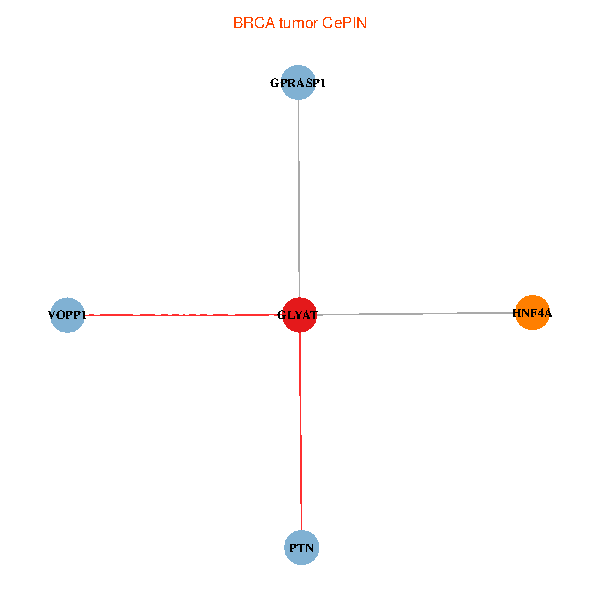
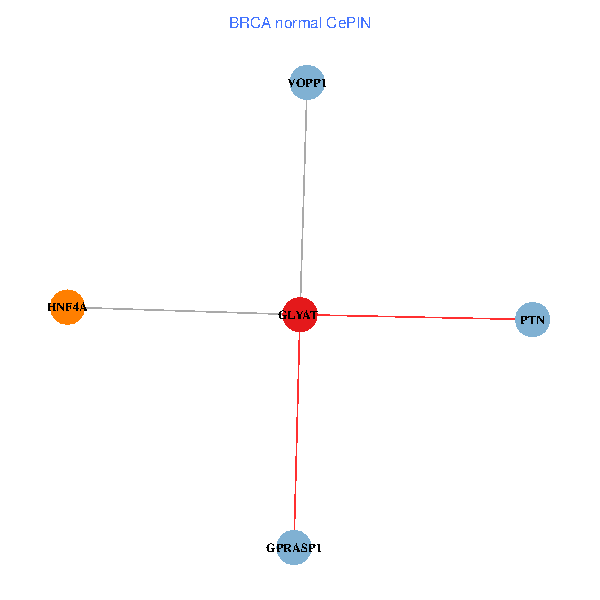
| BRCA (tumor) | BRCA (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
 |  |
|
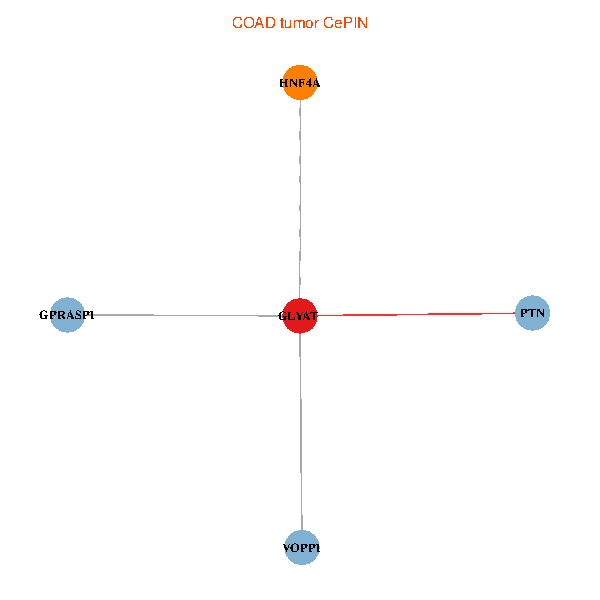
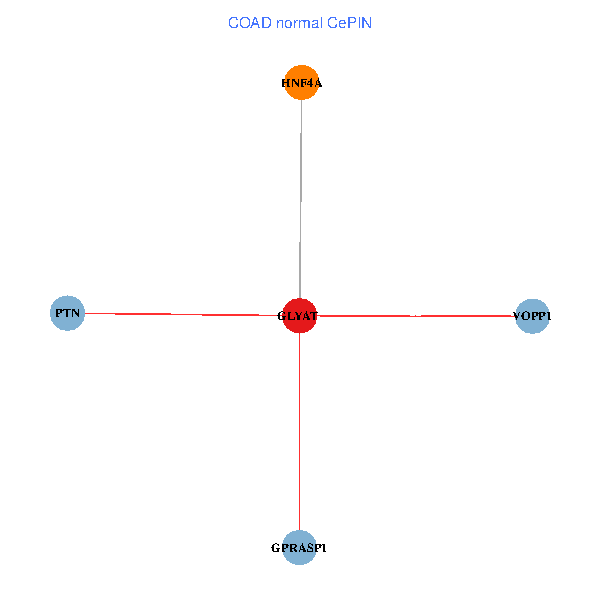
| COAD (tumor) | COAD (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
 |  |
|
| KICH (tumor) | KICH (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
 |  |
|
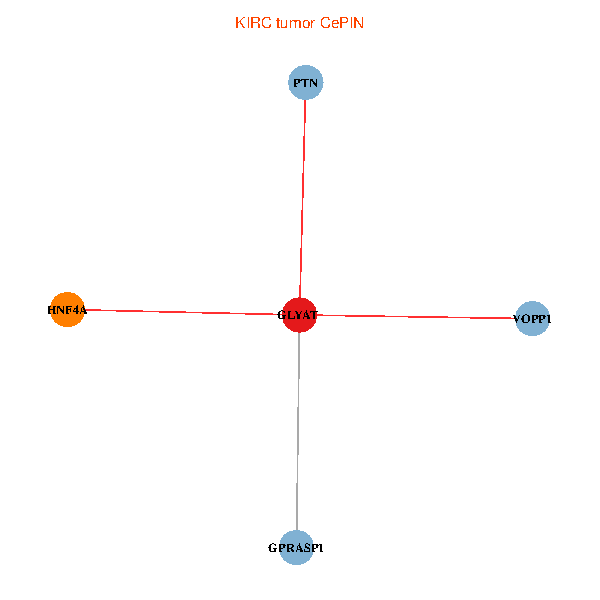
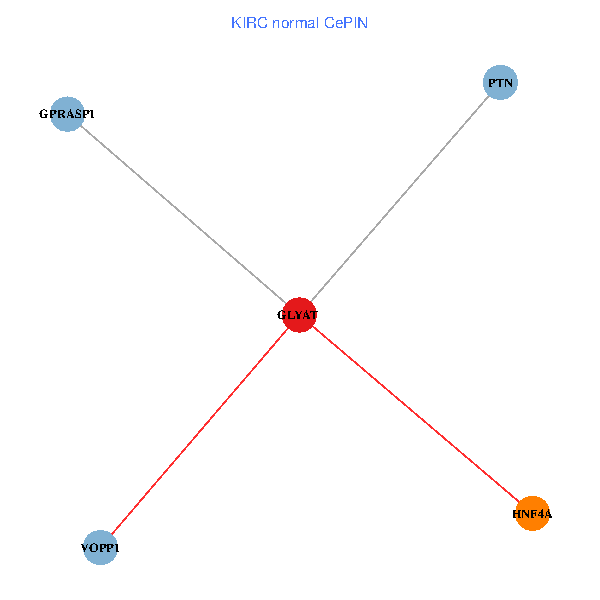
| KIRC (tumor) | KIRC (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
 |  |
|
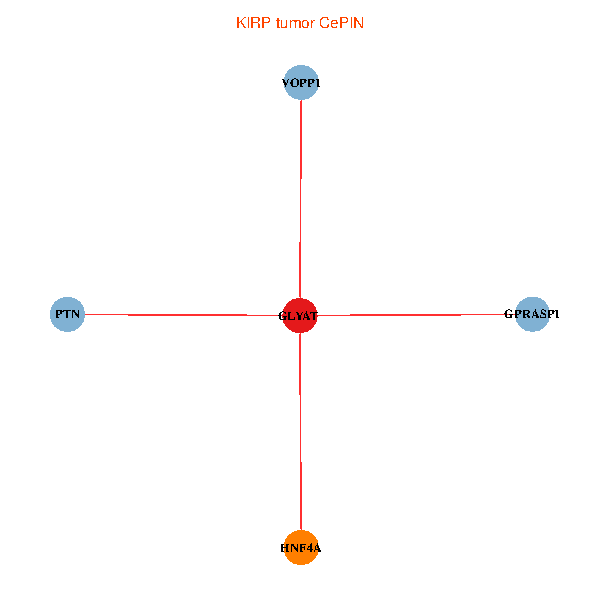
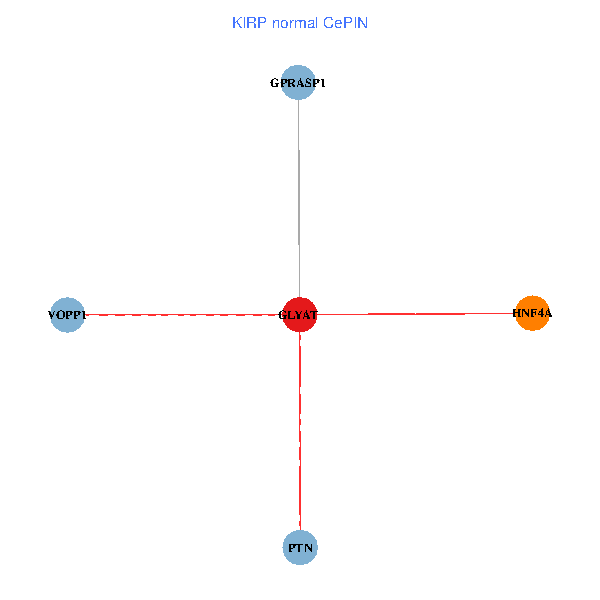
| KIRP (tumor) | KIRP (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
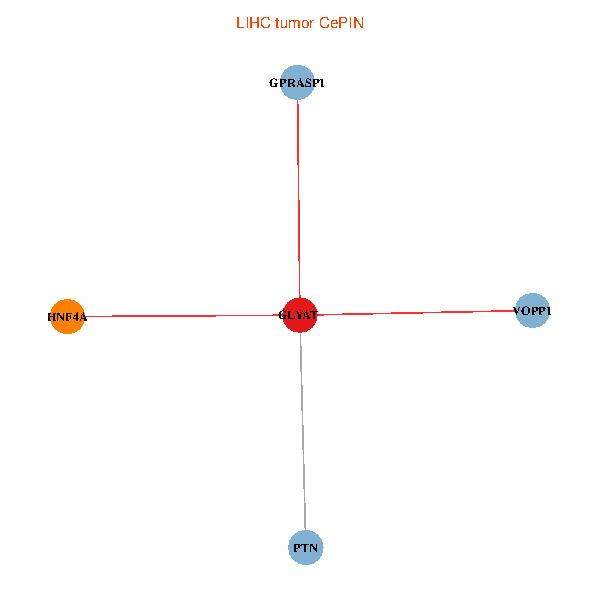 | 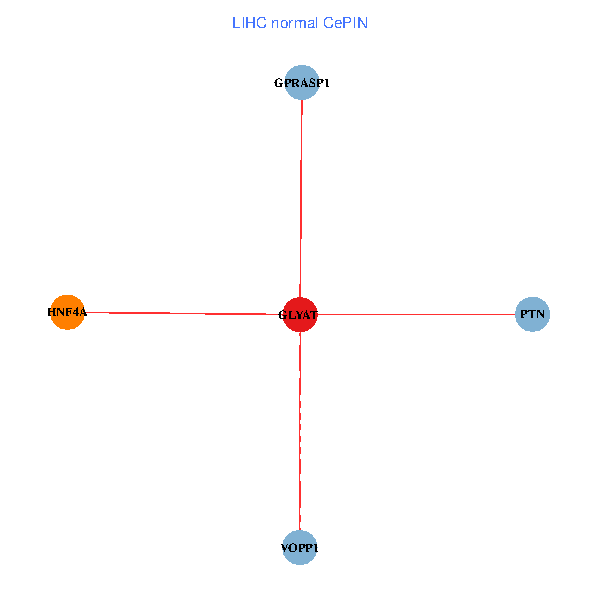 |
|
| LUAD (tumor) | LUAD (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
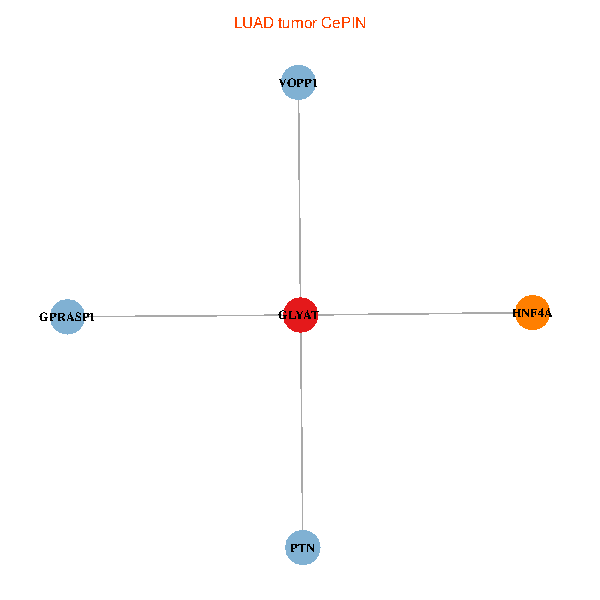 | 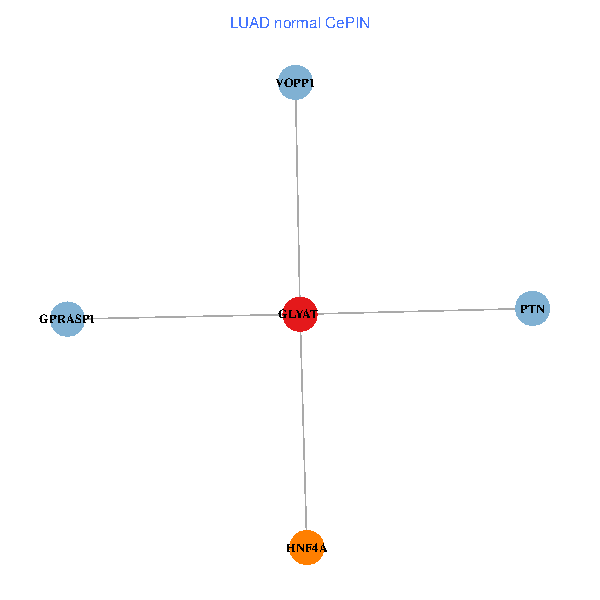 |
|
| LUSC (tumor) | LUSC (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
 |  |
|
| PRAD (tumor) | PRAD (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
 |  |
|
| STAD (tumor) | STAD (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
 |  |
|
| THCA (tumor) | THCA (normal) |
| GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (tumor) | GLYAT, HNF4A, PTN, GPRASP1, VOPP1 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0019693 | HIV Infections | 23 | BeFree |
| umls:C2239176 | Liver carcinoma | 19 | BeFree |
| umls:C0023903 | Liver neoplasms | 15 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 14 | BeFree |
| umls:C0678222 | Breast Carcinoma | 14 | BeFree |
| umls:C0027819 | Neuroblastoma | 5 | BeFree |
| umls:C0039730 | Thalassemia | 5 | BeFree |
| umls:C0600139 | Prostate carcinoma | 5 | BeFree |
| umls:C0700095 | Central neuroblastoma | 5 | BeFree |
| umls:C1458155 | Mammary Neoplasms | 5 | BeFree |
| umls:C0001418 | Adenocarcinoma | 4 | BeFree |
| umls:C0004096 | Asthma | 4 | BeFree |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 4 | BeFree |
| umls:C0206624 | Hepatoblastoma | 4 | BeFree |
| umls:C0235974 | Pancreatic carcinoma | 4 | BeFree |
| umls:C0376358 | Malignant neoplasm of prostate | 4 | BeFree |
| umls:C0752120 | Spinocerebellar Ataxia Type 1 | 4 | BeFree |
| umls:C0007102 | Malignant tumor of colon | 3 | BeFree |
| umls:C0024299 | Lymphoma | 3 | BeFree |
| umls:C0032002 | Pituitary Diseases | 3 | BeFree |
| umls:C0042900 | Vitiligo | 3 | BeFree |
| umls:C0043346 | Xeroderma Pigmentosum | 3 | BeFree |
| umls:C0242379 | Malignant neoplasm of lung | 3 | BeFree |
| umls:C0271980 | beta^0^ Thalassemia | 3 | BeFree |
| umls:C0302592 | Cervix carcinoma | 3 | BeFree |
| umls:C0476089 | Endometrial Carcinoma | 3 | BeFree |
| umls:C0596263 | Carcinogenesis | 3 | BeFree |
| umls:C0684249 | Carcinoma of lung | 3 | BeFree |
| umls:C0699790 | Colon Carcinoma | 3 | BeFree |
| umls:C1847835 | VITILIGO-ASSOCIATED MULTIPLE AUTOIMMUNE DISEASE SUSCEPTIBILITY 1 (finding) | 3 | BeFree |
| umls:C0007137 | Squamous cell carcinoma | 2 | BeFree |
| umls:C0008497 | Choriocarcinoma | 2 | BeFree |
| umls:C0009207 | Cockayne Syndrome | 2 | BeFree |
| umls:C0009402 | Colorectal Carcinoma | 2 | BeFree |
| umls:C0011854 | Diabetes Mellitus, Insulin-Dependent | 2 | BeFree |
| umls:C0011881 | Diabetic Nephropathy | 2 | BeFree |
| umls:C0017638 | Glioma | 2 | BeFree |
| umls:C0023493 | Adult T-Cell Lymphoma/Leukemia | 2 | BeFree |
| umls:C0025202 | melanoma | 2 | BeFree |
| umls:C0029408 | Degenerative polyarthritis | 2 | BeFree |
| umls:C0032461 | Polycythemia | 2 | BeFree |
| umls:C0162872 | Aortic Aneurysm, Thoracic | 2 | BeFree |
| umls:C0238461 | Anaplastic thyroid carcinoma | 2 | BeFree |
| umls:C0238463 | Papillary thyroid carcinoma | 2 | BeFree |
| umls:C0375023 | Respiratory syncytial virus (RSV) infection in conditions classified elsewhere and of unspecified site | 2 | BeFree |
| umls:C1527405 | Erythrocytosis | 2 | BeFree |
| umls:C1956346 | Coronary Artery Disease | 2 | BeFree |
| umls:C0001430 | Adenoma | 1 | BeFree |
| umls:C0001973 | Alcoholic Intoxication, Chronic | 1 | BeFree |
| umls:C0002736 | Amyotrophic Lateral Sclerosis | 1 | BeFree |
| umls:C0002894 | Refractory anaemia with excess blasts | 1 | BeFree |
| umls:C0003469 | Anxiety Disorders | 1 | BeFree |
| umls:C0004763 | Barrett Esophagus | 1 | BeFree |
| umls:C0005586 | Bipolar Disorder | 1 | BeFree |
| umls:C0007115 | Malignant neoplasm of thyroid | 1 | BeFree |
| umls:C0007130 | Mucinous Adenocarcinoma | 1 | BeFree |
| umls:C0007131 | Non-Small Cell Lung Carcinoma | 1 | BeFree |
| umls:C0007847 | Malignant tumor of cervix | 1 | BeFree |
| umls:C0008384 | Cholesterol Ester Storage Disease | 1 | BeFree |
| umls:C0008626 | Congenital chromosomal disease | 1 | BeFree |
| umls:C0010495 | Cutis Laxa | 1 | BeFree |
| umls:C0011389 | Dental Plaque | 1 | BeFree |
| umls:C0011615 | Dermatitis, Atopic | 1 | BeFree |
| umls:C0013595 | Eczema | 1 | BeFree |
| umls:C0016667 | Fragile X Syndrome | 1 | BeFree |
| umls:C0018784 | Sensorineural Hearing Loss (disorder) | 1 | BeFree |
| umls:C0018790 | Cardiac Arrest | 1 | BeFree |
| umls:C0019163 | Hepatitis B | 1 | BeFree |
| umls:C0019202 | Hepatolenticular Degeneration | 1 | BeFree |
| umls:C0019348 | Herpes Simplex Infections | 1 | BeFree |
| umls:C0021364 | Male infertility | 1 | BeFree |
| umls:C0021400 | Influenza | 1 | BeFree |
| umls:C0021670 | insulinoma | 1 | BeFree |
| umls:C0022610 | Kernicterus | 1 | BeFree |
| umls:C0023269 | leiomyosarcoma | 1 | BeFree |
| umls:C0023418 | leukemia | 1 | BeFree |
| umls:C0023524 | Leukoencephalopathy, Progressive Multifocal | 1 | BeFree |
| umls:C0024117 | Chronic Obstructive Airway Disease | 1 | BeFree |
| umls:C0024141 | Lupus Erythematosus, Systemic | 1 | BeFree |
| umls:C0024305 | Lymphoma, Non-Hodgkin | 1 | BeFree |
| umls:C0024530 | Malaria | 1 | BeFree |
| umls:C0024534 | Malaria, Cerebral | 1 | BeFree |
| umls:C0024623 | Malignant neoplasm of stomach | 1 | BeFree |
| umls:C0025007 | Measles | 1 | BeFree |
| umls:C0025362 | Mental Retardation | 1 | BeFree |
| umls:C0026499 | Monosomy | 1 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 1 | BeFree |
| umls:C0027708 | Nephroblastoma | 1 | BeFree |
| umls:C0029463 | Osteosarcoma | 1 | BeFree |
| umls:C0029925 | Ovarian Carcinoma | 1 | BeFree |
| umls:C0031511 | Pheochromocytoma | 1 | BeFree |
| umls:C0032019 | Pituitary Neoplasms | 1 | BeFree |
| umls:C0032131 | Plasmacytoma | 1 | BeFree |
| umls:C0033575 | Prostatic Diseases | 1 | BeFree |
| umls:C0037889 | Hereditary spherocytosis | 1 | BeFree |
| umls:C0038826 | Superinfection | 1 | BeFree |
| umls:C0042373 | Vascular Diseases | 1 | BeFree |
| umls:C0079474 | Hallopeau-Siemens Disease | 1 | BeFree |
| umls:C0085179 | Eosinophilia-Myalgia Syndrome | 1 | BeFree |
| umls:C0087012 | Ataxia, Spinocerebellar | 1 | BeFree |
| umls:C0149925 | Small cell carcinoma of lung | 1 | BeFree |
| umls:C0175754 | Agenesis of corpus callosum | 1 | BeFree |
| umls:C0206659 | Embryonal Carcinoma | 1 | BeFree |
| umls:C0206663 | Neuroectodermal Tumor, Primitive | 1 | BeFree |
| umls:C0206664 | Teratocarcinoma | 1 | BeFree |
| umls:C0206682 | Follicular thyroid carcinoma | 1 | BeFree |
| umls:C0234133 | Extrapyramidal sign | 1 | BeFree |
| umls:C0235031 | Neurologic Symptoms | 1 | BeFree |
| umls:C0235946 | Cerebral atrophy | 1 | BeFree |
| umls:C0238462 | Medullary carcinoma of thyroid | 1 | BeFree |
| umls:C0266929 | Chronic Periodontitis | 1 | BeFree |
| umls:C0268483 | Tyrosinemias | 1 | BeFree |
| umls:C0268540 | HHH syndrome | 1 | BeFree |
| umls:C0270814 | Spastic syndrome | 1 | BeFree |
| umls:C0272322 | Severe hereditary factor VIII deficiency disease | 1 | BeFree |
| umls:C0278837 | Stage IV Prostate Carcinoma | 1 | BeFree |
| umls:C0279980 | Extraosseous Ewings sarcoma-primitive neuroepithelial tumor | 1 | BeFree |
| umls:C0342471 | 3 beta-Hydroxysteroid dehydrogenase deficiency | 1 | BeFree |
| umls:C0343401 | MRSA - Methicillin resistant Staphylococcus aureus infection | 1 | BeFree |
| umls:C0345907 | Angiosarcoma of liver | 1 | BeFree |
| umls:C0346647 | Malignant neoplasm of pancreas | 1 | BeFree |
| umls:C0497406 | Overweight | 1 | BeFree |
| umls:C0524620 | Metabolic Syndrome X | 1 | BeFree |
| umls:C0549473 | Thyroid carcinoma | 1 | BeFree |
| umls:C0555232 | pseudohermaphrodite (non-specific) | 1 | BeFree |
| umls:C0585442 | Osteosarcoma of bone | 1 | BeFree |
| umls:C0684337 | Ewings sarcoma-primitive neuroectodermal tumor (PNET) | 1 | BeFree |
| umls:C0699791 | Stomach Carcinoma | 1 | BeFree |
| umls:C0872054 | latent infection | 1 | BeFree |
| umls:C0919267 | ovarian neoplasm | 1 | BeFree |
| umls:C1140680 | Malignant neoplasm of ovary | 1 | BeFree |
| umls:C1168401 | Squamous cell carcinoma of the head and neck | 1 | BeFree |
| umls:C1176475 | Ductal Carcinoma | 1 | BeFree |
| umls:C1258104 | Diffuse Scleroderma | 1 | BeFree |
| umls:C1333990 | Hereditary Nonpolyposis Colorectal Cancer | 1 | BeFree |
| umls:C1527249 | Colorectal Cancer | 1 | BeFree |
| umls:C1527349 | Ductal Breast Carcinoma | 1 | BeFree |
| umls:C1862941 | Amyotrophic Lateral Sclerosis, Sporadic | 1 | BeFree |
| umls:C2674218 | SPHEROCYTOSIS, TYPE 1 (disorder) | 1 | BeFree |
| umls:C3146264 | Stage IV Prostate Cancer AJCC v7 | 1 | BeFree |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))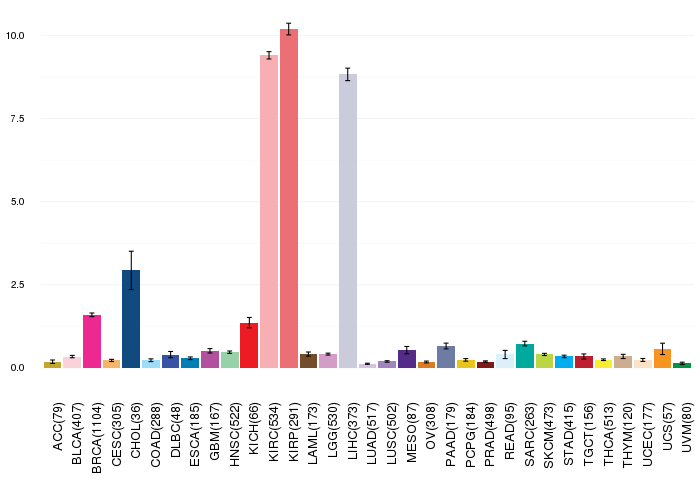
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))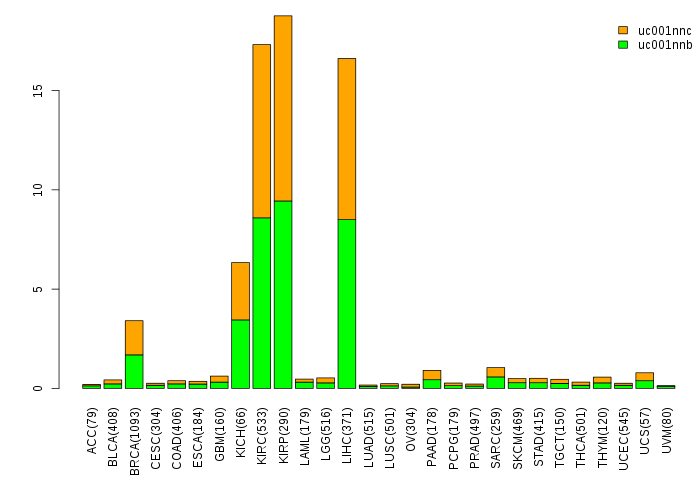
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data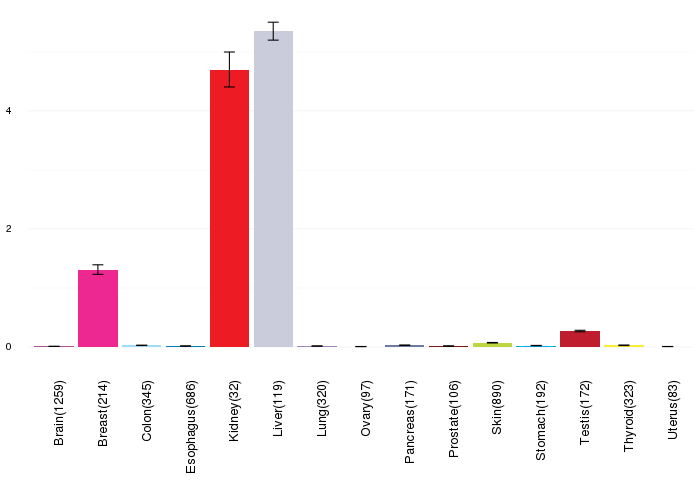
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))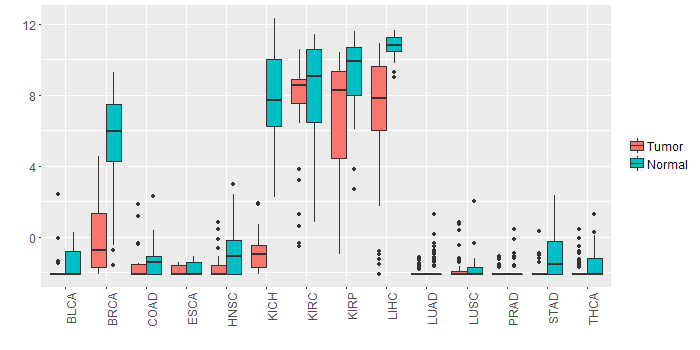
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.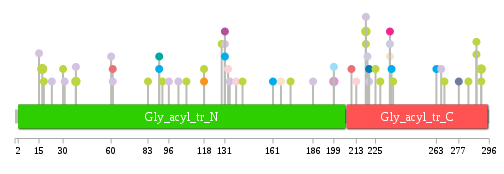

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 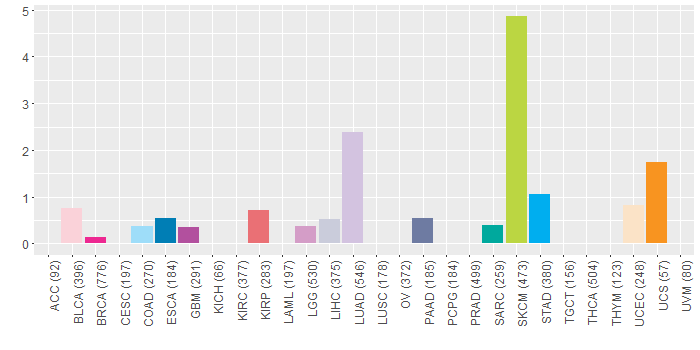
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)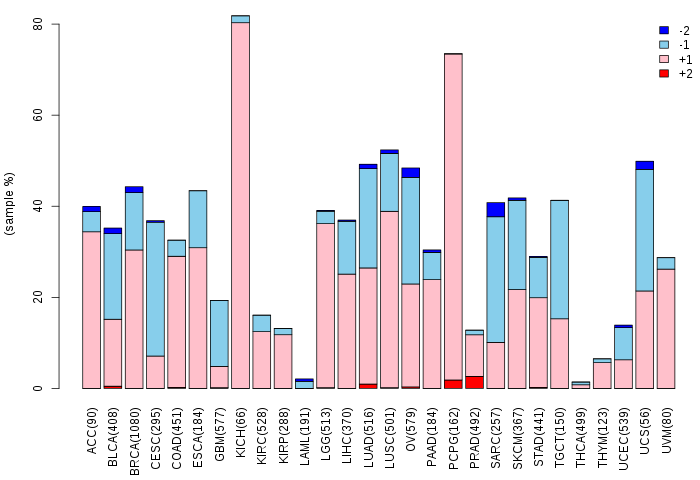
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 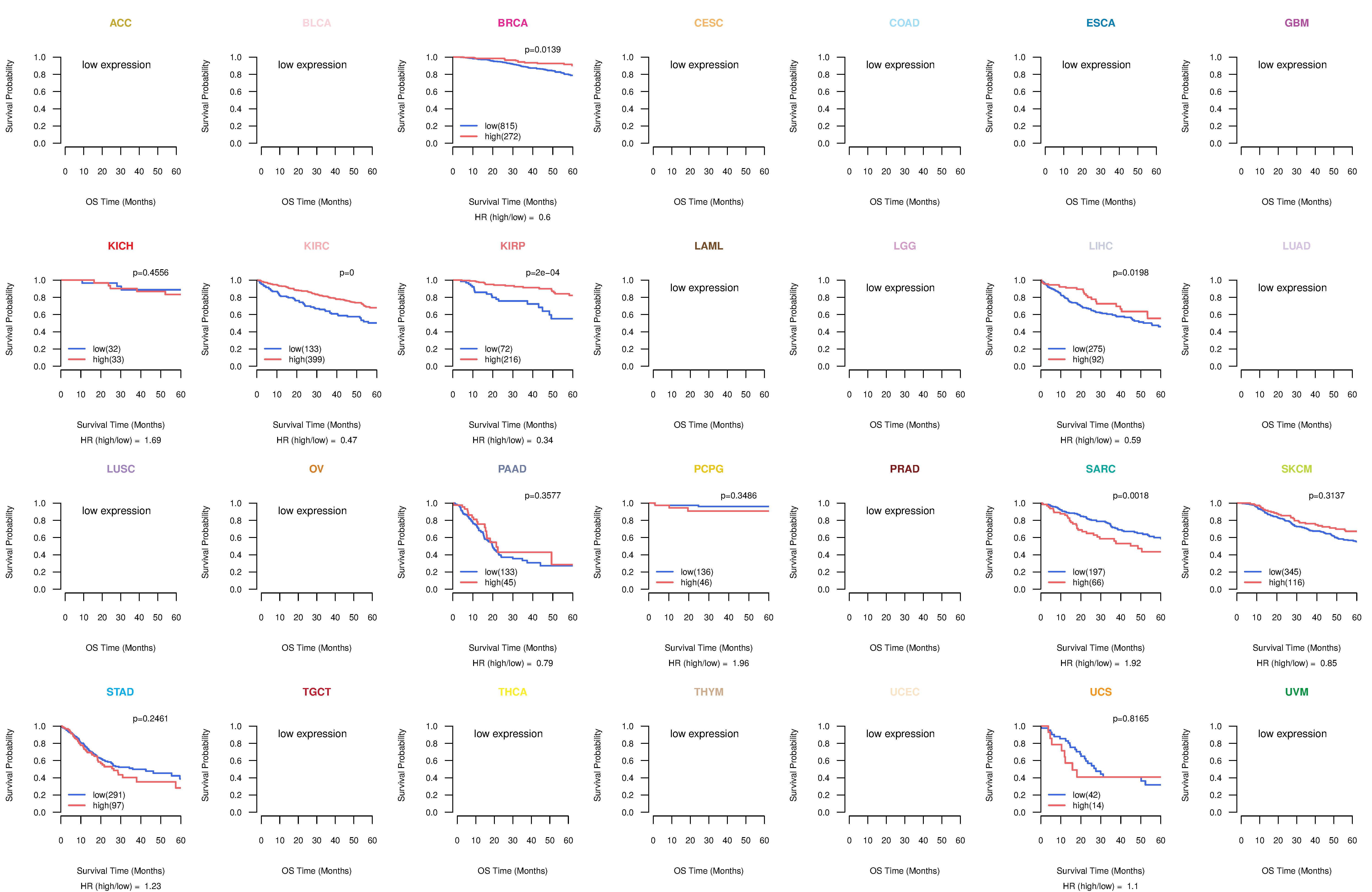
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 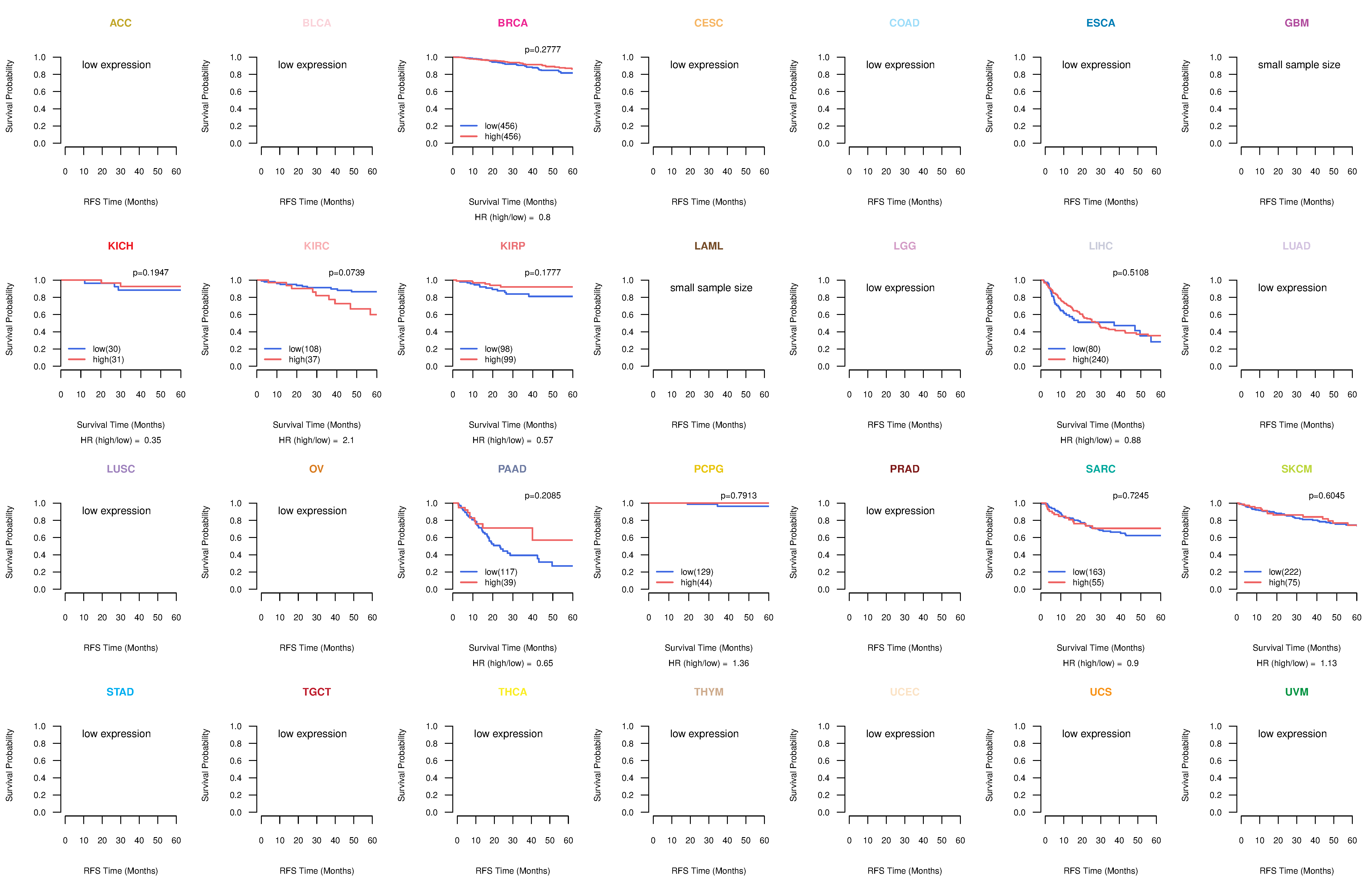
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 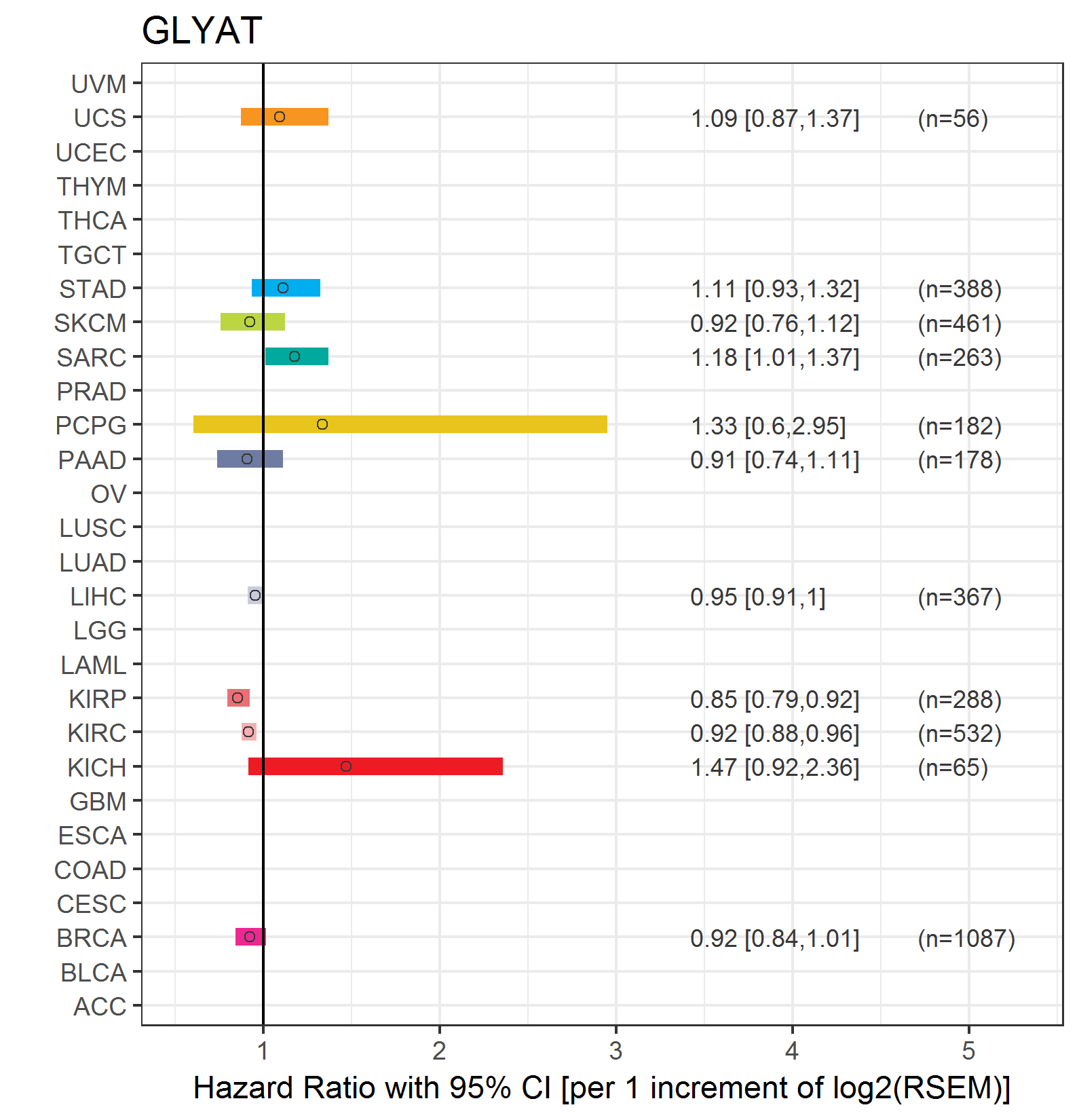
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 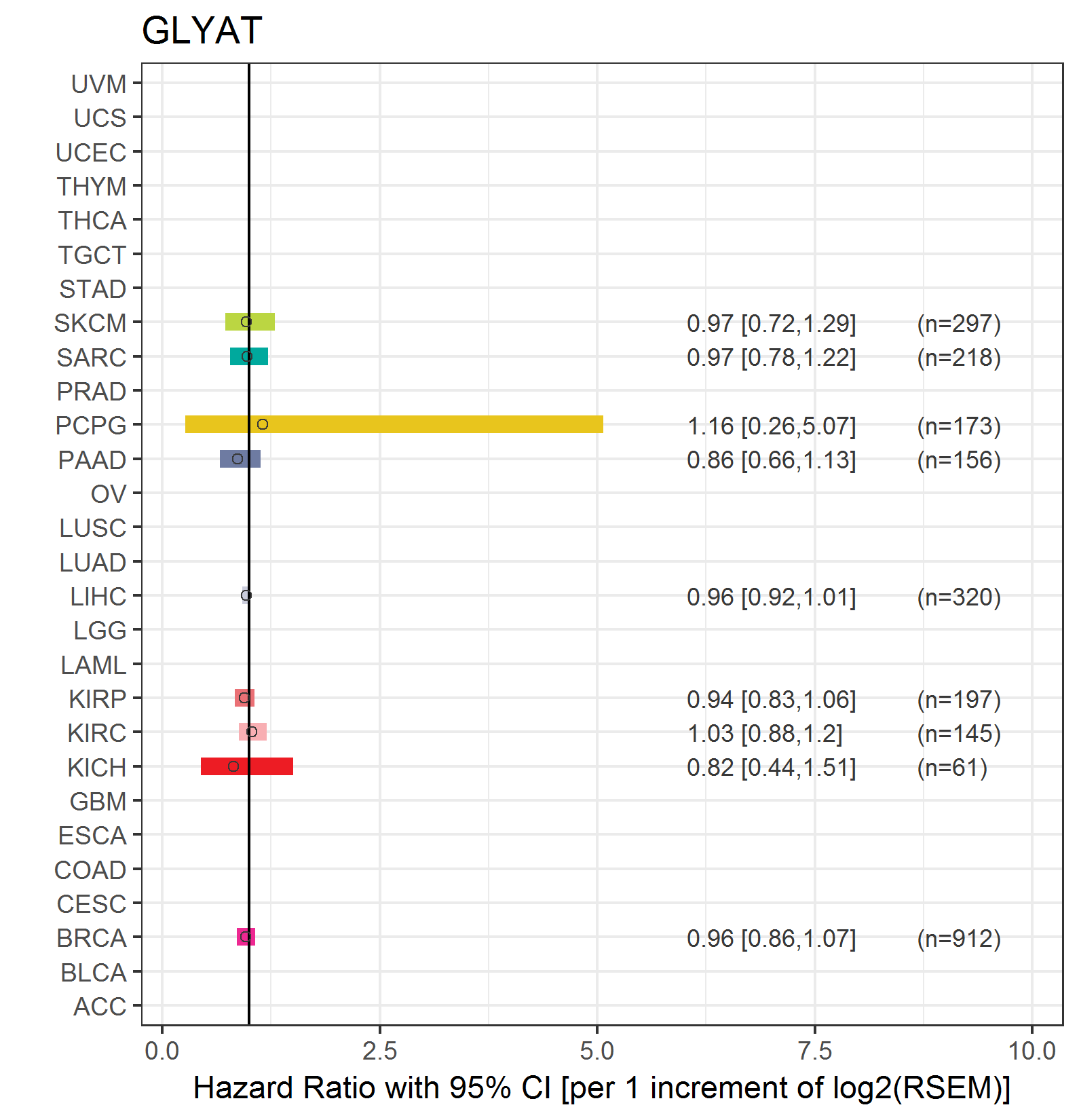
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























