Gene Page: NMUR2
Summary ?
| GeneID | 56923 |
| Symbol | NMUR2 |
| Synonyms | FM-4|FM4|NMU-R2|NMU2R|TGR-1|TGR1 |
| Description | neuromedin U receptor 2 |
| Reference | MIM:605108|HGNC:HGNC:16454|Ensembl:ENSG00000132911|HPRD:12003|Vega:OTTHUMG00000130131 |
| Gene type | protein-coding |
| Map location | 5q33.1 |
| Pascal p-value | 0.005 |
| Fetal beta | -0.169 |
| eGene | Cortex Frontal Cortex BA9 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs79212538 | chr5 | 151993104 | TG | 3.843E-8 | intergenic | NMUR2,LINC01470 | dist=208264;dist=5421 |
Section II. Transcriptome annotation
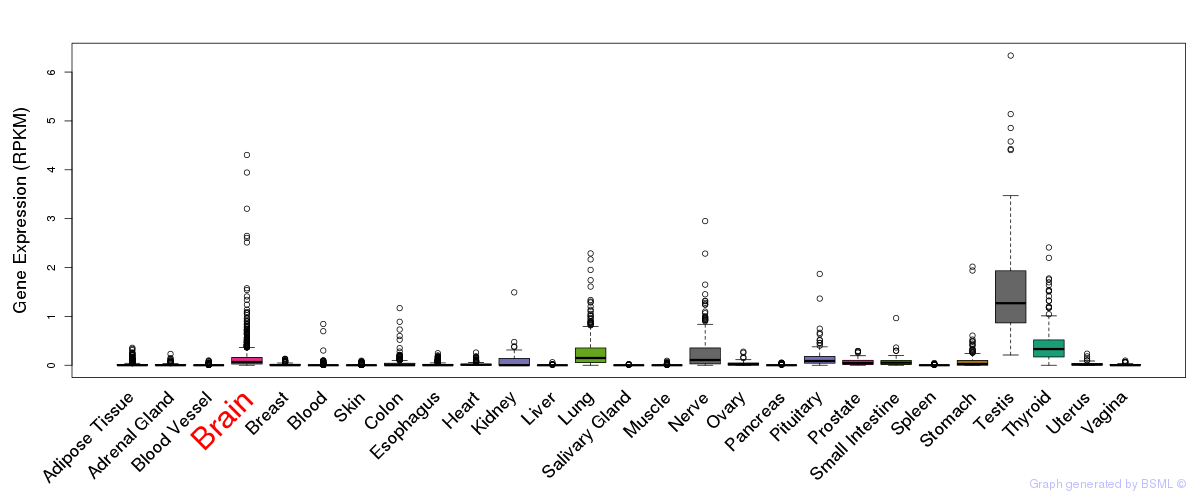
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001607 | neuromedin U receptor activity | IDA | 10887190 | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005525 | GTP binding | IDA | 10899166 | |
| GO:0005229 | intracellular calcium activated chloride channel activity | IDA | 10899166 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | IEP | Brain (GO term level: 6) | 10899166 |
| GO:0007218 | neuropeptide signaling pathway | TAS | Neurotransmitter (GO term level: 8) | 10894543 |
| GO:0019226 | transmission of nerve impulse | IEP | neuron (GO term level: 5) | 10899166 |
| GO:0007200 | G-protein signaling, coupled to IP3 second messenger (phospholipase C activating) | IDA | 10899166 | |
| GO:0007204 | elevation of cytosolic calcium ion concentration | IDA | 10899166 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0048016 | inositol phosphate-mediated signaling | IDA | 10899166 | |
| GO:0007631 | feeding behavior | TAS | 10894543 | |
| GO:0006816 | calcium ion transport | IDA | 10899166 | |
| GO:0006940 | regulation of smooth muscle contraction | NAS | 10887190 | |
| GO:0043006 | calcium-dependent phospholipase A2 activation | IDA | 10899166 | |
| GO:0050482 | arachidonic acid secretion | IDA | 10887190 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016021 | integral to membrane | IDA | 10899166 | |
| GO:0005886 | plasma membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |