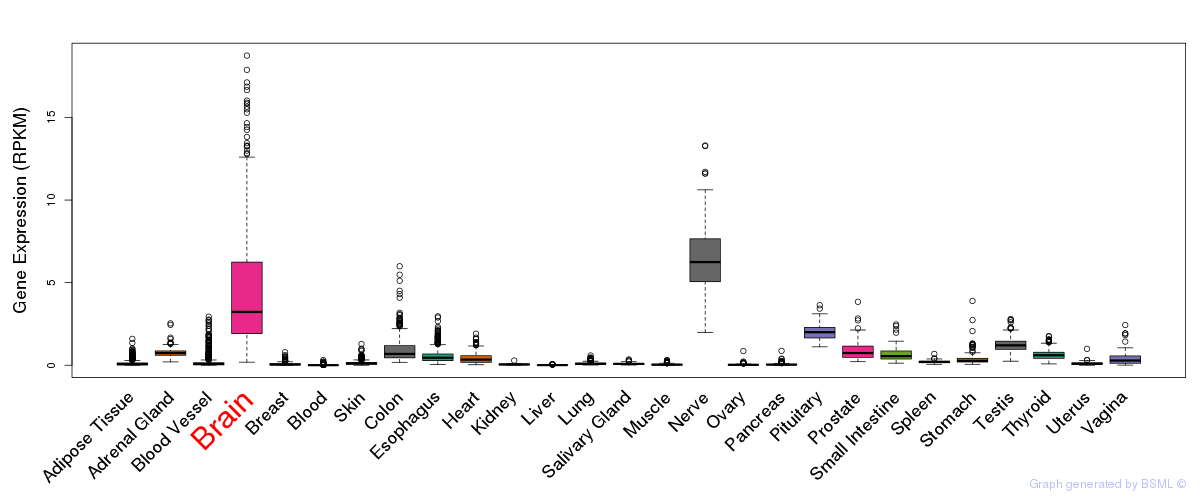
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BRD1 | 0.98 | 0.97 |
| MEX3B | 0.97 | 0.94 |
| ZSCAN2 | 0.97 | 0.95 |
| GPR161 | 0.96 | 0.92 |
| EHMT1 | 0.96 | 0.93 |
| BEND3 | 0.96 | 0.95 |
| BRD3 | 0.96 | 0.97 |
| AL033532.1 | 0.96 | 0.91 |
| USP42 | 0.96 | 0.97 |
| C10orf2 | 0.96 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.70 | -0.81 |
| HLA-F | -0.70 | -0.78 |
| AIFM3 | -0.69 | -0.77 |
| ALDOC | -0.69 | -0.73 |
| FBXO2 | -0.68 | -0.66 |
| S100B | -0.67 | -0.87 |
| TSC22D4 | -0.67 | -0.80 |
| CLU | -0.67 | -0.71 |
| PTH1R | -0.67 | -0.72 |
| LHPP | -0.67 | -0.61 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA UP | 98 | 64 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP | 148 | 96 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| MCCABE HOXC6 TARGETS DN | 21 | 15 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS WITH HCP H3K27ME3 | 102 | 76 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |