Gene Page: HDAC4
Summary ?
| GeneID | 9759 |
| Symbol | HDAC4 |
| Synonyms | AHO3|BDMR|HA6116|HD4|HDAC-4|HDAC-A|HDACA |
| Description | histone deacetylase 4 |
| Reference | MIM:605314|HGNC:HGNC:14063|Ensembl:ENSG00000068024|HPRD:05610|Vega:OTTHUMG00000133344 |
| Gene type | protein-coding |
| Map location | 2q37.3 |
| Pascal p-value | 0.717 |
| Sherlock p-value | 0.376 |
| Fetal beta | 0.386 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 4 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 4 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15929228 | 2 | 240323491 | HDAC4 | 1.976E-4 | -0.296 | 0.035 | DMG:Wockner_2014 |
| cg17356718 | 2 | 240230587 | HDAC4 | 3.267E-4 | 0.33 | 0.04 | DMG:Wockner_2014 |
| cg14273620 | 2 | 240170857 | HDAC4 | 3.668E-4 | 0.305 | 0.042 | DMG:Wockner_2014 |
| cg15142485 | 2 | 240323713 | HDAC4 | 7.55E-9 | -0.01 | 3.72E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17007452 | chr12 | 81243404 | HDAC4 | 9759 | 0.17 | trans |
Section II. Transcriptome annotation
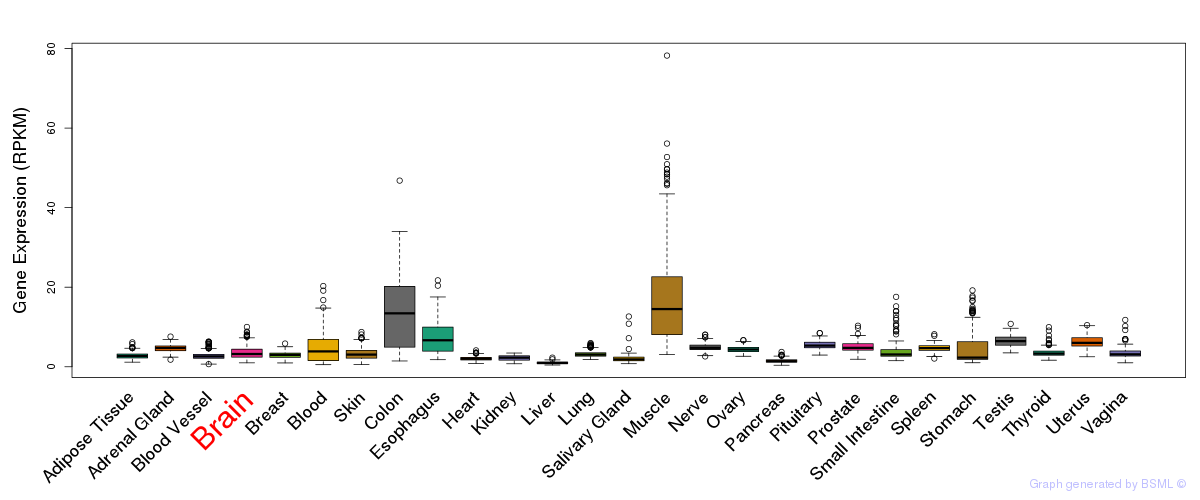
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AATK | 0.87 | 0.75 |
| TMEM184B | 0.86 | 0.85 |
| FAM78A | 0.85 | 0.82 |
| ABCA2 | 0.85 | 0.81 |
| TNK2 | 0.84 | 0.78 |
| NAT8L | 0.84 | 0.80 |
| NCKIPSD | 0.83 | 0.79 |
| IPO13 | 0.83 | 0.83 |
| MAN2A2 | 0.83 | 0.76 |
| SLC6A8 | 0.83 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ST20 | -0.45 | -0.52 |
| C9orf46 | -0.44 | -0.50 |
| RP9P | -0.43 | -0.56 |
| FAM36A | -0.42 | -0.41 |
| RPL35 | -0.42 | -0.49 |
| RPL24 | -0.42 | -0.49 |
| RPL31 | -0.41 | -0.48 |
| GTF3C6 | -0.41 | -0.42 |
| SNRPG | -0.41 | -0.53 |
| RPL13AP22 | -0.41 | -0.62 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0004407 | histone deacetylase activity | NAS | 10523670 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008134 | transcription factor binding | TAS | 12711221 | |
| GO:0016564 | transcription repressor activity | IEA | - | |
| GO:0016564 | transcription repressor activity | TAS | 10523670 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 12711221 |
| GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0001501 | skeletal system development | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0007049 | cell cycle | NAS | - | |
| GO:0008285 | negative regulation of cell proliferation | IEA | - | |
| GO:0006954 | inflammatory response | TAS | 12711221 | |
| GO:0007275 | multicellular organismal development | NAS | - | |
| GO:0016568 | chromatin modification | TAS | 12711221 | |
| GO:0030183 | B cell differentiation | TAS | 12711221 | |
| GO:0045843 | negative regulation of striated muscle development | TAS | 12711221 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000118 | histone deacetylase complex | TAS | 12711221 | |
| GO:0005634 | nucleus | NAS | - | |
| GO:0005737 | cytoplasm | TAS | 12711221 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANKRD11 | ANCO-1 | LZ16 | T13 | ankyrin repeat domain 11 | - | HPRD | 15184363 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Reconstituted Complex | BioGRID | 12943985 |
| BCL6 | BCL5 | BCL6A | LAZ3 | ZBTB27 | ZNF51 | B-cell CLL/lymphoma 6 | - | HPRD,BioGRID | 11929873 |
| BCOR | ANOP2 | FLJ20285 | FLJ38041 | KIAA1575 | MAA2 | MCOPS2 | MGC131961 | MGC71031 | BCL6 co-repressor | - | HPRD,BioGRID | 10898795 |
| CBX5 | HP1 | HP1A | chromobox homolog 5 (HP1 alpha homolog, Drosophila) | - | HPRD,BioGRID | 12242305 |
| CTBP1 | BARS | MGC104684 | C-terminal binding protein 1 | Reconstituted Complex | BioGRID | 11022042 |
| EVI1 | AML1-EVI-1 | EVI-1 | MDS1-EVI1 | MGC163392 | PRDM3 | ecotropic viral integration site 1 | - | HPRD,BioGRID | 11568182 |
| GATA1 | ERYF1 | GF-1 | GF1 | NFE1 | GATA binding protein 1 (globin transcription factor 1) | Affinity Capture-Western Reconstituted Complex | BioGRID | 14668799 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | - | HPRD | 11804585 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | - | HPRD,BioGRID | 11466315 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | Reconstituted Complex | BioGRID | 11486037 |
| HDAC9 | DKFZp779K1053 | HD7 | HDAC | HDAC7 | HDAC7B | HDAC9B | HDAC9FL | HDRP | KIAA0744 | MITR | histone deacetylase 9 | - | HPRD,BioGRID | 12590135 |
| IKZF1 | Hs.54452 | IK1 | IKAROS | LYF1 | PRO0758 | ZNFN1A1 | hIk-1 | IKAROS family zinc finger 1 (Ikaros) | Affinity Capture-Western | BioGRID | 12015313 |
| IKZF2 | HELIOS | MGC34330 | ZNF1A2 | ZNFN1A2 | IKAROS family zinc finger 2 (Helios) | Reconstituted Complex | BioGRID | 12943985 |
| IKZF3 | AIO | AIOLOS | ZNFN1A3 | IKAROS family zinc finger 3 (Aiolos) | Affinity Capture-Western | BioGRID | 12015313 |
| IKZF4 | EOS | KIAA1782 | ZNFN1A4 | IKAROS family zinc finger 4 (Eos) | Affinity Capture-Western | BioGRID | 12015313 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | - | HPRD,BioGRID | 11114188 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD,BioGRID | 11114188 |
| MEF2A | ADCAD1 | RSRFC4 | RSRFC9 | myocyte enhancer factor 2A | - | HPRD,BioGRID | 10487761 |
| MEF2C | - | myocyte enhancer factor 2C | - | HPRD | 10523670 |11504882 |
| MEF2C | - | myocyte enhancer factor 2C | Affinity Capture-Western Reconstituted Complex | BioGRID | 10523670 |11486037 |
| MEF2D | DKFZp686I1536 | myocyte enhancer factor 2D | - | HPRD,BioGRID | 10523670 |
| MEGF10 | DKFZp781K1852 | FLJ41574 | KIAA1780 | multiple EGF-like-domains 10 | - | HPRD | 12421765 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | - | HPRD | 10944117 |11804585 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10640275 |11804585 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD,BioGRID | 11804585 |
| NR2C1 | TR2 | nuclear receptor subfamily 2, group C, member 1 | - | HPRD,BioGRID | 11463856 |12943985 |
| NR2C2 | TAK1 | TR2R1 | TR4 | hTAK1 | nuclear receptor subfamily 2, group C, member 2 | Reconstituted Complex | BioGRID | 12943985 |
| PPARD | FAAR | MGC3931 | NR1C2 | NUC1 | NUCI | NUCII | PPAR-beta | PPARB | peroxisome proliferator-activated receptor delta | Reconstituted Complex | BioGRID | 12943985 |
| PPARG | NR1C3 | PPARG1 | PPARG2 | PPARgamma | peroxisome proliferator-activated receptor gamma | Reconstituted Complex | BioGRID | 12943985 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | Reconstituted Complex | BioGRID | 12943985 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | Affinity Capture-Western | BioGRID | 10220385 |
| RUNX3 | AML2 | CBFA3 | FLJ34510 | MGC16070 | PEBP2aC | runt-related transcription factor 3 | - | HPRD | 15138260 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | RUVBL2 (reptin) interacts with HDAC4. | BIND | 15829968 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | Reconstituted Complex | BioGRID | 12943985 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| SUMO2 | HSMT3 | MGC117191 | SMT3B | SMT3H2 | SMT3 suppressor of mif two 3 homolog 2 (S. cerevisiae) | - | HPRD | 11451954 |
| SUV39H1 | KMT1A | MG44 | SUV39H | suppressor of variegation 3-9 homolog 1 (Drosophila) | Affinity Capture-Western | BioGRID | 12242305 |
| TNFRSF14 | ATAR | HVEA | HVEM | LIGHTR | TR2 | tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | - | HPRD | 11463856 |
| TNFSF10 | APO2L | Apo-2L | CD253 | TL2 | TRAIL | tumor necrosis factor (ligand) superfamily, member 10 | HDAC4 interacts with the TNFSF10 promoter. | BIND | 15619633 |
| UBE2I | C358B7.1 | P18 | UBC9 | ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) | Biochemical Activity | BioGRID | 18691969 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | Affinity Capture-Western | BioGRID | 10869435 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | Affinity Capture-Western Reconstituted Complex | BioGRID | 10869435 |11504882 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | - | HPRD | 11486037 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | Reconstituted Complex | BioGRID | 11504882 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Reconstituted Complex | BioGRID | 11504882 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD | 11486037 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD,BioGRID | 11929873 |15467736 |
| ZNF354A | EZNF | HKL1 | KID-1 | KID1 | TCF17 | zinc finger protein 354A | Reconstituted Complex | BioGRID | 12943985 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA CARM ER PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSII PATHWAY | 34 | 27 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSIII PATHWAY | 25 | 20 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID RANBP2 PATHWAY | 11 | 11 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | 46 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| ZHONG RESPONSE TO AZACITIDINE AND TSA DN | 70 | 38 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| CASORELLI APL SECONDARY VS DE NOVO UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP | 108 | 66 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| PEART HDAC PROLIFERATION CLUSTER DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| MEDINA SMARCA4 TARGETS | 44 | 29 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| BERNARD PPAPDC1B TARGETS DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 3547 | 3553 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA | ||||
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-10 | 2434 | 2441 | 1A,m8 | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-122 | 2929 | 2935 | 1A | hsa-miR-122a | UGGAGUGUGACAAUGGUGUUUGU |
| miR-124.1 | 734 | 741 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 734 | 740 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA | ||||
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-138 | 3280 | 3287 | 1A,m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-140 | 453 | 459 | m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-141/200a | 1056 | 1063 | 1A,m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-142-5p | 3692 | 3699 | 1A,m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-155 | 2955 | 2961 | m8 | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-17-5p/20/93.mr/106/519.d | 3169 | 3176 | 1A,m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-19 | 3712 | 3718 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-200bc/429 | 1890 | 1896 | 1A | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-205 | 3822 | 3828 | 1A | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-22 | 3728 | 3734 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-29 | 410 | 416 | m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-299-5p | 451 | 458 | 1A,m8 | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
| miR-320 | 3730 | 3736 | m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-365 | 3591 | 3597 | m8 | hsa-miR-365 | UAAUGCCCCUAAAAAUCCUUAU |
| miR-370 | 3283 | 3289 | m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-409-3p | 3513 | 3519 | m8 | hsa-miR-409-3p | CGAAUGUUGCUCGGUGAACCCCU |
| miR-455 | 1477 | 1483 | 1A | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| miR-485-3p | 1534 | 1541 | 1A,m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| miR-9 | 2064 | 2070 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 3168 | 3174 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.