Gene Page: NR1H4
Summary ?
| GeneID | 9971 |
| Symbol | NR1H4 |
| Synonyms | BAR|FXR|HRR-1|HRR1|RIP14 |
| Description | nuclear receptor subfamily 1 group H member 4 |
| Reference | MIM:603826|HGNC:HGNC:7967|Ensembl:ENSG00000012504|HPRD:04827|Vega:OTTHUMG00000170359 |
| Gene type | protein-coding |
| Map location | 12q23.1 |
| Pascal p-value | 0.017 |
| Fetal beta | -0.131 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
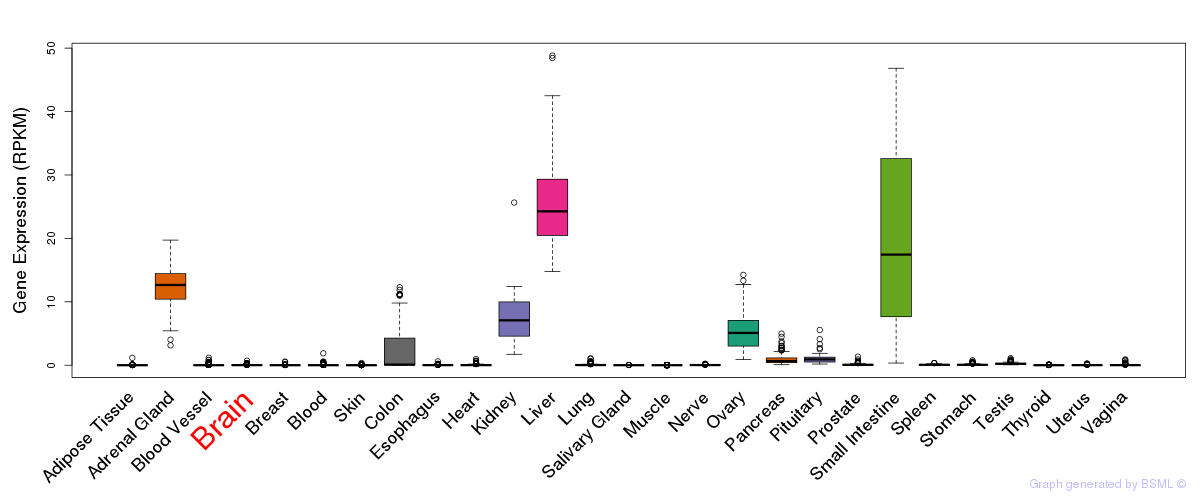
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| YTHDF3 | 0.96 | 0.98 |
| SPIN1 | 0.95 | 0.96 |
| MTMR9 | 0.95 | 0.97 |
| KPNA1 | 0.94 | 0.97 |
| C5orf24 | 0.94 | 0.96 |
| ETNK1 | 0.94 | 0.96 |
| SCAMP1 | 0.94 | 0.96 |
| AVL9 | 0.94 | 0.96 |
| NAT12 | 0.94 | 0.96 |
| FAM160B1 | 0.94 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.71 | -0.80 |
| MT-CO2 | -0.68 | -0.79 |
| AF347015.31 | -0.67 | -0.77 |
| HIGD1B | -0.66 | -0.78 |
| HSD17B14 | -0.66 | -0.72 |
| AF347015.33 | -0.65 | -0.74 |
| AC018755.7 | -0.65 | -0.70 |
| MT-CYB | -0.65 | -0.75 |
| AF347015.8 | -0.65 | -0.77 |
| TLCD1 | -0.65 | -0.74 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA NUCLEARRS PATHWAY | 15 | 12 | All SZGR 2.0 genes in this pathway |
| PID RXR VDR PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES DN | 145 | 91 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | 261 | 153 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA DN | 52 | 33 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER WITH H3K27ME3 | 196 | 93 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER DN | 101 | 65 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |