Gene Page: CNTN4
Summary ?
| GeneID | 152330 |
| Symbol | CNTN4 |
| Synonyms | AXCAM|BIG-2 |
| Description | contactin 4 |
| Reference | MIM:607280|HGNC:HGNC:2174|Ensembl:ENSG00000144619|HPRD:16239|Vega:OTTHUMG00000119031 |
| Gene type | protein-coding |
| Map location | 3p26.3 |
| Fetal beta | 1.652 |
| eGene | Anterior cingulate cortex BA24 Myers' cis & trans |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 9 |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs17194490 | chr3 | 2547786 | TG | 4.865E-11 | intronic | CNTN4 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs7370564 | 0 | CNTN4 | 152330 | 0.09 | trans | |||
| rs13068096 | 3 | 2518282 | CNTN4 | ENSG00000144619.10 | 1.20428E-6 | 0.04 | 377785 | gtex_brain_ba24 |
| rs34163547 | 3 | 2519380 | CNTN4 | ENSG00000144619.10 | 1.22317E-6 | 0.04 | 378883 | gtex_brain_ba24 |
| rs11706456 | 3 | 2522747 | CNTN4 | ENSG00000144619.10 | 7.67373E-7 | 0.04 | 382250 | gtex_brain_ba24 |
Section II. Transcriptome annotation
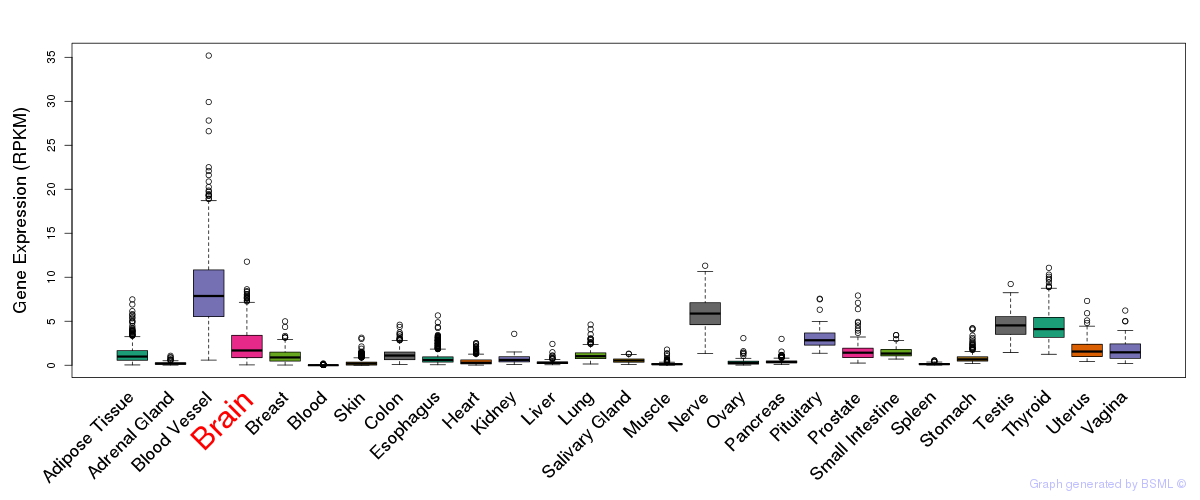
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | ISS | Brain (GO term level: 7) | - |
| GO:0007413 | axonal fasciculation | TAS | neuron, axon (GO term level: 13) | 15106122 |
| GO:0007411 | axon guidance | TAS | axon (GO term level: 13) | 15106122 |
| GO:0031175 | neurite development | ISS | neuron, axon, neurite, dendrite (GO term level: 10) | - |
| GO:0048167 | regulation of synaptic plasticity | TAS | Synap (GO term level: 8) | 15106122 |
| GO:0007158 | neuron adhesion | TAS | neuron (GO term level: 5) | 14571131 |
| GO:0045665 | negative regulation of neuron differentiation | IMP | neuron (GO term level: 10) | 14571131 |
| GO:0007399 | nervous system development | IMP | neurite (GO term level: 5) | 15106122 |
| GO:0007155 | cell adhesion | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0030424 | axon | NAS | neuron, axon, Neurotransmitter (GO term level: 6) | 14571131 |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005886 | plasma membrane | NAS | 15106122 | |
| GO:0031225 | anchored to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA LOH REGIONS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| TAYLOR METHYLATED IN ACUTE LYMPHOBLASTIC LEUKEMIA | 77 | 52 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 DN | 45 | 30 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 1590 | 1597 | 1A,m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-142-5p | 1168 | 1174 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-145 | 107 | 113 | 1A | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-148/152 | 208 | 215 | 1A,m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-155 | 785 | 791 | m8 | hsa-miR-155 | UUAAUGCUAAUCGUGAUAGGGG |
| miR-181 | 913 | 920 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-200bc/429 | 1516 | 1522 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-25/32/92/363/367 | 703 | 710 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-30-5p | 246 | 252 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-33 | 205 | 211 | m8 | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-362 | 463 | 469 | m8 | hsa-miR-362 | AAUCCUUGGAACCUAGGUGUGAGU |
| miR-365 | 142 | 148 | 1A | hsa-miR-365 | UAAUGCCCCUAAAAAUCCUUAU |
| miR-369-3p | 276 | 282 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 276 | 282 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-376c | 1560 | 1566 | m8 | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-410 | 278 | 284 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-448 | 262 | 268 | m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-9 | 237 | 243 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.