Gene Page: DAZAP2
Summary ?
| GeneID | 9802 |
| Symbol | DAZAP2 |
| Synonyms | PRTB |
| Description | DAZ associated protein 2 |
| Reference | MIM:607431|HGNC:HGNC:2684|Ensembl:ENSG00000183283|HPRD:08463|Vega:OTTHUMG00000169649 |
| Gene type | protein-coding |
| Map location | 12q12 |
| Pascal p-value | 0.093 |
| Sherlock p-value | 0.614 |
| Fetal beta | -0.791 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.01 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17322163 | 12 | 51632344 | DAZAP2 | 1.476E-4 | -0.324 | 0.032 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
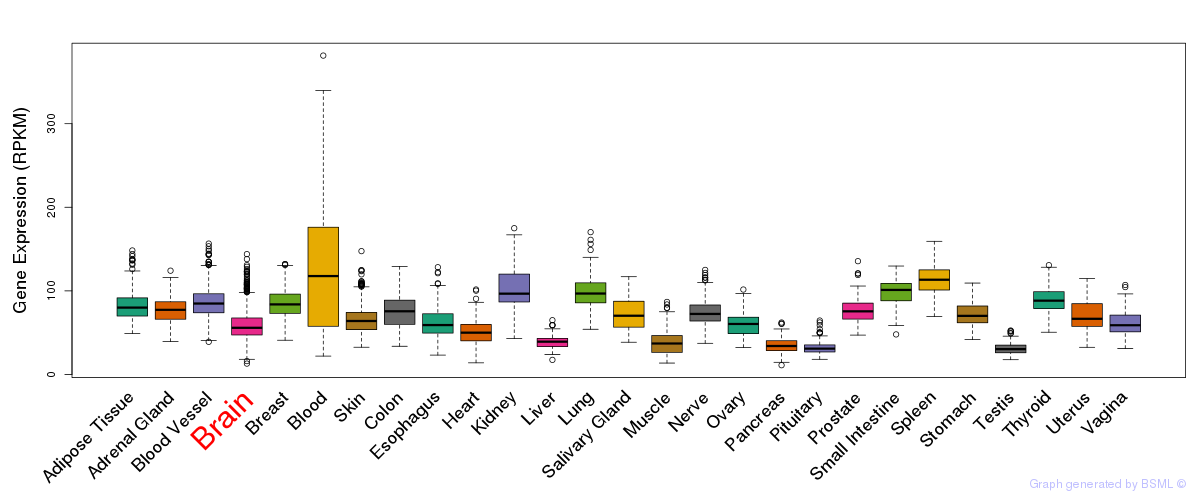
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CCNH | 0.82 | 0.81 |
| ATP5C1 | 0.81 | 0.81 |
| PSMA6 | 0.81 | 0.80 |
| C3orf14 | 0.81 | 0.84 |
| NFU1 | 0.80 | 0.81 |
| CHMP5 | 0.80 | 0.81 |
| MRPL1 | 0.79 | 0.81 |
| PSMC6 | 0.79 | 0.83 |
| TXNDC9 | 0.79 | 0.82 |
| PARL | 0.79 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DOCK6 | -0.58 | -0.63 |
| ENG | -0.56 | -0.58 |
| NOS3 | -0.52 | -0.56 |
| FAM38A | -0.51 | -0.57 |
| LIMS2 | -0.51 | -0.56 |
| GATA2 | -0.50 | -0.54 |
| MTSS1L | -0.50 | -0.51 |
| SEMA3G | -0.49 | -0.52 |
| AF347015.18 | -0.48 | -0.46 |
| LAMB2 | -0.48 | -0.53 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 16713569 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AATF | CHE-1 | CHE1 | DED | apoptosis antagonizing transcription factor | Two-hybrid | BioGRID | 16189514 |
| BAG3 | BAG-3 | BIS | CAIR-1 | MGC104307 | BCL2-associated athanogene 3 | Two-hybrid | BioGRID | 16189514 |
| BATF2 | MGC20410 | basic leucine zipper transcription factor, ATF-like 2 | Two-hybrid | BioGRID | 16189514 |
| C1orf94 | MGC15882 | chromosome 1 open reading frame 94 | Two-hybrid | BioGRID | 16189514 |
| CALCOCO2 | MGC17318 | NDP52 | calcium binding and coiled-coil domain 2 | Two-hybrid | BioGRID | 16189514 |
| CCDC120 | JM11 | coiled-coil domain containing 120 | Two-hybrid | BioGRID | 16189514 |
| DAZ1 | DAZ | SPGY | deleted in azoospermia 1 | - | HPRD,BioGRID | 10857750 |
| DAZAP2 | KIAA0058 | MGC14319 | MGC766 | PRTB | DAZ associated protein 2 | Two-hybrid | BioGRID | 16189514 |
| DAZL | DAZH | DAZL1 | DAZLA | MGC26406 | SPGYLA | deleted in azoospermia-like | - | HPRD,BioGRID | 10857750 |
| DCUN1D1 | DCUN1L1 | RP42 | SCCRO | SCRO | Tes3 | DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| FAM46C | FLJ20202 | family with sequence similarity 46, member C | Two-hybrid | BioGRID | 16189514 |
| FLJ20433 | FLJ30442 | MGC131904 | MGC74981 | hypothetical protein FLJ20433 | Two-hybrid | BioGRID | 16189514 |
| GPSM1 | AGS3 | DKFZp727I051 | G-protein signaling modulator 1 (AGS3-like, C. elegans) | Two-hybrid | BioGRID | 16189514 |
| HGS | HRS | ZFYVE8 | hepatocyte growth factor-regulated tyrosine kinase substrate | Two-hybrid | BioGRID | 16189514 |
| LMO2 | RBTN2 | RBTNL1 | RHOM2 | TTG2 | LIM domain only 2 (rhombotin-like 1) | Two-hybrid | BioGRID | 16189514 |
| LOC83459 | MGC88636 | hypothetical protein LOC83459 | Two-hybrid | BioGRID | 16189514 |
| NDUFA5 | B13 | CI-13KD-B | DKFZp781K1356 | FLJ12147 | NUFM | UQOR13 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa | Two-hybrid | BioGRID | 16189514 |
| NEDD4 | KIAA0093 | MGC176705 | NEDD4-1 | RPF1 | neural precursor cell expressed, developmentally down-regulated 4 | - | HPRD | 11342538 |
| NOC4L | MGC3162 | NOC4 | nucleolar complex associated 4 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| PLSCR1 | MMTRA1B | phospholipid scramblase 1 | Two-hybrid | BioGRID | 16189514 |
| POLI | RAD30B | RAD3OB | polymerase (DNA directed) iota | Two-hybrid | BioGRID | 16189514 |
| RBM9 | FOX2 | Fox-2 | HNRBP2 | HRNBP2 | RTA | dJ106I20.3 | fxh | RNA binding motif protein 9 | Two-hybrid | BioGRID | 16189514 |
| RBPMS | HERMES | RNA binding protein with multiple splicing | Two-hybrid | BioGRID | 16189514 |
| RHOXF2 | PEPP-2 | PEPP2 | THG1 | Rhox homeobox family, member 2 | Two-hybrid | BioGRID | 16189514 |
| RPS27A | CEP80 | HUBCEP80 | UBA80 | UBCEP1 | UBCEP80 | ribosomal protein S27a | Two-hybrid | BioGRID | 16189514 |
| SEPT2 | DIFF6 | KIAA0158 | NEDD5 | Pnutl3 | hNedd5 | septin 2 | Two-hybrid | BioGRID | 16189514 |
| STAM2 | DKFZp564C047 | Hbp | STAM2A | STAM2B | signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | Two-hybrid | BioGRID | 16189514 |
| TOLLIP | FLJ33531 | IL-1RAcPIP | toll interacting protein | Two-hybrid | BioGRID | 16189514 |
| UBAC1 | GBDR1 | RP11-432J22.3 | UBADC1 | UBA domain containing 1 | Two-hybrid | BioGRID | 16189514 |
| UBB | FLJ25987 | MGC8385 | ubiquitin B | Two-hybrid | BioGRID | 16189514 |
| UBC | HMG20 | ubiquitin C | Two-hybrid | BioGRID | 16189514 |
| WWP1 | AIP5 | DKFZp434D2111 | Tiul1 | hSDRP1 | WW domain containing E3 ubiquitin protein ligase 1 | Two-hybrid | BioGRID | 16189514 |
| ZFAND2B | - | zinc finger, AN1-type domain 2B | Two-hybrid | BioGRID | 16189514 |
| ZNF581 | FLJ22550 | HSPC189 | zinc finger protein 581 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| PROVENZANI METASTASIS UP | 194 | 112 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION DN | 98 | 67 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE DN | 106 | 68 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| ELLWOOD MYC TARGETS DN | 40 | 27 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| IVANOVSKA MIR106B TARGETS | 90 | 56 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 1075 | 1081 | 1A | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-128 | 1233 | 1239 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-132/212 | 1195 | 1202 | 1A,m8 | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-17-5p/20/93.mr/106/519.d | 143 | 149 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-181 | 231 | 237 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-182 | 367 | 374 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-452 | 808 | 814 | m8 | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
| miR-496 | 168 | 174 | m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 1242 | 1248 | 1A | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
| hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG | ||||
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
| miR-96 | 368 | 374 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.