Gene Page: CDC27
Summary ?
| GeneID | 996 |
| Symbol | CDC27 |
| Synonyms | ANAPC3|APC3|CDC27Hs|D0S1430E|D17S978E|H-NUC|HNUC|NUC2 |
| Description | cell division cycle 27 |
| Reference | MIM:116946|HGNC:HGNC:1728|Ensembl:ENSG00000004897|HPRD:00304|Vega:OTTHUMG00000166429 |
| Gene type | protein-coding |
| Map location | 17q21.32 |
| Pascal p-value | 0.174 |
| Sherlock p-value | 0.015 |
| Fetal beta | -0.463 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11051641 | chr12 | 32020255 | CDC27 | 996 | 0.16 | trans |
Section II. Transcriptome annotation
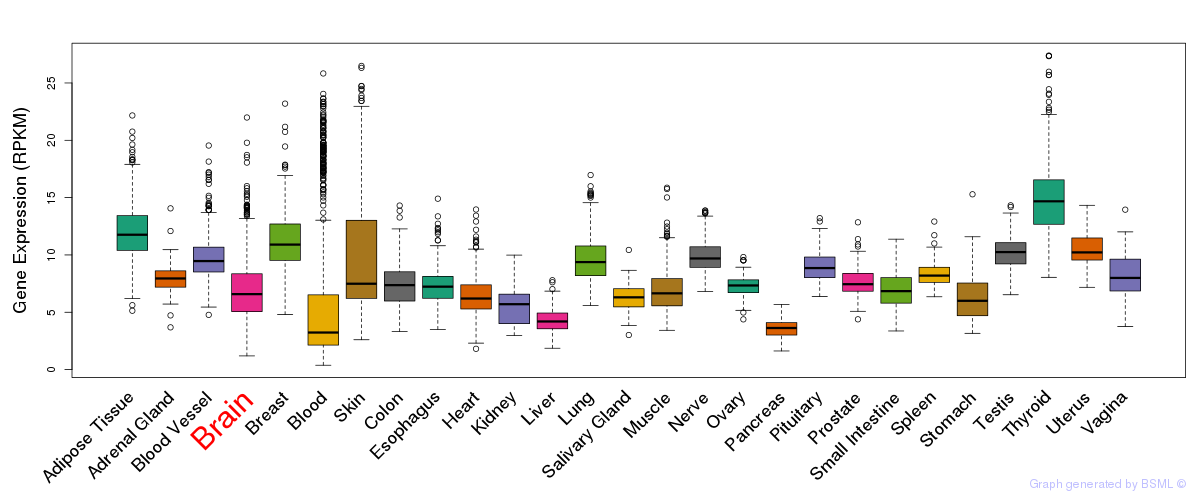
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ST3GAL1 | 0.82 | 0.84 |
| SSBP3 | 0.82 | 0.79 |
| TP53I11 | 0.80 | 0.93 |
| AC127496.3 | 0.80 | 0.83 |
| BCL7A | 0.78 | 0.90 |
| SEZ6 | 0.78 | 0.92 |
| C14orf37 | 0.77 | 0.81 |
| SOBP | 0.77 | 0.90 |
| SRGAP2 | 0.77 | 0.89 |
| ATXN7L1 | 0.77 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINB6 | -0.60 | -0.75 |
| HEPN1 | -0.57 | -0.75 |
| S100B | -0.57 | -0.83 |
| AP003117.1 | -0.56 | -0.68 |
| AF347015.27 | -0.56 | -0.85 |
| TSC22D4 | -0.56 | -0.77 |
| HLA-F | -0.56 | -0.75 |
| AIFM3 | -0.56 | -0.75 |
| AF347015.31 | -0.56 | -0.84 |
| AF347015.33 | -0.56 | -0.85 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANAPC1 | APC1 | MCPR | TSG24 | anaphase promoting complex subunit 1 | Affinity Capture-Western Co-purification | BioGRID | 11076961 |12956947 |
| ANAPC10 | APC10 | DKFZp564L0562 | DOC1 | anaphase promoting complex subunit 10 | Affinity Capture-Western | BioGRID | 12956947 |
| ANAPC11 | APC11 | Apc11p | HSPC214 | MGC882 | anaphase promoting complex subunit 11 | Affinity Capture-Western | BioGRID | 12956947 |
| ANAPC4 | APC4 | anaphase promoting complex subunit 4 | Affinity Capture-Western | BioGRID | 12956947 |
| ANAPC5 | APC5 | anaphase promoting complex subunit 5 | - | HPRD,BioGRID | 10922056 |
| ANAPC7 | APC7 | anaphase promoting complex subunit 7 | Affinity Capture-Western | BioGRID | 12956947 |
| APC2 | APCL | adenomatosis polyposis coli 2 | Affinity Capture-Western | BioGRID | 12956947 |
| BUB1B | BUB1beta | BUBR1 | Bub1A | MAD3L | SSK1 | hBUBR1 | budding uninhibited by benzimidazoles 1 homolog beta (yeast) | - | HPRD,BioGRID | 10477750 |
| CDC16 | APC6 | cell division cycle 16 homolog (S. cerevisiae) | - | HPRD,BioGRID | 9405394 |
| CDC20 | CDC20A | MGC102824 | bA276H19.3 | p55CDC | cell division cycle 20 homolog (S. cerevisiae) | Affinity Capture-Western Reconstituted Complex | BioGRID | 9628895 |9736712 |9811605 |12196507 |12956947 |14561775 |
| CDC20 | CDC20A | MGC102824 | bA276H19.3 | p55CDC | cell division cycle 20 homolog (S. cerevisiae) | - | HPRD | 9736712 |12196507 |
| CDC23 | APC8 | cell division cycle 23 homolog (S. cerevisiae) | Affinity Capture-Western | BioGRID | 12956947 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12956947 |
| FZR1 | CDC20C | CDH1 | FZR | FZR2 | HCDH | HCDH1 | KIAA1242 | fizzy/cell division cycle 20 related 1 (Drosophila) | Cdh1 interacts with Cdc27. | BIND | 15916961 |
| FZR1 | CDC20C | CDH1 | FZR | FZR2 | HCDH | HCDH1 | KIAA1242 | fizzy/cell division cycle 20 related 1 (Drosophila) | - | HPRD,BioGRID | 12797865 |
| HOXC10 | HOX3I | MGC5259 | homeobox C10 | - | HPRD | 12853486 |
| MAD2L1 | HSMAD2 | MAD2 | MAD2 mitotic arrest deficient-like 1 (yeast) | - | HPRD,BioGRID | 9736712 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | - | HPRD,BioGRID | 9499405 |10939594 |
| PIN1 | DOD | UBL5 | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 | Pin1 interacts with Cdc27. | BIND | 9499405 |
| PPP5C | FLJ36922 | PP5 | PPP5 | protein phosphatase 5, catalytic subunit | - | HPRD,BioGRID | 9405394 |
| SKIL | SNO | SnoA | SnoN | SKI-like oncogene | - | HPRD | 11691834 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | - | HPRD,BioGRID | 11691834 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD,BioGRID | 11691834 |
| SPATC1 | MGC61633 | SPATA15 | SPERIOLIN | spermatogenesis and centriole associated 1 | Affinity Capture-Western | BioGRID | 15280373 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL CYCLE | 128 | 84 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| KEGG PROGESTERONE MEDIATED OOCYTE MATURATION | 86 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | 24 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF MITOTIC CELL CYCLE | 85 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | 72 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | 73 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | 64 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | 26 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | 22 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHORYLATION OF THE APC C | 23 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | 28 | 12 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS UP | 114 | 66 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| WAESCH ANAPHASE PROMOTING COMPLEX | 12 | 7 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER SOX9 TARGETS IN PROSTATE DEVELOPMENT DN | 45 | 33 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 HELA | 69 | 47 | All SZGR 2.0 genes in this pathway |
| SAKAI TUMOR INFILTRATING MONOCYTES DN | 81 | 51 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION UP | 74 | 40 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 M | 216 | 124 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |