Gene Page: MAGI2
Summary ?
| GeneID | 9863 |
| Symbol | MAGI2 |
| Synonyms | ACVRIP1|AIP-1|AIP1|ARIP1|MAGI-2|SSCAM |
| Description | membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| Reference | MIM:606382|HGNC:HGNC:18957|Ensembl:ENSG00000187391|HPRD:16212|Vega:OTTHUMG00000130697 |
| Gene type | protein-coding |
| Map location | 7q21 |
| Fetal beta | 0.238 |
| DMG | 2 (# studies) |
| Support | CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 3 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13051700 | 7 | 79050007 | MAGI2 | 8.29E-5 | -0.549 | 0.026 | DMG:Wockner_2014 |
| cg06809544 | 7 | 79081666 | MAGI2 | 1.19E-8 | -0.013 | 4.92E-6 | DMG:Jaffe_2016 |
| cg05270344 | 7 | 79083635 | MAGI2 | 1E-7 | -0.007 | 2.21E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
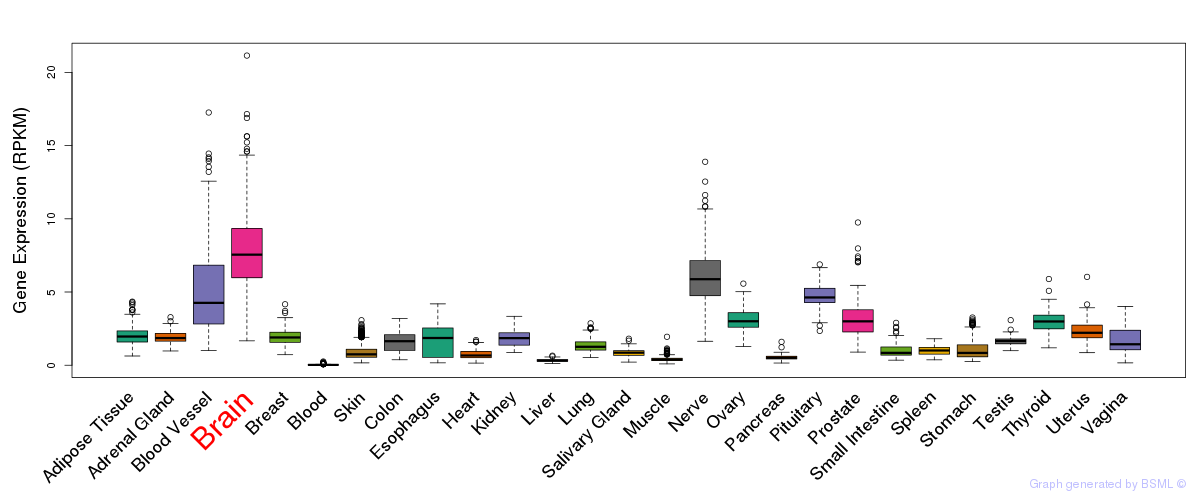
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| POLR2C | 0.90 | 0.88 |
| CIAO1 | 0.88 | 0.87 |
| PRPF19 | 0.85 | 0.85 |
| TMED4 | 0.85 | 0.82 |
| KLHL12 | 0.84 | 0.83 |
| TOX4 | 0.84 | 0.82 |
| C1orf109 | 0.83 | 0.81 |
| SURF4 | 0.83 | 0.82 |
| PI4KB | 0.83 | 0.81 |
| CNBP | 0.83 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.8 | -0.65 | -0.63 |
| AF347015.21 | -0.65 | -0.64 |
| MT-CO2 | -0.65 | -0.61 |
| AF347015.26 | -0.63 | -0.61 |
| MT-CYB | -0.63 | -0.61 |
| AF347015.31 | -0.63 | -0.60 |
| AF347015.18 | -0.62 | -0.70 |
| AF347015.2 | -0.62 | -0.56 |
| AF347015.15 | -0.62 | -0.60 |
| AF347015.33 | -0.61 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 9647693 | |
| GO:0019902 | phosphatase binding | IPI | 10760291 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME NEPHRIN INTERACTIONS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE DN | 98 | 59 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR DN | 86 | 62 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| MOLENAAR TARGETS OF CCND1 AND CDK4 UP | 67 | 48 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 2025 | 2031 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-101 | 1858 | 1865 | 1A,m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| hsa-miR-101 | UACAGUACUGUGAUAACUGAAG | ||||
| miR-124.1 | 402 | 408 | 1A | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-134 | 2109 | 2115 | 1A | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-141/200a | 2046 | 2052 | 1A | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-144 | 2191 | 2197 | 1A | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-145 | 390 | 397 | 1A,m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-19 | 1936 | 1943 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-205 | 325 | 332 | 1A,m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-218 | 1711 | 1718 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-27 | 2054 | 2060 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-30-5p | 2187 | 2193 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-320 | 286 | 292 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-339 | 503 | 509 | m8 | hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA |
| miR-485-3p | 2194 | 2200 | m8 | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| miR-485-5p | 1641 | 1647 | 1A | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
| miR-539 | 1030 | 1037 | 1A,m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.