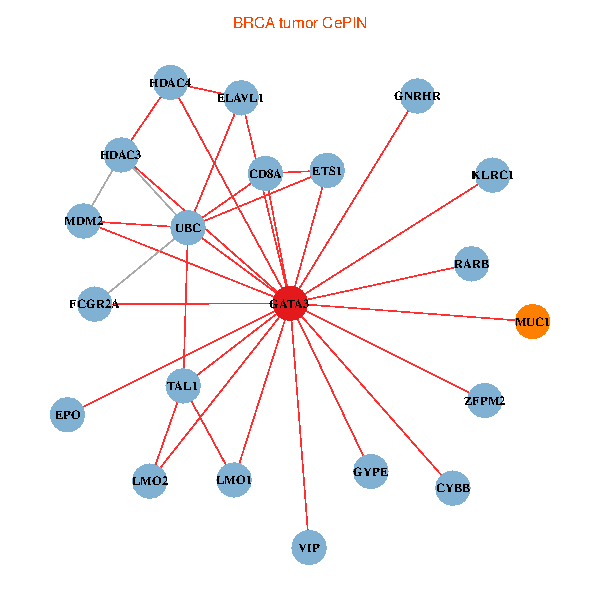
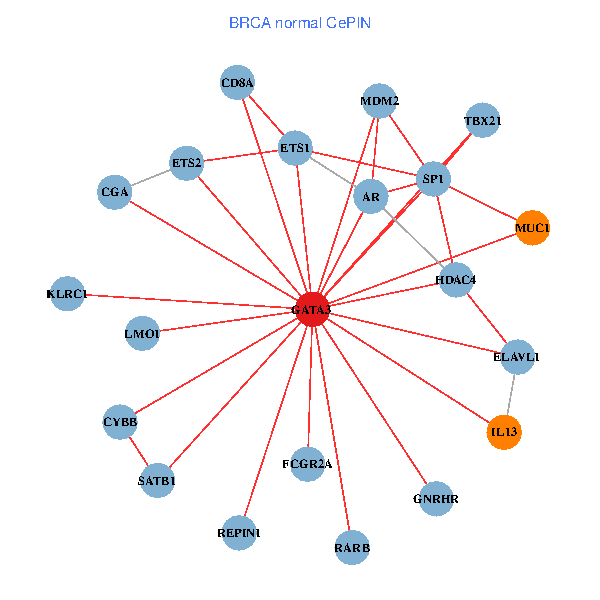
| BRCA (tumor) | BRCA (normal) |
| GATA3, ELAVL1, UBC, MDM2, ETS1, HDAC3, VIP, RARB, CYBB, HDAC4, LMO2, EPO, CD8A, MUC1, ZFPM2, TAL1, GNRHR, FCGR2A, GYPE, KLRC1, LMO1 (tumor) | GATA3, ELAVL1, SP1, MDM2, SATB1, ETS2, ETS1, AR, RARB, CYBB, HDAC4, CD8A, MUC1, IL13, GNRHR, CGA, FCGR2A, REPIN1, TBX21, KLRC1, LMO1 (normal) |
 |  |
|
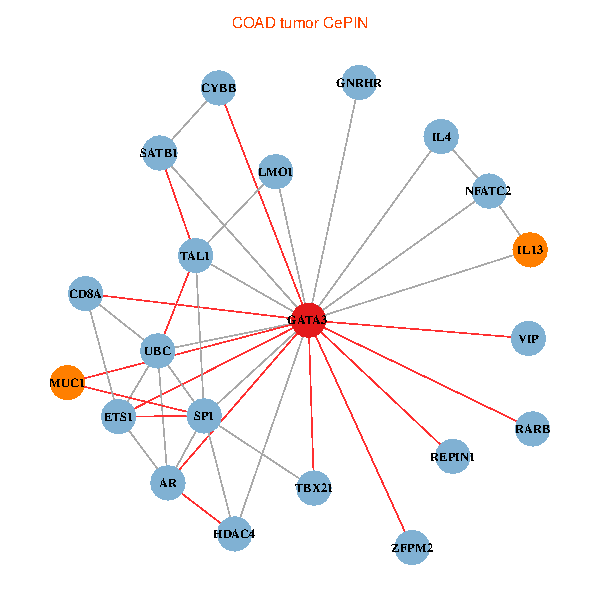
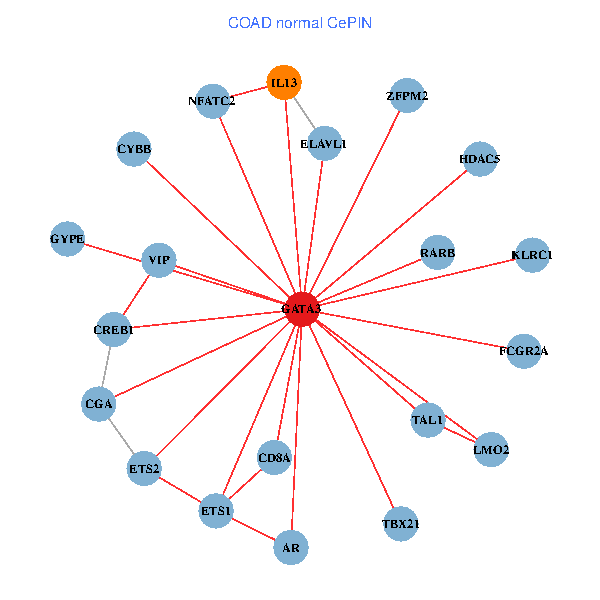
| COAD (tumor) | COAD (normal) |
| GATA3, UBC, SP1, SATB1, ETS1, VIP, AR, RARB, CYBB, HDAC4, NFATC2, IL4, CD8A, MUC1, IL13, ZFPM2, TAL1, GNRHR, REPIN1, TBX21, LMO1 (tumor) | GATA3, ELAVL1, ETS2, ETS1, CREB1, HDAC5, VIP, AR, RARB, CYBB, NFATC2, LMO2, CD8A, IL13, ZFPM2, TAL1, CGA, FCGR2A, TBX21, GYPE, KLRC1 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| GATA3, UBC, MDM2, SATB1, ETS2, ETS1, HDAC3, RARB, CYBB, HDAC4, NFATC2, EPO, CD8A, GNRHR, CGA, FCGR2A, REPIN1, TBX21, HSD17B1, GYPE, KLRC1 (tumor) | GATA3, ELAVL1, SATB1, ETS1, AR, RARB, CYBB, NFATC2, LMO2, IL5, CD8A, MED1, IL13, ZFPM2, TAL1, CGA, FCGR2A, TBX21, HSD17B1, GYPE, KLRC1 (normal) |
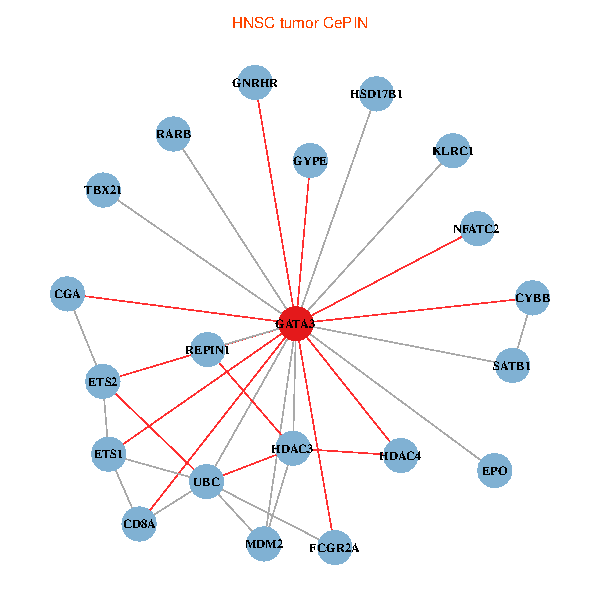 | 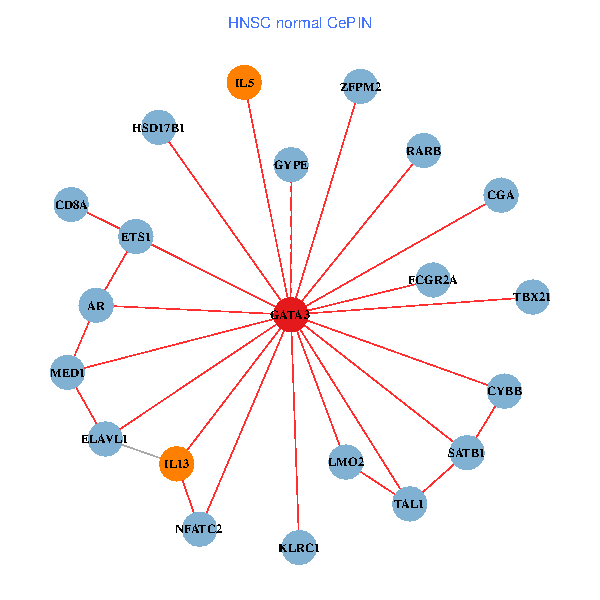 |
|
| KICH (tumor) | KICH (normal) |
| GATA3, SATB1, ETS2, ETS1, HDAC3, VIP, AR, RARB, CYBB, HDAC4, LMO2, CD8A, IL13, ZFPM2, TAL1, GNRHR, FCGR2A, REPIN1, TBX21, HSD17B1, LMO1 (tumor) | GATA3, SP1, ETS1, HDAC3, CREB1, VIP, RARB, CYBB, LMO2, IL4, CD8A, MUC1, ZFPM2, TAL1, GNRHR, FCGR2A, TBX21, HSD17B1, GYPE, KLRC1, LMO1 (normal) |
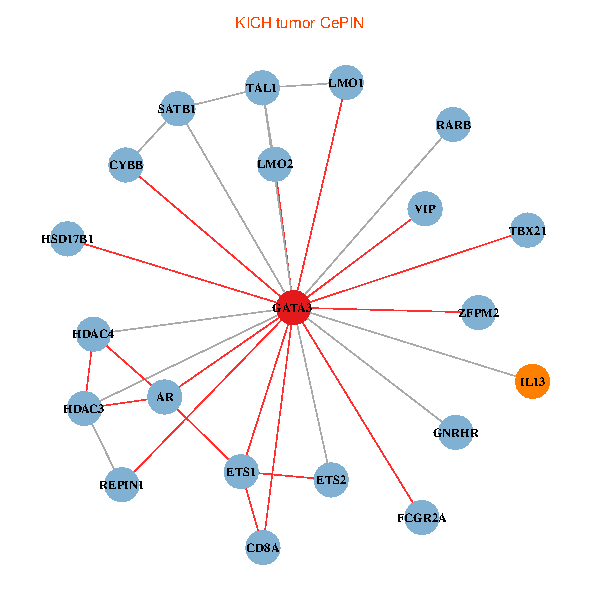 | 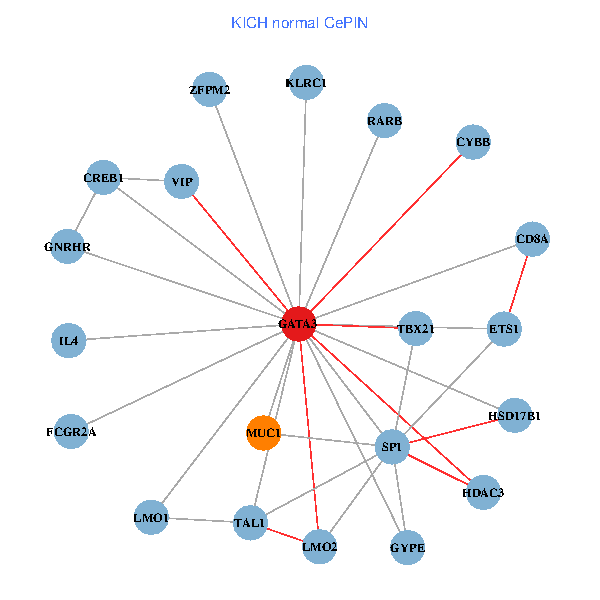 |
|
| KIRC (tumor) | KIRC (normal) |
| GATA3, ELAVL1, SATB1, ETS1, HDAC5, VIP, AR, RARB, CYBB, EPO, IL5, CD8A, MED1, MUC1, ZFPM2, FCGR2A, REPIN1, TBX21, HSD17B1, KLRC1, LMO1 (tumor) | GATA3, ELAVL1, SP1, MDM2, ETS2, HDAC3, HDAC5, AR, EPO, IL4, IL5, CD8A, MED1, MUC1, IL13, ZFPM2, CGA, TBX21, HSD17B1, KLRC1, LMO1 (normal) |
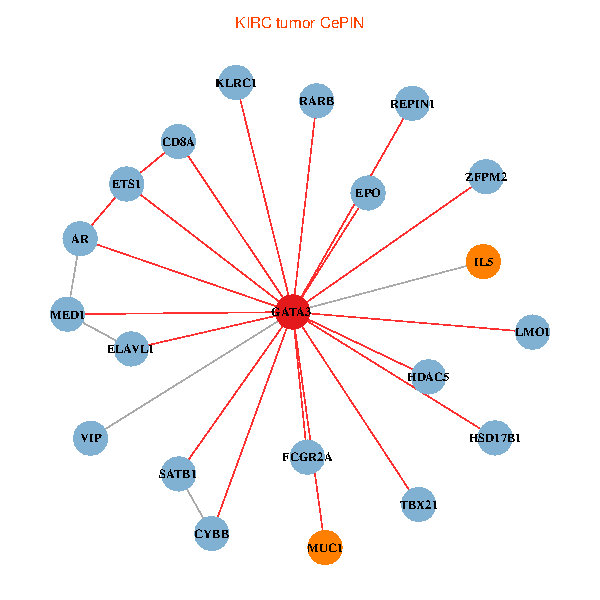 | 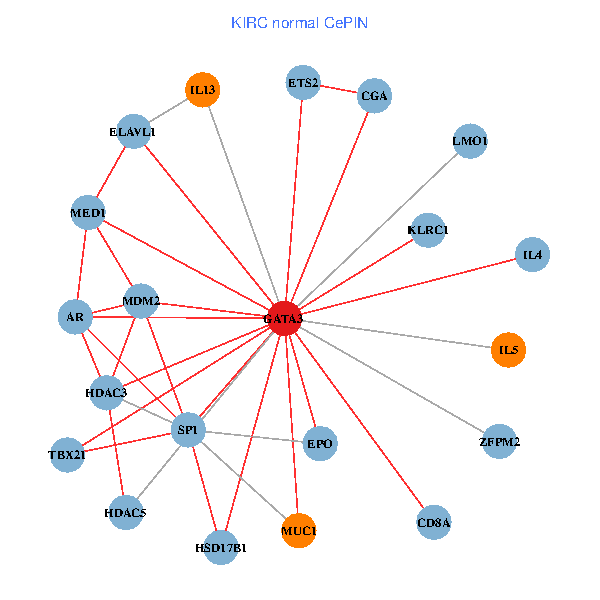 |
|
| KIRP (tumor) | KIRP (normal) |
| GATA3, ELAVL1, ETS2, ETS1, HDAC3, HDAC5, VIP, AR, RARB, CYBB, LMO2, EPO, MUC1, IL13, ZFPM2, CGA, FCGR2A, REPIN1, HSD17B1, KLRC1, LMO1 (tumor) | GATA3, UBC, SP1, ETS2, ETS1, CREB1, VIP, AR, HDAC4, NFATC2, LMO2, CD8A, MUC1, TAL1, CGA, FCGR2A, REPIN1, TBX21, GYPE, KLRC1, LMO1 (normal) |
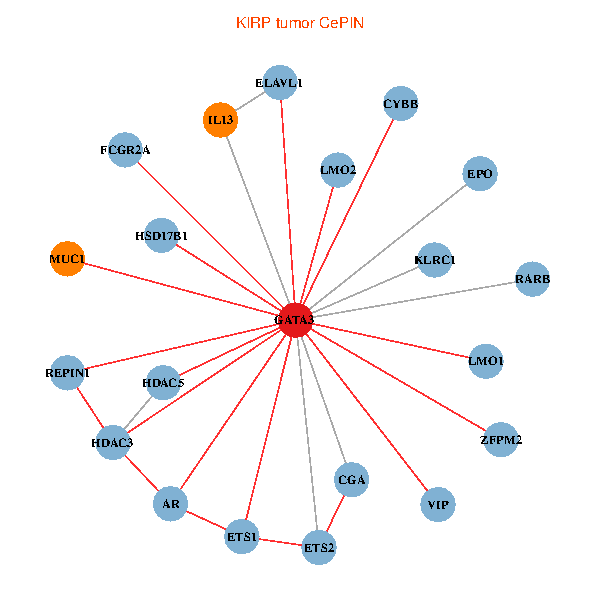 | 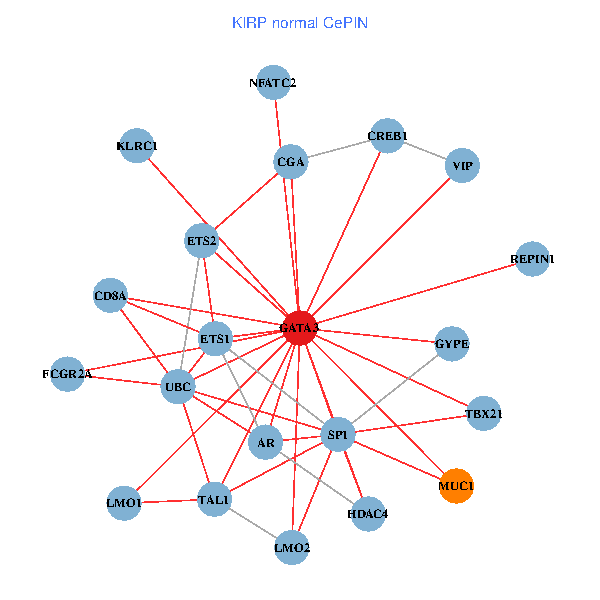 |
|
| LIHC (tumor) | LIHC (normal) |
| GATA3, SP1, SATB1, ETS1, HDAC3, CREB1, HDAC5, RARB, CYBB, NFATC2, LMO2, CD8A, MED1, MUC1, ZFPM2, TAL1, GNRHR, FCGR2A, REPIN1, TBX21, GYPE (tumor) | GATA3, SATB1, ETS1, CREB1, VIP, AR, RARB, CYBB, NFATC2, LMO2, EPO, IL5, CD8A, MUC1, ZFPM2, TAL1, FCGR2A, REPIN1, TBX21, GYPE, KLRC1 (normal) |
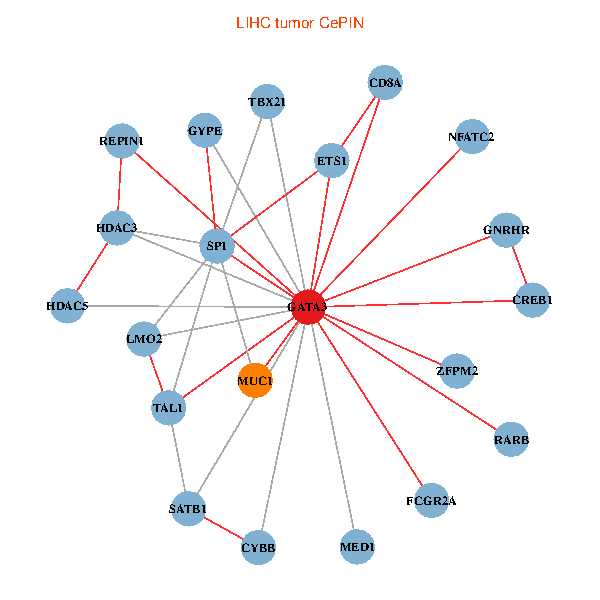 | 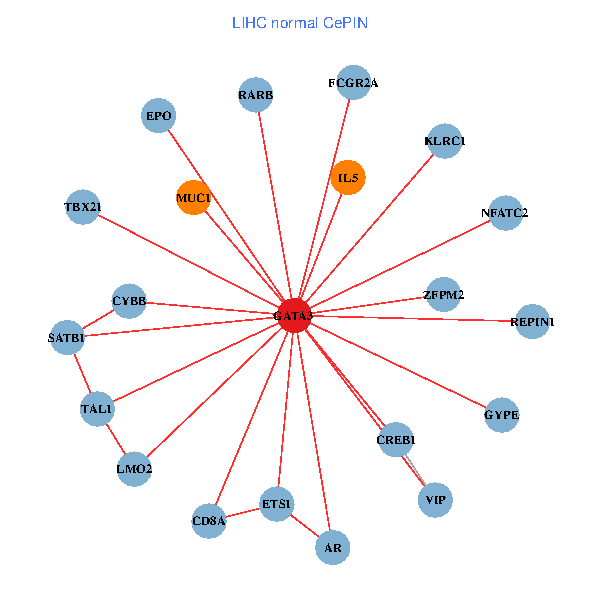 |
|
| LUAD (tumor) | LUAD (normal) |
| GATA3, UBC, SP1, MDM2, SATB1, ETS2, CREB1, HDAC5, VIP, AR, RARB, NFATC2, LMO2, MUC1, IL13, TAL1, CGA, FCGR2A, REPIN1, TBX21, GYPE (tumor) | GATA3, UBC, SP1, SATB1, ETS1, RARB, CYBB, NFATC2, LMO2, EPO, CD8A, MUC1, IL13, ZFPM2, TAL1, FCGR2A, REPIN1, TBX21, HSD17B1, GYPE, KLRC1 (normal) |
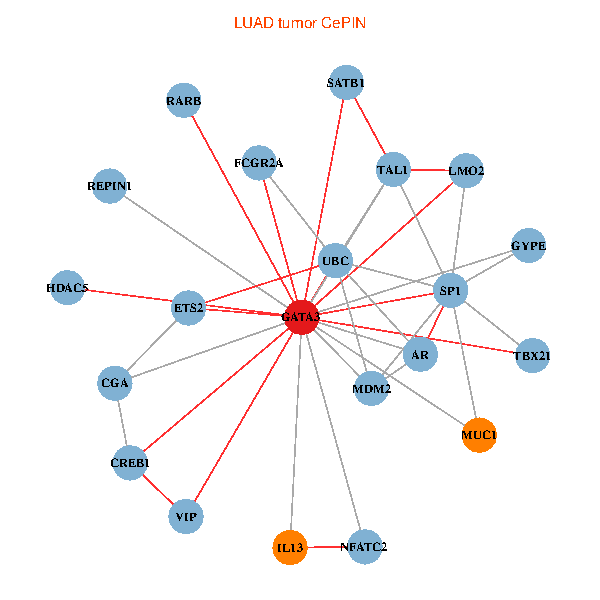 | 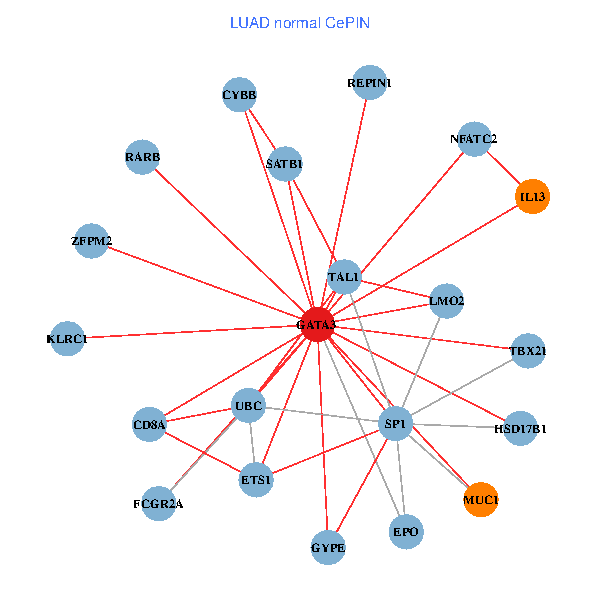 |
|
| LUSC (tumor) | LUSC (normal) |
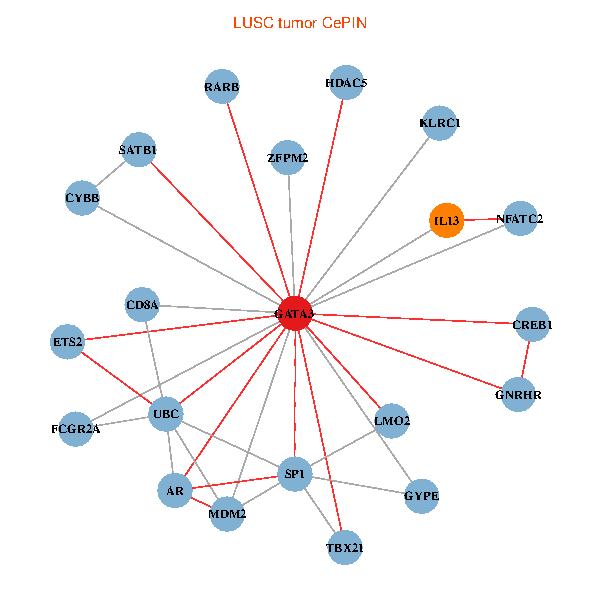
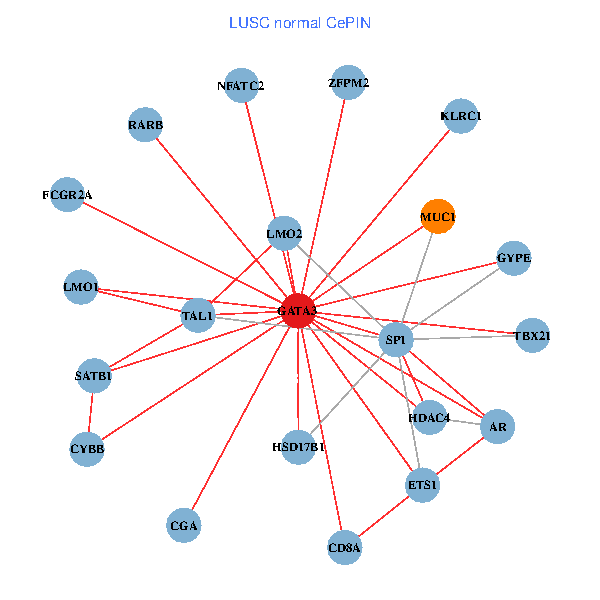
| GATA3, UBC, SP1, MDM2, SATB1, ETS2, CREB1, HDAC5, AR, RARB, CYBB, NFATC2, LMO2, CD8A, IL13, ZFPM2, GNRHR, FCGR2A, TBX21, GYPE, KLRC1 (tumor) | GATA3, SP1, SATB1, ETS1, AR, RARB, CYBB, HDAC4, NFATC2, LMO2, CD8A, MUC1, ZFPM2, TAL1, CGA, FCGR2A, TBX21, HSD17B1, GYPE, KLRC1, LMO1 (normal) |
 |  |
|
| PRAD (tumor) | PRAD (normal) |
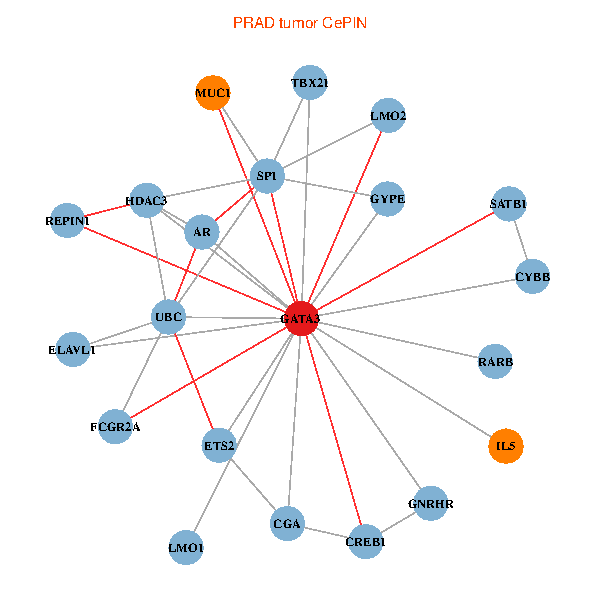
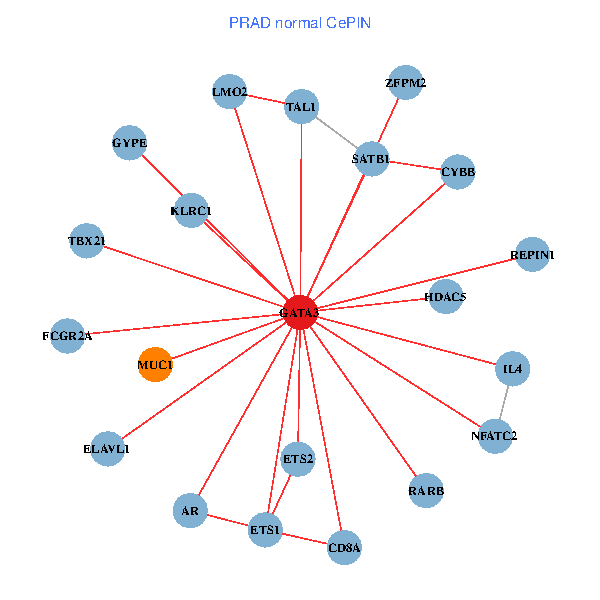
| GATA3, ELAVL1, UBC, SP1, SATB1, ETS2, HDAC3, CREB1, AR, RARB, CYBB, LMO2, IL5, MUC1, GNRHR, CGA, FCGR2A, REPIN1, TBX21, GYPE, LMO1 (tumor) | GATA3, ELAVL1, SATB1, ETS2, ETS1, HDAC5, AR, RARB, CYBB, NFATC2, LMO2, IL4, CD8A, MUC1, ZFPM2, TAL1, FCGR2A, REPIN1, TBX21, GYPE, KLRC1 (normal) |
 |  |
|
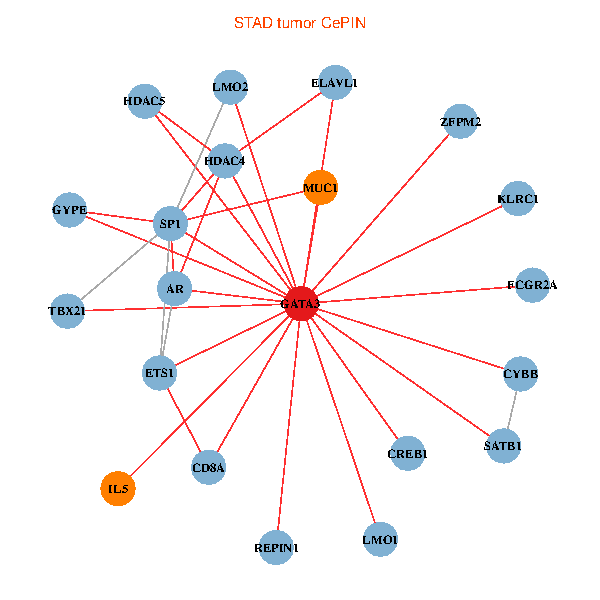
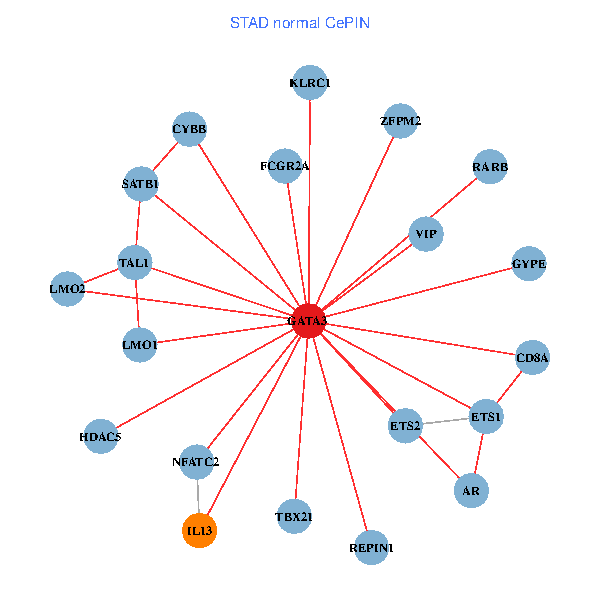
| STAD (tumor) | STAD (normal) |
| GATA3, ELAVL1, SP1, SATB1, ETS1, CREB1, HDAC5, AR, CYBB, HDAC4, LMO2, IL5, CD8A, MUC1, ZFPM2, FCGR2A, REPIN1, TBX21, GYPE, KLRC1, LMO1 (tumor) | GATA3, SATB1, ETS2, ETS1, HDAC5, VIP, AR, RARB, CYBB, NFATC2, LMO2, CD8A, IL13, ZFPM2, TAL1, FCGR2A, REPIN1, TBX21, GYPE, KLRC1, LMO1 (normal) |
 |  |
|
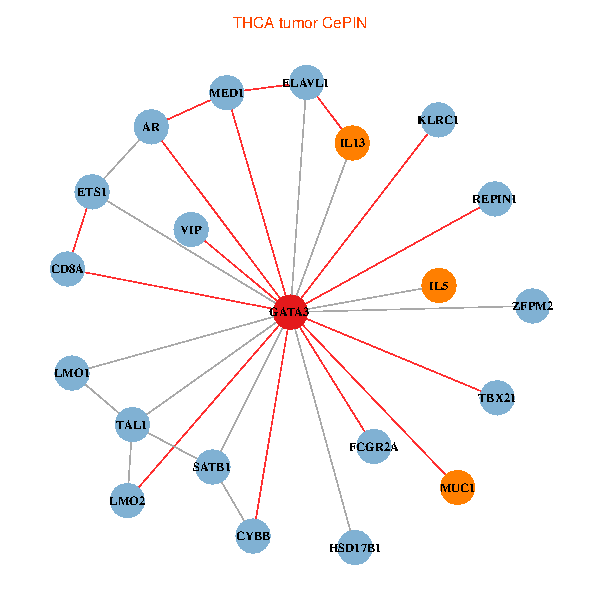
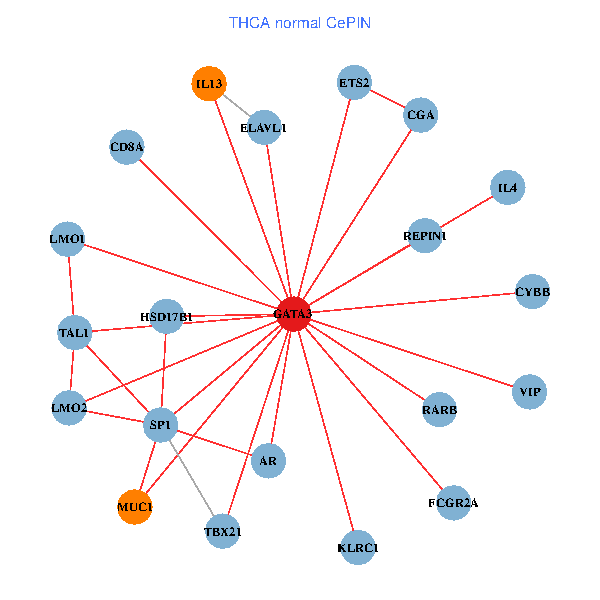
| THCA (tumor) | THCA (normal) |
| GATA3, ELAVL1, SATB1, ETS1, VIP, AR, CYBB, LMO2, IL5, CD8A, MED1, MUC1, IL13, ZFPM2, TAL1, FCGR2A, REPIN1, TBX21, HSD17B1, KLRC1, LMO1 (tumor) | GATA3, ELAVL1, SP1, ETS2, VIP, AR, RARB, CYBB, LMO2, IL4, CD8A, MUC1, IL13, TAL1, CGA, FCGR2A, REPIN1, TBX21, HSD17B1, KLRC1, LMO1 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0006142 | Malignant neoplasm of breast | 40 | BeFree,GAD |
| umls:C0678222 | Breast Carcinoma | 36 | BeFree |
| umls:C1840333 | Barakat syndrome | 24 | BeFree,CLINVAR,CTD_human,MGD,ORPHANET,UNIPROT |
| umls:C0004096 | Asthma | 22 | BeFree,GAD,RGD |
| umls:C0020626 | Hypoparathyroidism | 22 | BeFree,LHGDN |
| umls:C1458155 | Mammary Neoplasms | 14 | BeFree,LHGDN |
| umls:C0235831 | Renal Cell Dysplasia | 13 | BeFree |
| umls:C3536714 | Renal dysplasia | 13 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 11 | BeFree |
| umls:C0596263 | Carcinogenesis | 9 | BeFree |
| umls:C0019829 | Hodgkin Disease | 5 | BeFree,CTD_human,GAD,GWASCAT |
| umls:C2607914 | Allergic rhinitis (disorder) | 5 | BeFree,GAD |
| umls:C0003873 | Rheumatoid Arthritis | 4 | BeFree,CTD_human,GWASCAT |
| umls:C0011053 | Deafness | 4 | LHGDN |
| umls:C0011615 | Dermatitis, Atopic | 4 | BeFree,GAD,LHGDN |
| umls:C0018784 | Sensorineural Hearing Loss (disorder) | 4 | BeFree |
| umls:C0155877 | Allergic asthma | 4 | BeFree |
| umls:C0178874 | Tumor Progression | 4 | BeFree |
| umls:C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 4 | GAD,GWASCAT |
| umls:C0011570 | Mental Depression | 3 | BeFree |
| umls:C0011581 | Depressive disorder | 3 | BeFree |
| umls:C0013595 | Eczema | 3 | BeFree |
| umls:C0021053 | Immune System Diseases | 3 | BeFree |
| umls:C0024141 | Lupus Erythematosus, Systemic | 3 | BeFree,LHGDN |
| umls:C0266292 | Congenital anomaly of the kidney | 3 | BeFree |
| umls:C0000768 | Congenital Abnormality | 2 | BeFree |
| umls:C0002874 | Aplastic Anemia | 2 | BeFree,LHGDN |
| umls:C0005684 | Malignant neoplasm of urinary bladder | 2 | BeFree,GAD |
| umls:C0007134 | Renal Cell Carcinoma | 2 | BeFree |
| umls:C0007138 | Carcinoma, Transitional Cell | 2 | BeFree |
| umls:C0009324 | Ulcerative Colitis | 2 | BeFree |
| umls:C0011603 | Dermatitis | 2 | BeFree |
| umls:C0011849 | Diabetes Mellitus | 2 | BeFree,GAD |
| umls:C0015625 | Fanconi Anemia | 2 | BeFree |
| umls:C0020523 | Immediate hypersensitivity | 2 | GAD |
| umls:C0021367 | Mammary Ductal Carcinoma | 2 | BeFree |
| umls:C0024299 | Lymphoma | 2 | BeFree,LHGDN |
| umls:C0027819 | Neuroblastoma | 2 | BeFree |
| umls:C0033860 | Psoriasis | 2 | BeFree |
| umls:C0035455 | Rhinitis | 2 | BeFree,GAD,LHGDN |
| umls:C0036421 | Systemic Scleroderma | 2 | BeFree |
| umls:C0079774 | Peripheral T-Cell Lymphoma | 2 | BeFree |
| umls:C0085129 | Bronchial Hyperreactivity | 2 | GAD,RGD |
| umls:C0153676 | Secondary malignant neoplasm of lung | 2 | BeFree |
| umls:C0155552 | Hearing Loss, Mixed Conductive-Sensorineural | 2 | BeFree |
| umls:C0242379 | Malignant neoplasm of lung | 2 | BeFree,GAD |
| umls:C0302592 | Cervix carcinoma | 2 | BeFree |
| umls:C0376358 | Malignant neoplasm of prostate | 2 | BeFree |
| umls:C0521158 | Recurrent tumor | 2 | BeFree |
| umls:C0600139 | Prostate carcinoma | 2 | BeFree |
| umls:C0684249 | Carcinoma of lung | 2 | BeFree |
| umls:C0686619 | Secondary malignant neoplasm of lymph node | 2 | BeFree |
| umls:C0700095 | Central neuroblastoma | 2 | BeFree |
| umls:C0853879 | Invasive breast carcinoma | 2 | BeFree |
| umls:C2145472 | Urothelial Carcinoma | 2 | BeFree |
| umls:C3469521 | FANCONI ANEMIA, COMPLEMENTATION GROUP A (disorder) | 2 | BeFree |
| umls:C0002171 | Alopecia Areata | 1 | BeFree |
| umls:C0002622 | Amnesia | 1 | BeFree |
| umls:C0004943 | Behcet Syndrome | 1 | BeFree |
| umls:C0006274 | Bronchiolitis, Viral | 1 | GAD |
| umls:C0007112 | Adenocarcinoma of prostate | 1 | BeFree |
| umls:C0007137 | Squamous cell carcinoma | 1 | BeFree |
| umls:C0007570 | Celiac Disease | 1 | BeFree |
| umls:C0007787 | Transient Ischemic Attack | 1 | BeFree |
| umls:C0009319 | Colitis | 1 | BeFree |
| umls:C0010278 | Craniosynostosis | 1 | BeFree |
| umls:C0011311 | Dengue Fever | 1 | BeFree |
| umls:C0011847 | Diabetes | 1 | BeFree |
| umls:C0011854 | Diabetes Mellitus, Insulin-Dependent | 1 | BeFree |
| umls:C0014072 | Experimental Autoimmune Encephalomyelitis | 1 | BeFree |
| umls:C0018213 | Graves Disease | 1 | BeFree |
| umls:C0019100 | Severe Dengue | 1 | BeFree |
| umls:C0020522 | Delayed Hypersensitivity | 1 | RGD |
| umls:C0021368 | Inflammation | 1 | CTD_human |
| umls:C0021400 | Influenza | 1 | BeFree |
| umls:C0022104 | Irritable Bowel Syndrome | 1 | BeFree |
| umls:C0023452 | Leukemia, Lymphocytic, Acute, L1 | 1 | BeFree |
| umls:C0023485 | Precursor B-Cell Lymphoblastic Leukemia-Lymphoma | 1 | CTD_human |
| umls:C0023493 | Adult T-Cell Lymphoma/Leukemia | 1 | CTD_human |
| umls:C0024117 | Chronic Obstructive Airway Disease | 1 | GAD |
| umls:C0024131 | Lupus Vulgaris | 1 | BeFree |
| umls:C0024138 | Lupus Erythematosus, Discoid | 1 | BeFree |
| umls:C0024143 | Lupus Nephritis | 1 | BeFree |
| umls:C0024667 | Animal Mammary Neoplasms | 1 | CTD_human |
| umls:C0024668 | Mammary Neoplasms, Experimental | 1 | CTD_human |
| umls:C0026691 | Mucocutaneous Lymph Node Syndrome | 1 | BeFree |
| umls:C0027430 | Nasal Polyps | 1 | BeFree |
| umls:C0027697 | Nephritis | 1 | BeFree |
| umls:C0030297 | Pancreatic Neoplasm | 1 | LHGDN |
| umls:C0031099 | Periodontitis | 1 | BeFree |
| umls:C0031106 | Periodontitis, Juvenile | 1 | BeFree |
| umls:C0031154 | Peritonitis | 1 | BeFree |
| umls:C0032285 | Pneumonia | 1 | BeFree |
| umls:C0033578 | Prostatic Neoplasms | 1 | BeFree |
| umls:C0035235 | Respiratory Syncytial Virus Infections | 1 | GAD |
| umls:C0035457 | Rhinitis, Allergic, Perennial | 1 | GAD |
| umls:C0037274 | Dermatologic disorders | 1 | BeFree |
| umls:C0038356 | Stomach Neoplasms | 1 | LHGDN |
| umls:C0038454 | Cerebrovascular accident | 1 | BeFree,GAD |
| umls:C0042065 | Genitourinary Neoplasms | 1 | BeFree |
| umls:C0042170 | Uveomeningoencephalitic Syndrome | 1 | BeFree |
| umls:C0079772 | T-Cell Lymphoma | 1 | BeFree |
| umls:C0085859 | Polyglandular Type I Autoimmune Syndrome | 1 | BeFree |
| umls:C0152013 | Adenocarcinoma of lung (disorder) | 1 | BeFree |
| umls:C0206180 | Ki-1+ Anaplastic Large Cell Lymphoma | 1 | BeFree |
| umls:C0206708 | Cervical Intraepithelial Neoplasia | 1 | BeFree |
| umls:C0220647 | Carcinoma of unknown primary | 1 | BeFree |
| umls:C0240035 | Interstitial fibrosis | 1 | BeFree |
| umls:C0262985 | Psoriasiform eczema | 1 | BeFree |
| umls:C0266295 | Congenital hypoplasia of kidney | 1 | BeFree |
| umls:C0270823 | Petit mal status | 1 | BeFree |
| umls:C0275522 | Asymptomatic Infections | 1 | BeFree |
| umls:C0279000 | Liver and Intrahepatic Biliary Tract Carcinoma | 1 | BeFree |
| umls:C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 1 | BeFree |
| umls:C0345904 | Malignant neoplasm of liver | 1 | BeFree |
| umls:C0346153 | Breast Cancer, Familial | 1 | BeFree |
| umls:C0349458 | Cervical intraepithelial neoplasia grade 1 | 1 | BeFree |
| umls:C0349459 | Cervical intraepithelial neoplasia grade 2 | 1 | BeFree |
| umls:C0375023 | Respiratory syncytial virus (RSV) infection in conditions classified elsewhere and of unspecified site | 1 | BeFree |
| umls:C0403367 | proliferative nephritis unspecified | 1 | BeFree |
| umls:C0403601 | Transplant glomerulopathy | 1 | BeFree |
| umls:C0409974 | Lupus Erythematosus | 1 | BeFree |
| umls:C0432306 | Ichthyosis Bullosa of Siemens | 1 | BeFree |
| umls:C0476227 | pricking of skin | 1 | BeFree |
| umls:C0598766 | Leukemogenesis | 1 | BeFree |
| umls:C0598935 | Tumor Initiation | 1 | BeFree |
| umls:C0699885 | Carcinoma of bladder | 1 | BeFree |
| umls:C0729552 | Genital infection | 1 | BeFree |
| umls:C0751295 | Memory Loss | 1 | BeFree |
| umls:C0795836 | Chromosome 10, monosomy 10p | 1 | BeFree |
| umls:C0851140 | Carcinoma in situ of uterine cervix | 1 | BeFree |
| umls:C1290884 | Inflammatory disorder | 1 | BeFree |
| umls:C1332225 | Aggressive Non-Hodgkin Lymphoma | 1 | BeFree |
| umls:C1333064 | Classical Hodgkin's Lymphoma | 1 | BeFree |
| umls:C1336745 | Thymic Lymphoma | 1 | BeFree |
| umls:C1527249 | Colorectal Cancer | 1 | GWASCAT |
| umls:C1561535 | Creatinine finding | 1 | GAD |
| umls:C1709246 | Non-Neoplastic Disorder | 1 | BeFree |
| umls:C1854365 | BREAST CANCER 3 | 1 | BeFree |
| umls:C1961099 | Precursor T-Cell Lymphoblastic Leukemia-Lymphoma | 1 | BeFree |
| umls:C2239176 | Liver carcinoma | 1 | BeFree |
| umls:C3539878 | Triple Negative Breast Neoplasms | 1 | BeFree |
| umls:C3809874 | LEUKEMIA, ACUTE LYMPHOBLASTIC, SUSCEPTIBILITY TO, 3 | 0 | ORPHANET |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))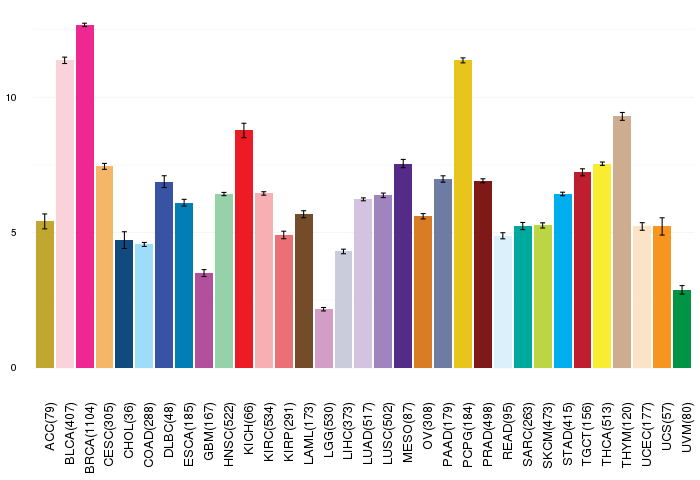
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))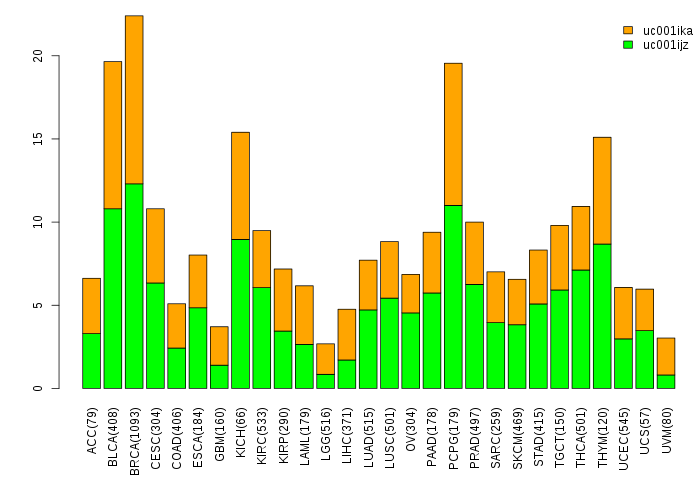
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data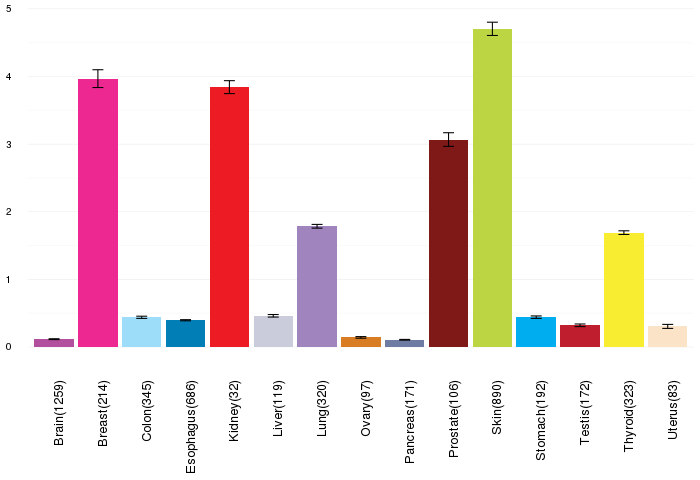
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))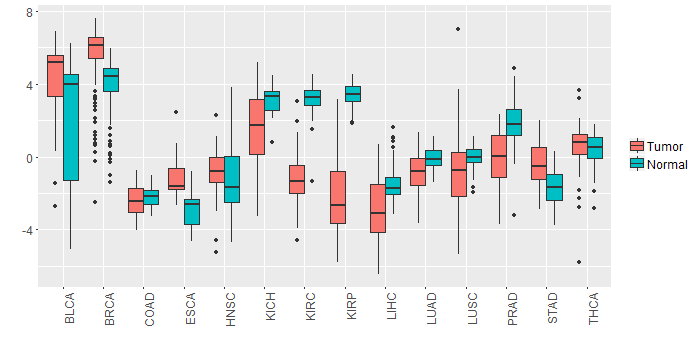
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.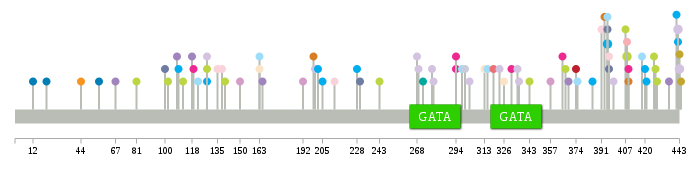

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 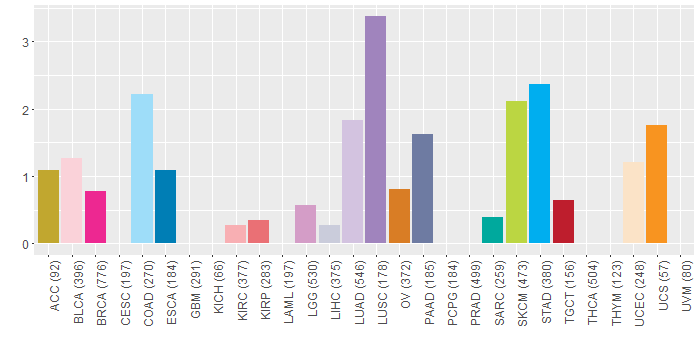
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)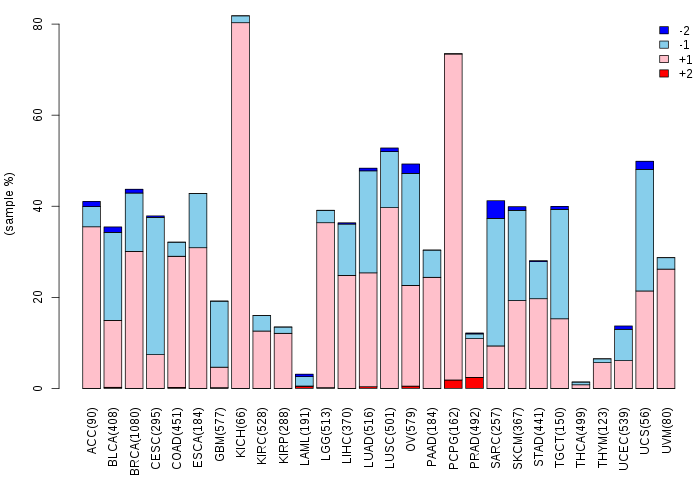
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 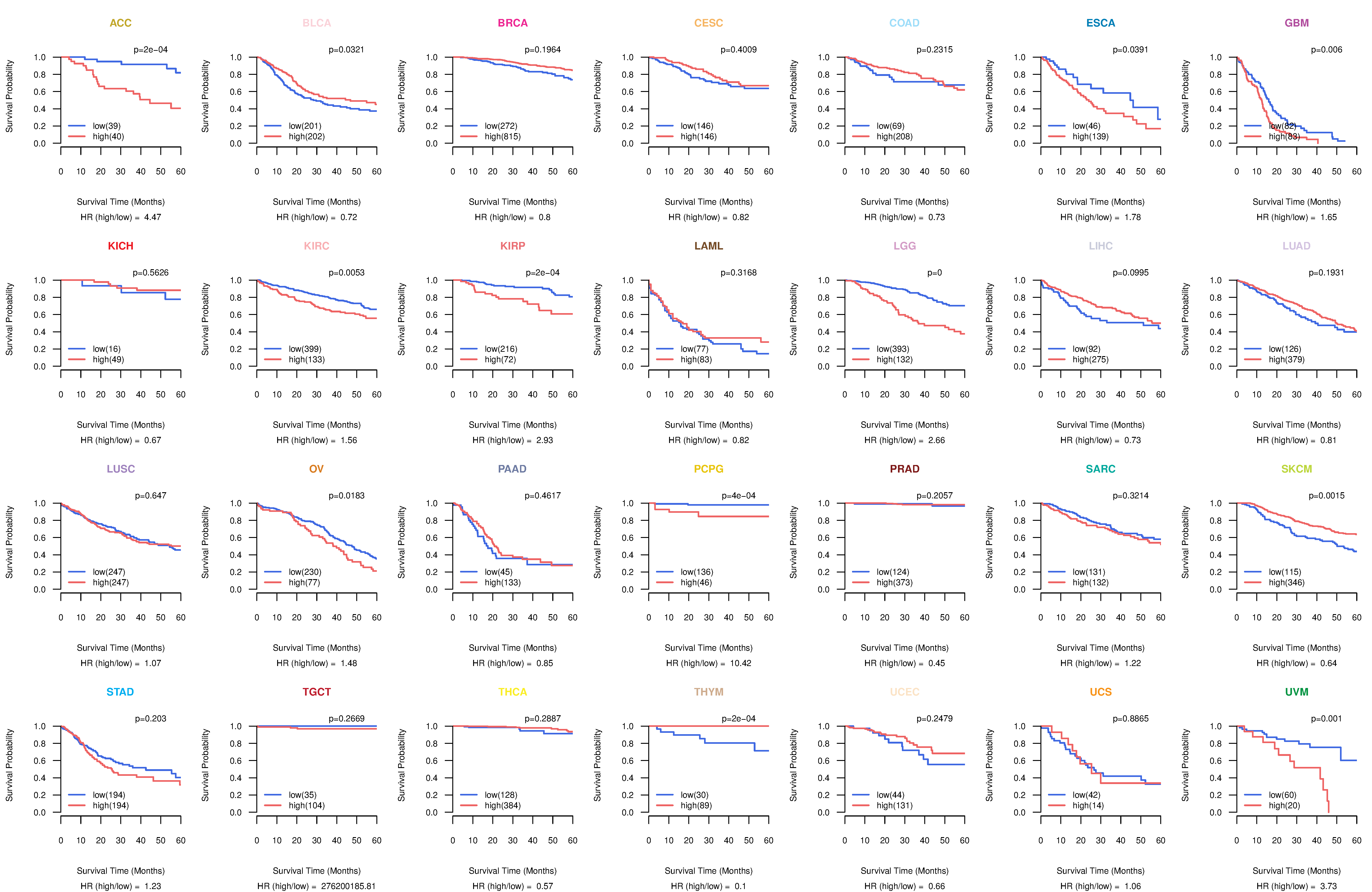
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 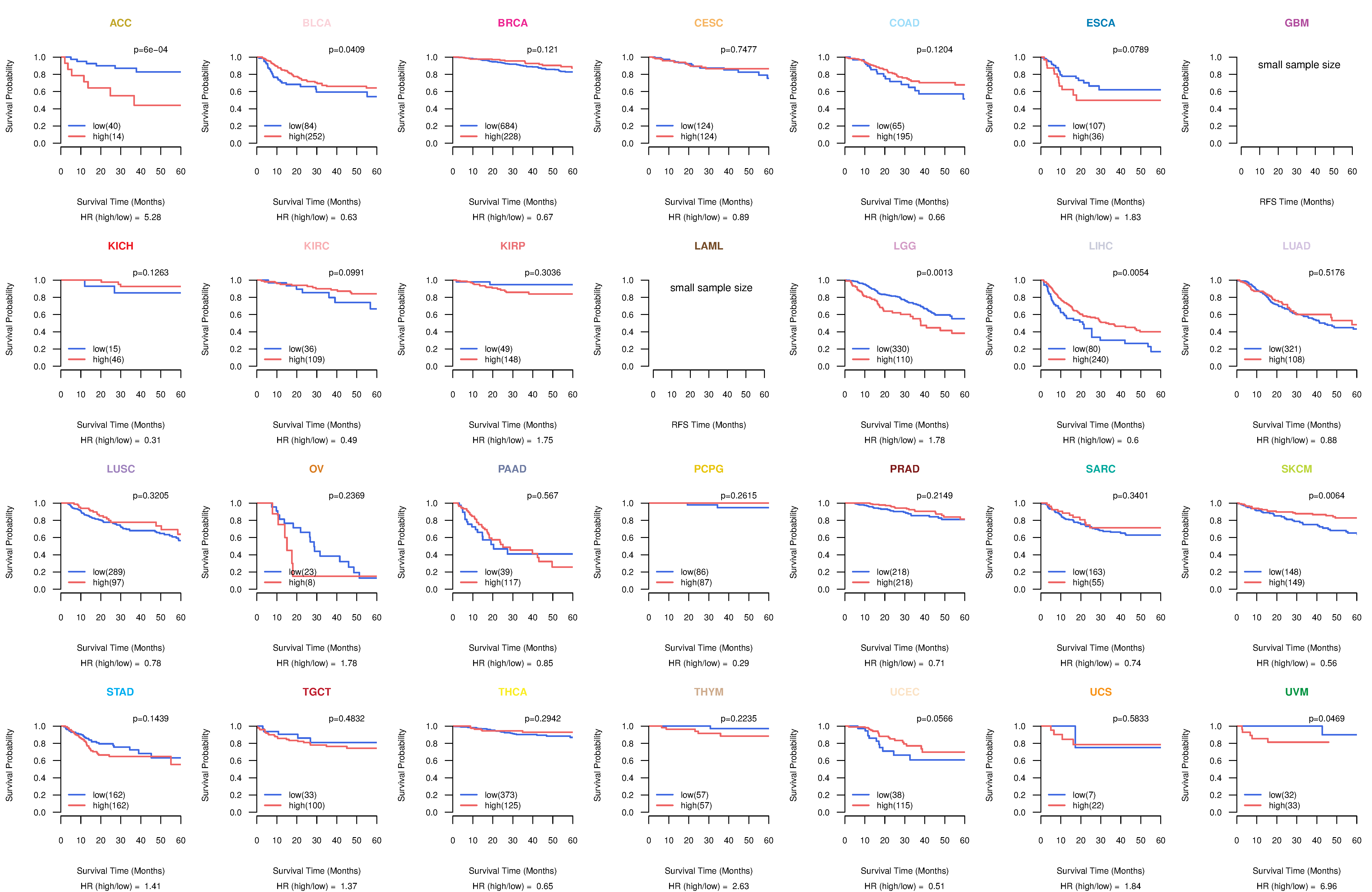
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 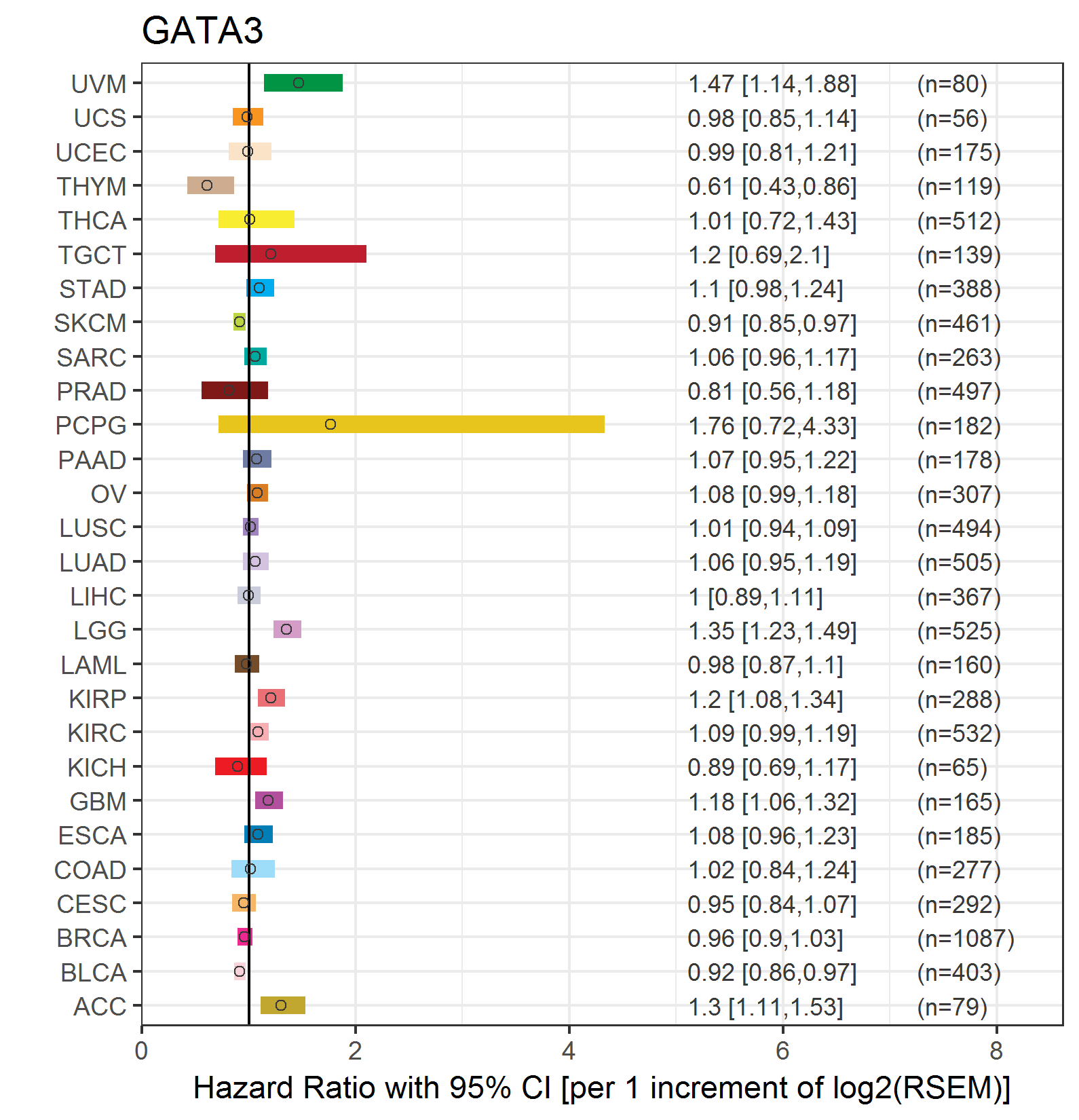
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 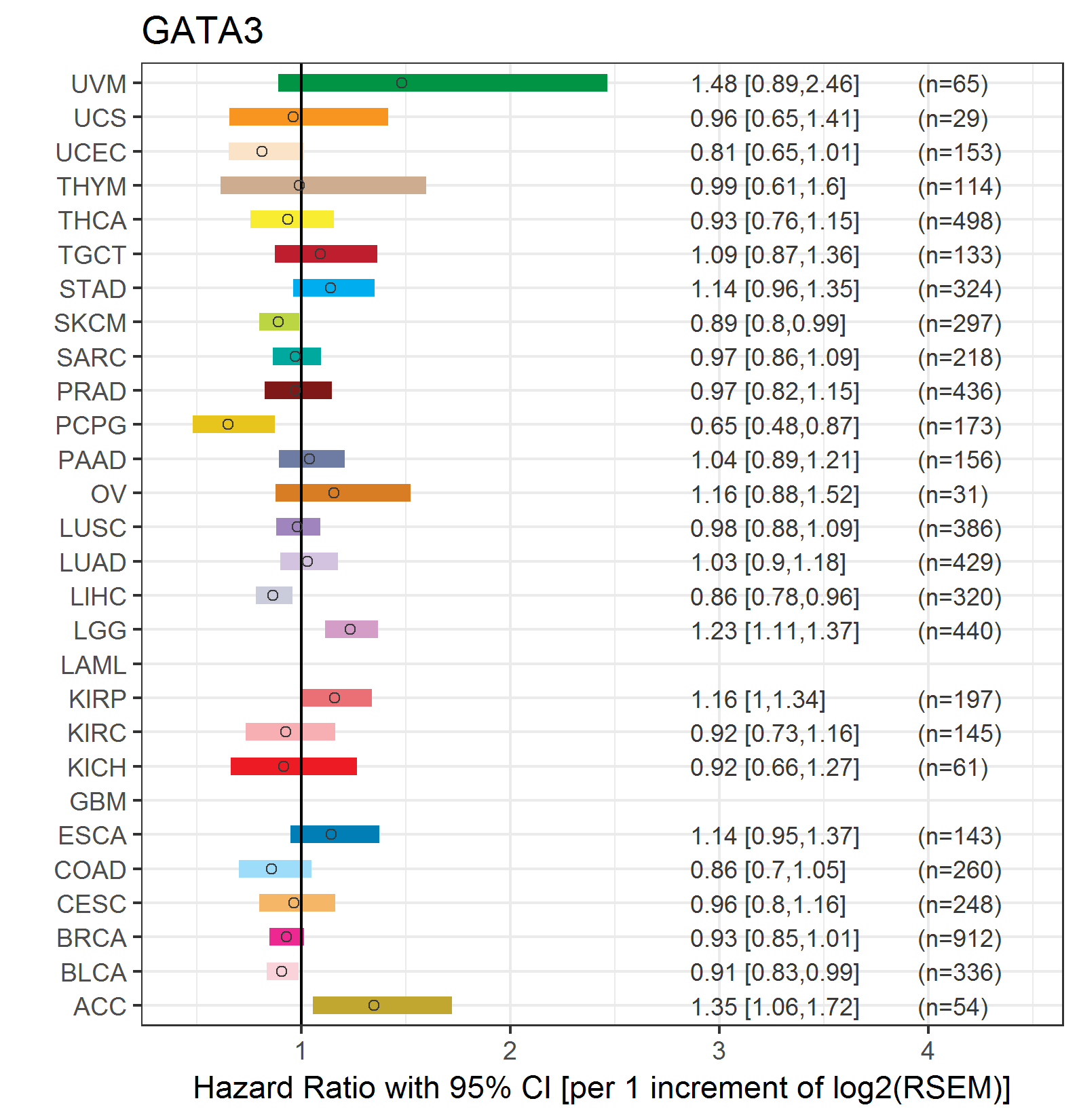
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























