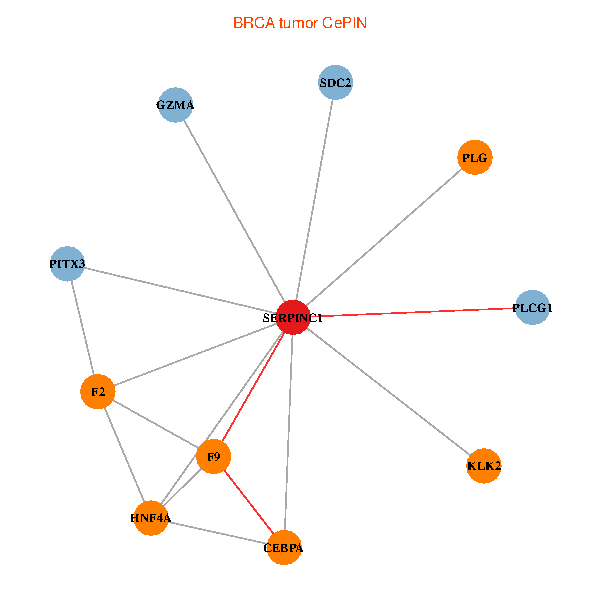
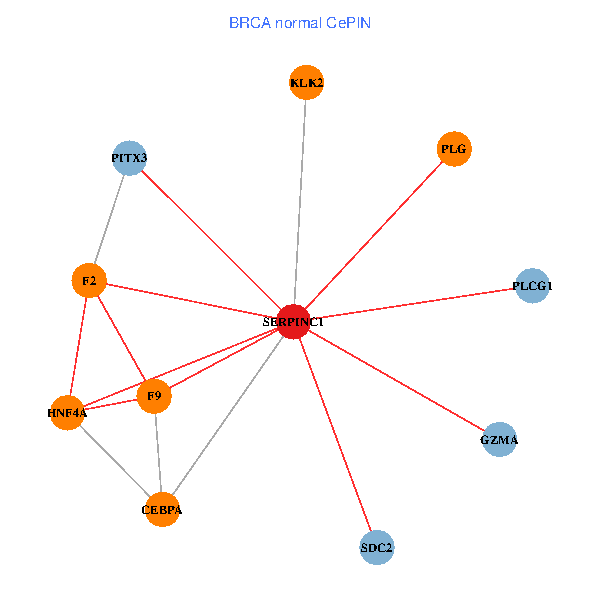
| BRCA (tumor) | BRCA (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
 |  |
|
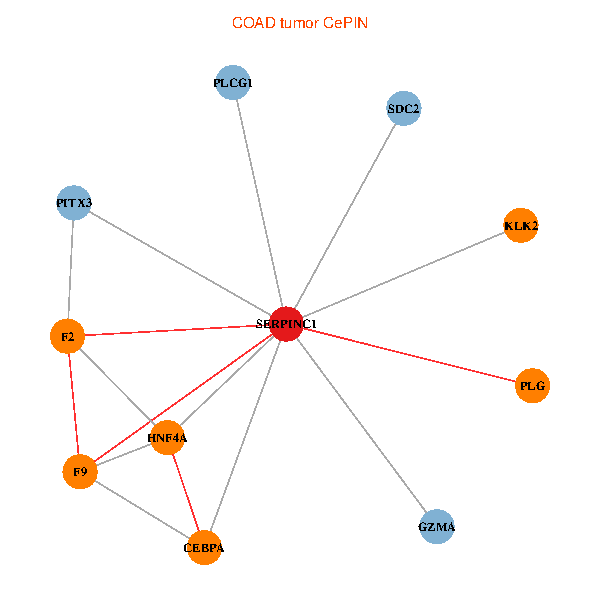
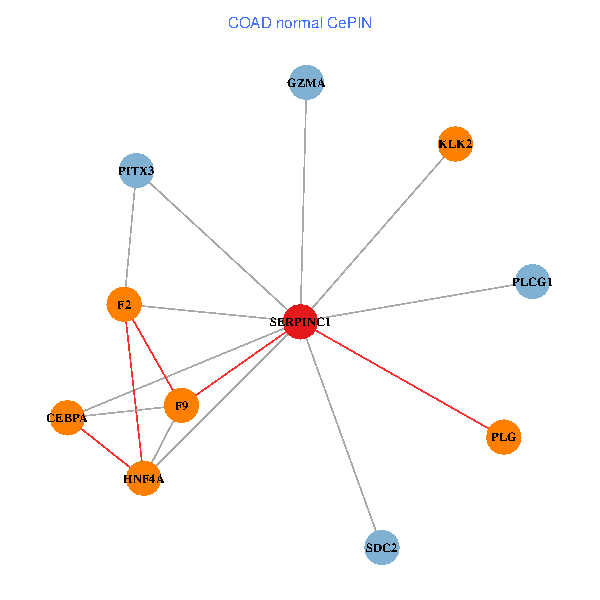
| COAD (tumor) | COAD (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
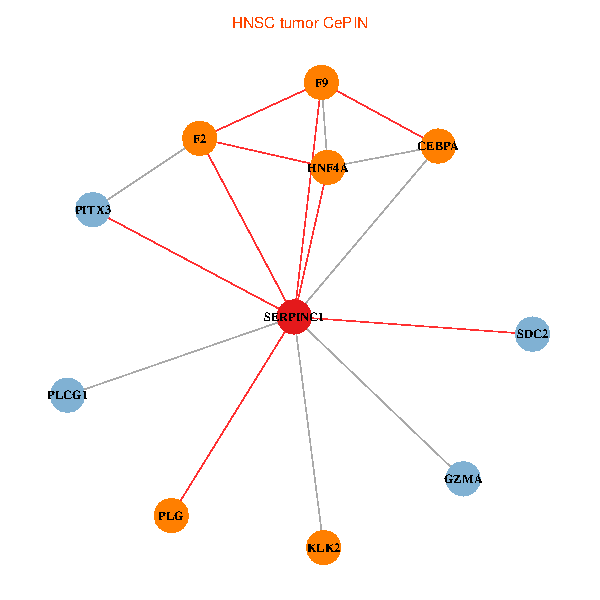 | 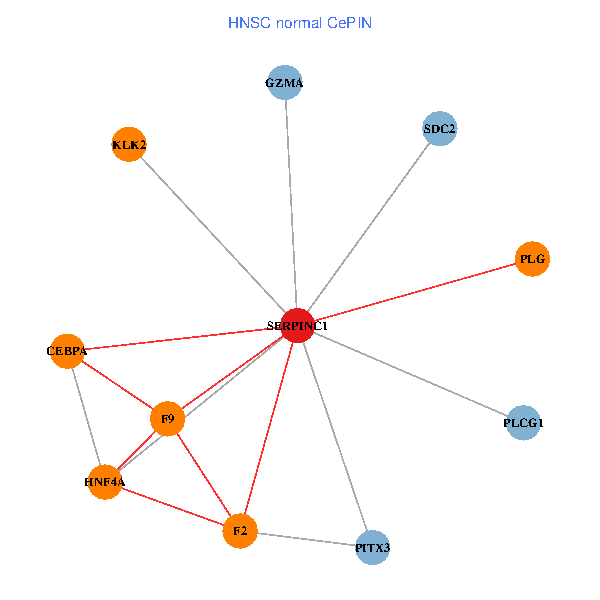 |
|
| KICH (tumor) | KICH (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
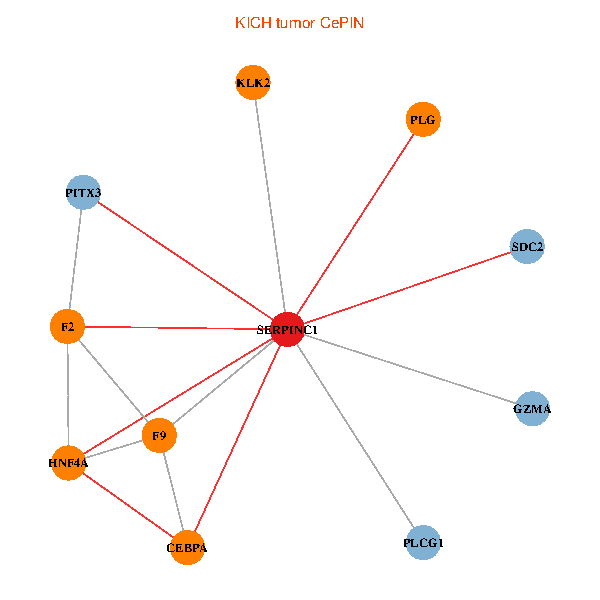 | 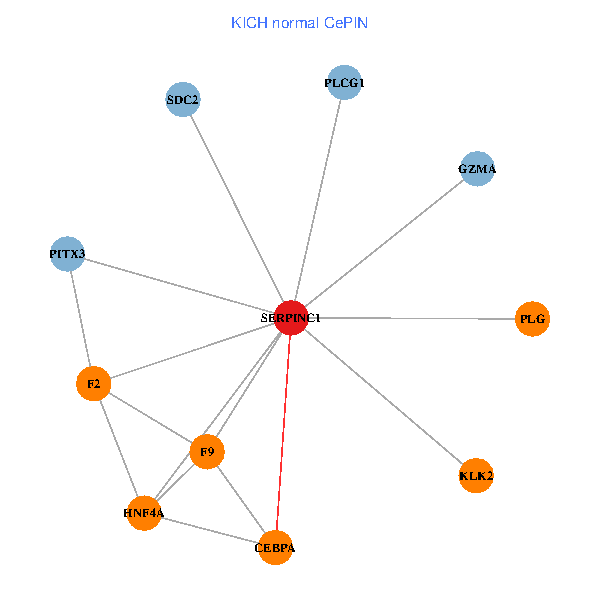 |
|
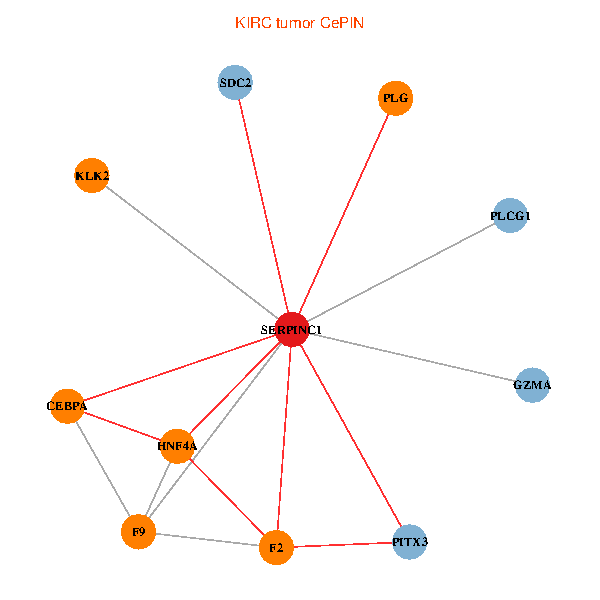
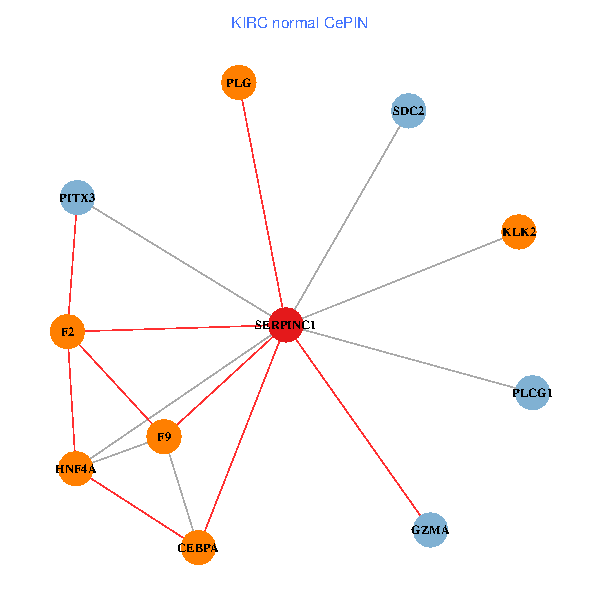
| KIRC (tumor) | KIRC (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
 |  |
|
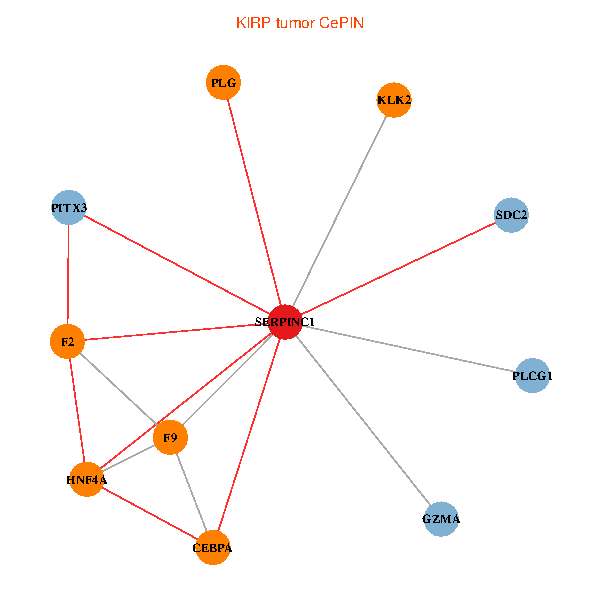
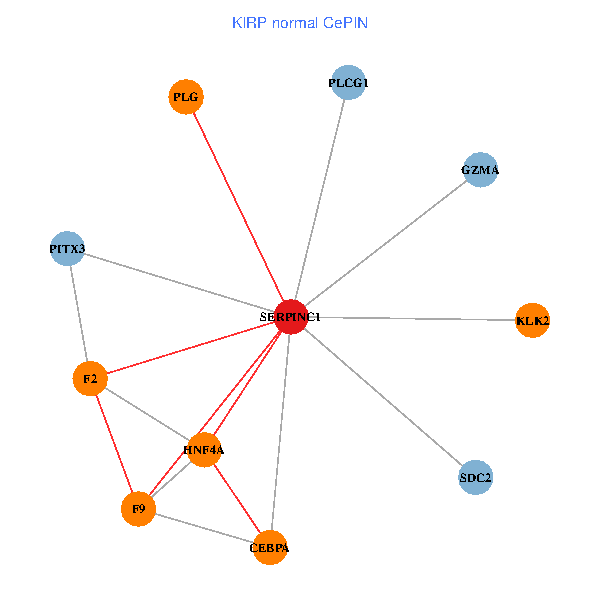
| KIRP (tumor) | KIRP (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
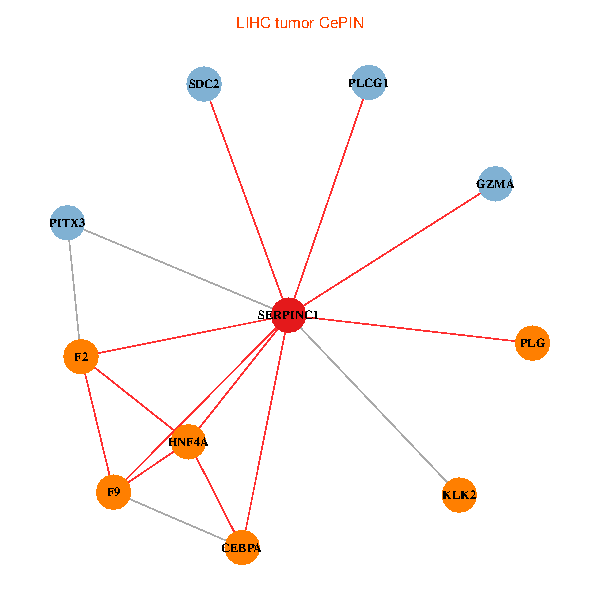 | 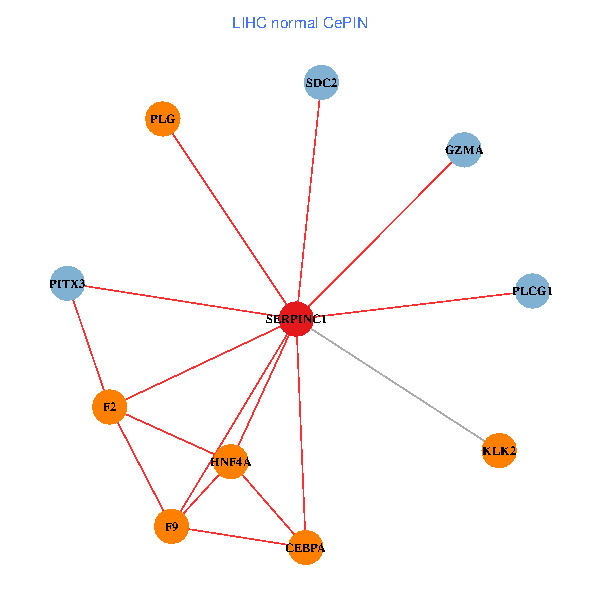 |
|
| LUAD (tumor) | LUAD (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
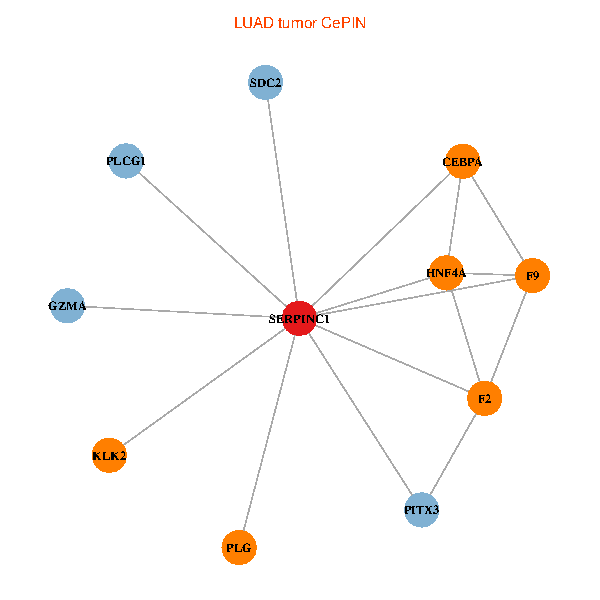 | 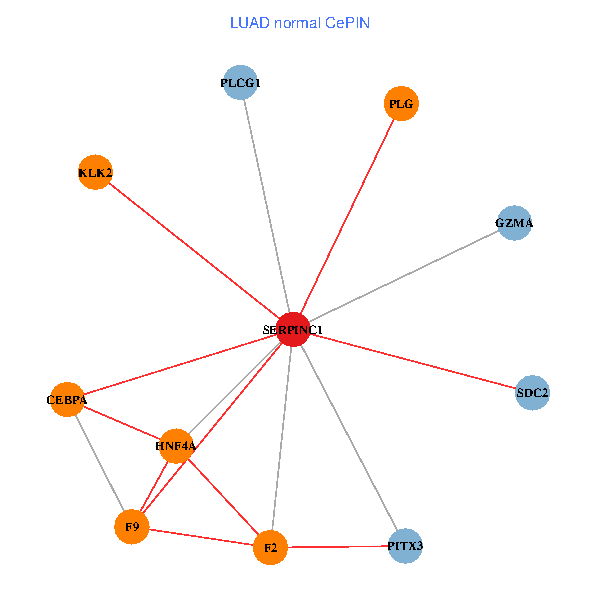 |
|
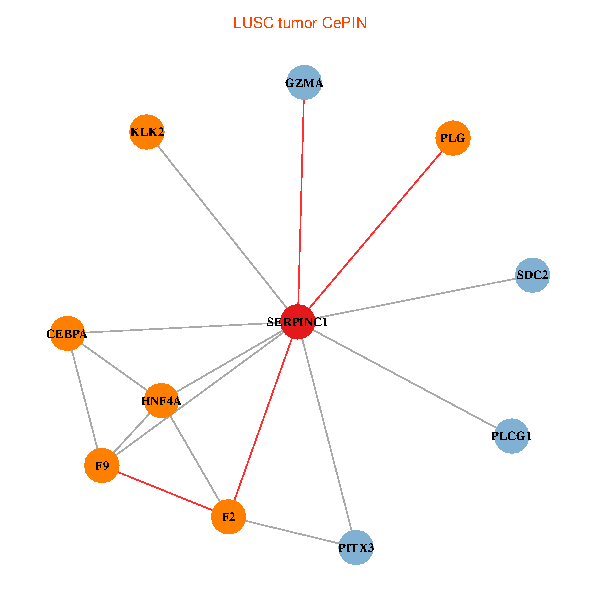
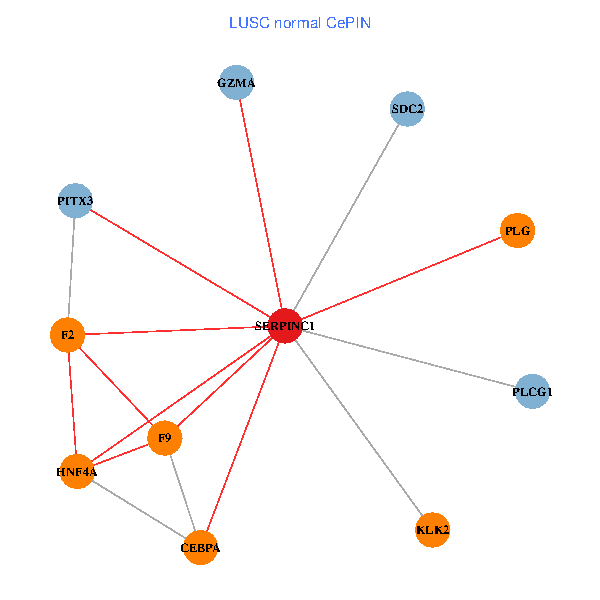
| LUSC (tumor) | LUSC (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
 |  |
|
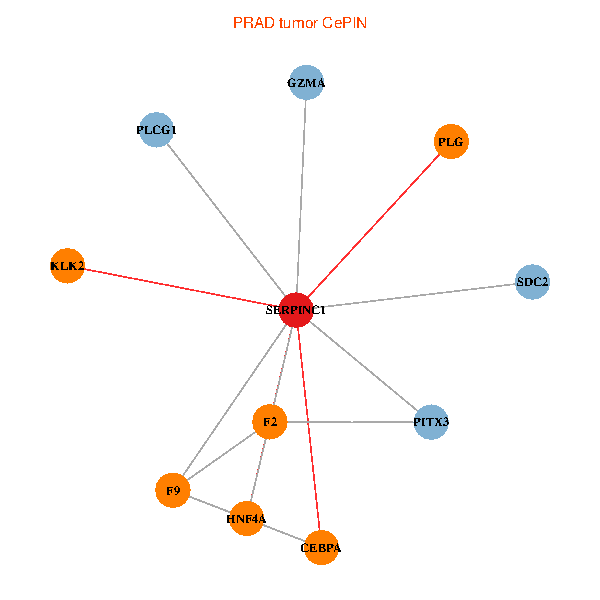
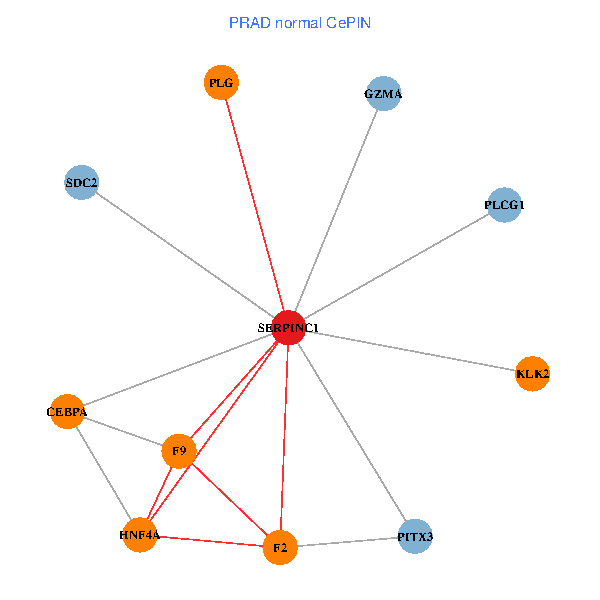
| PRAD (tumor) | PRAD (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
 |  |
|
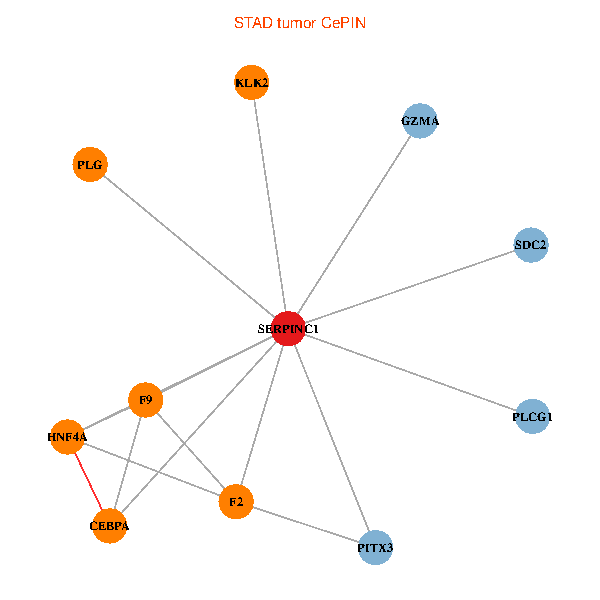
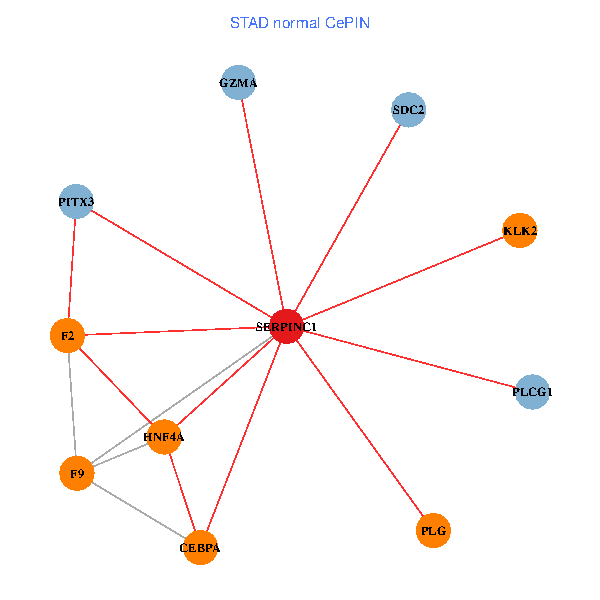
| STAD (tumor) | STAD (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
 |  |
|
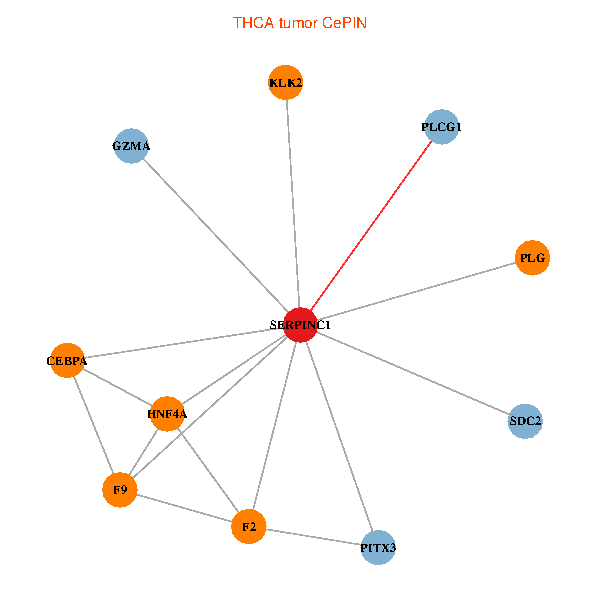
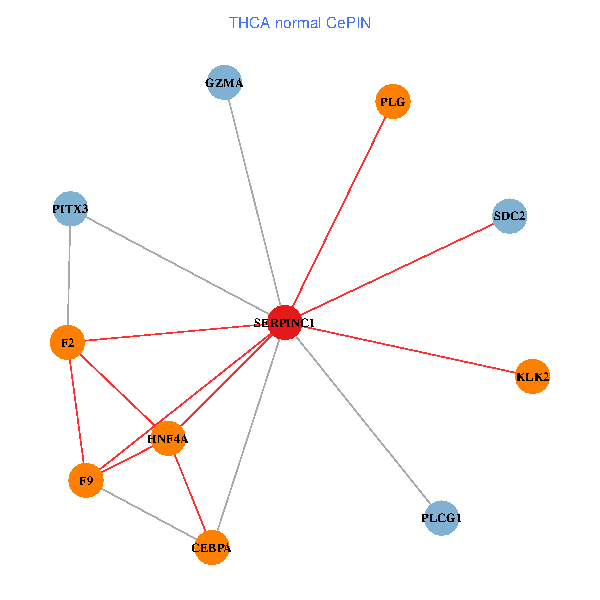
| THCA (tumor) | THCA (normal) |
| SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (tumor) | SERPINC1, HNF4A, PLCG1, SDC2, PITX3, PLG, CEBPA, F2, GZMA, KLK2, F9 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0272375 | Antithrombin III Deficiency | 45 | BeFree,CLINVAR,CTD_human,GAD,MGD,ORPHANET,UNIPROT |
| umls:C0600433 | Activated Protein C Resistance | 15 | BeFree |
| umls:C0398623 | Thrombophilia | 13 | BeFree,GAD |
| umls:C0042487 | Venous Thrombosis | 12 | CTD_human,GAD,LHGDN |
| umls:C0242666 | Protein S Deficiency | 11 | BeFree |
| umls:C1861171 | THROMBOPHILIA DUE TO ACTIVATED PROTEIN C RESISTANCE (disorder) | 11 | BeFree |
| umls:C0040038 | Thromboembolism | 10 | BeFree,GAD |
| umls:C0311370 | Lupus anticoagulant disorder | 9 | BeFree |
| umls:C0584960 | Factor V Leiden mutation | 7 | BeFree |
| umls:C0000768 | Congenital Abnormality | 6 | BeFree |
| umls:C0040053 | Thrombosis | 6 | CTD_human,GAD,LHGDN |
| umls:C0149871 | Deep Vein Thrombosis | 6 | BeFree |
| umls:C3658294 | Hereditary Antithrombin Deficiency | 6 | BeFree |
| umls:C0398625 | Protein C Deficiency | 5 | BeFree |
| umls:C0012739 | Disseminated Intravascular Coagulation | 4 | BeFree,CTD_human |
| umls:C0948008 | Ischemic stroke | 4 | BeFree |
| umls:C3272363 | Ischemic Cerebrovascular Accident | 4 | BeFree |
| umls:C0005779 | Blood Coagulation Disorders | 3 | BeFree,CTD_human |
| umls:C0007785 | Cerebral Infarction | 3 | BeFree,GAD |
| umls:C0034065 | Pulmonary Embolism | 3 | BeFree |
| umls:C0038454 | Cerebrovascular accident | 3 | BeFree |
| umls:C0340708 | Deep vein thrombosis of lower limb | 3 | BeFree |
| umls:C0398621 | Hypoplasminogenemia | 3 | BeFree |
| umls:C2239176 | Liver carcinoma | 3 | BeFree |
| umls:C0010068 | Coronary heart disease | 2 | BeFree |
| umls:C0019880 | Homocystinuria | 2 | BeFree |
| umls:C0020538 | Hypertensive disease | 2 | BeFree,GAD |
| umls:C0023903 | Liver neoplasms | 2 | BeFree |
| umls:C0040046 | Thrombophlebitis | 2 | BeFree |
| umls:C0151950 | Deep thrombophlebitis | 2 | BeFree |
| umls:C0162429 | Malnutrition | 2 | BeFree |
| umls:C0237997 | ANTITHROMBIN III DEFICIENCY, FAMILIAL | 2 | BeFree |
| umls:C0267412 | Mesenteric Venous Thrombosis | 2 | BeFree |
| umls:C0338573 | Cerebral venous sinus thrombosis | 2 | BeFree |
| umls:C0376618 | Endotoxemia | 2 | RGD |
| umls:C1861172 | Venous Thromboembolism | 2 | CTD_human,GAD |
| umls:C3495426 | Homocysteinemia | 2 | BeFree |
| umls:C0002395 | Alzheimer's Disease | 1 | GAD |
| umls:C0002726 | Amyloidosis | 1 | BeFree |
| umls:C0002874 | Aplastic Anemia | 1 | BeFree |
| umls:C0003850 | Arteriosclerosis | 1 | BeFree |
| umls:C0003873 | Rheumatoid Arthritis | 1 | BeFree |
| umls:C0004153 | Atherosclerosis | 1 | BeFree |
| umls:C0004364 | Autoimmune Diseases | 1 | BeFree |
| umls:C0007959 | Charcot-Marie-Tooth Disease | 1 | BeFree |
| umls:C0008495 | Chorioamnionitis | 1 | GAD |
| umls:C0010072 | Coronary Thrombosis | 1 | GAD |
| umls:C0011847 | Diabetes | 1 | BeFree |
| umls:C0011849 | Diabetes Mellitus | 1 | BeFree |
| umls:C0011854 | Diabetes Mellitus, Insulin-Dependent | 1 | BeFree |
| umls:C0013080 | Down Syndrome | 1 | BeFree |
| umls:C0014859 | Esophageal Neoplasms | 1 | BeFree |
| umls:C0015944 | Fetal Membranes, Premature Rupture | 1 | GAD |
| umls:C0019154 | Hepatic Vein Thrombosis | 1 | BeFree |
| umls:C0020544 | Renal hypertension | 1 | LHGDN |
| umls:C0022346 | Icterus | 1 | RGD |
| umls:C0022658 | Kidney Diseases | 1 | BeFree |
| umls:C0022876 | Premature Obstetric Labor | 1 | GAD |
| umls:C0023234 | Legg-Calve-Perthes Disease | 1 | BeFree |
| umls:C0023449 | Acute lymphocytic leukemia | 1 | BeFree |
| umls:C0023452 | Leukemia, Lymphocytic, Acute, L1 | 1 | BeFree |
| umls:C0024131 | Lupus Vulgaris | 1 | BeFree |
| umls:C0024138 | Lupus Erythematosus, Discoid | 1 | BeFree |
| umls:C0024141 | Lupus Erythematosus, Systemic | 1 | BeFree |
| umls:C0024299 | Lymphoma | 1 | LHGDN |
| umls:C0024790 | Paroxysmal nocturnal hemoglobinuria | 1 | BeFree |
| umls:C0027051 | Myocardial Infarction | 1 | GAD |
| umls:C0027726 | Nephrotic Syndrome | 1 | CTD_human |
| umls:C0029408 | Degenerative polyarthritis | 1 | BeFree |
| umls:C0032460 | Polycystic Ovary Syndrome | 1 | BeFree |
| umls:C0032914 | Pre-Eclampsia | 1 | GAD |
| umls:C0033578 | Prostatic Neoplasms | 1 | LHGDN |
| umls:C0035066 | Renal Artery Obstruction | 1 | LHGDN |
| umls:C0035222 | Respiratory Distress Syndrome, Adult | 1 | CTD_human |
| umls:C0035309 | Retinal Diseases | 1 | BeFree |
| umls:C0039240 | Supraventricular tachycardia | 1 | BeFree |
| umls:C0079102 | Cerebral Thrombosis | 1 | BeFree |
| umls:C0085400 | Neurofibrillary degeneration (morphologic abnormality) | 1 | BeFree |
| umls:C0085580 | Essential Hypertension | 1 | BeFree |
| umls:C0151526 | Premature Birth | 1 | GAD |
| umls:C0152018 | Esophageal carcinoma | 1 | BeFree |
| umls:C0162557 | Liver Failure, Acute | 1 | CTD_human |
| umls:C0272363 | anticoagulant disorders | 1 | BeFree |
| umls:C0279000 | Liver and Intrahepatic Biliary Tract Carcinoma | 1 | BeFree |
| umls:C0345904 | Malignant neoplasm of liver | 1 | BeFree |
| umls:C0409974 | Lupus Erythematosus | 1 | BeFree |
| umls:C0546837 | Malignant neoplasm of esophagus | 1 | BeFree |
| umls:C0553681 | Hypofibrinogenemia | 1 | BeFree |
| umls:C0687675 | Pregnancy loss | 1 | GAD |
| umls:C0742472 | Central nervous system lymphoma | 1 | BeFree |
| umls:C0752143 | Intracranial Thrombosis | 1 | CTD_human |
| umls:C0856761 | Budd-Chiari Syndrome | 1 | BeFree |
| umls:C0948089 | Acute Coronary Syndrome | 1 | CTD_human |
| umls:C1261473 | Sarcoma | 1 | BeFree |
| umls:C1519666 | Tumor-Associated Vasculature | 1 | BeFree |
| umls:C1865145 | Congenital disorder of glycosylation type 1B | 1 | BeFree |
| umls:C1959635 | Parvovirus B19 (disease) | 1 | BeFree |
| umls:C2584409 | Prothrombin G20210A mutation | 1 | BeFree |
| umls:C2586031 | Hereditary antithrombin III deficiency | 1 | BeFree |
| umls:C2919032 | Infection of amniotic sac and membranes, unspecified, unspecified trimester, not applicable or unspecified | 1 | GAD |
| umls:C2936349 | Plaque, Amyloid | 1 | BeFree |
| umls:C2937358 | Cerebral Hemorrhage | 1 | CTD_human |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))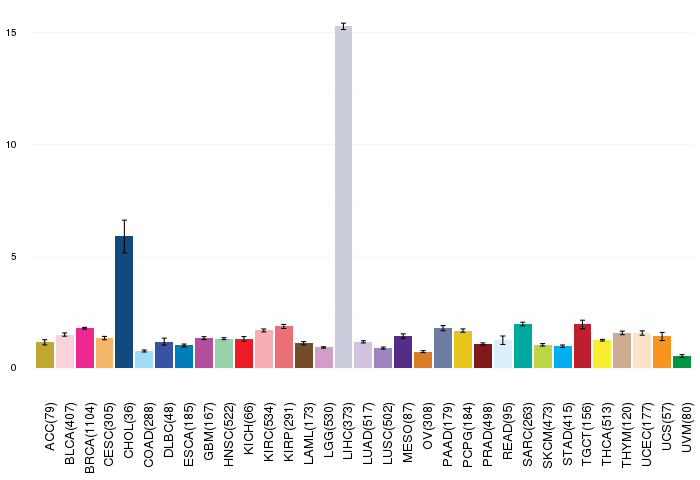
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))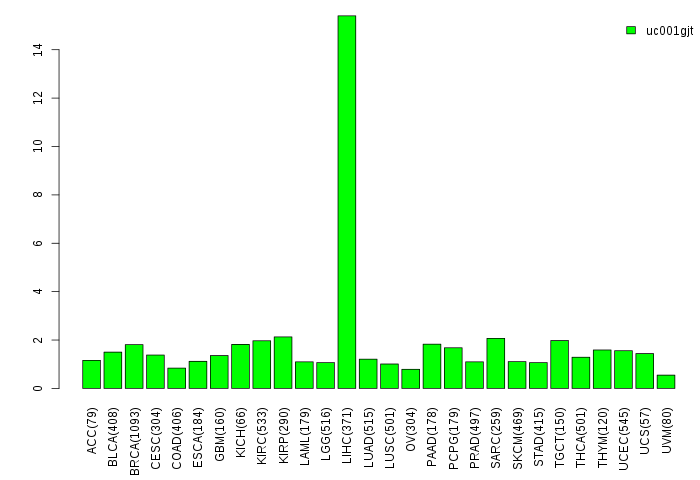
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data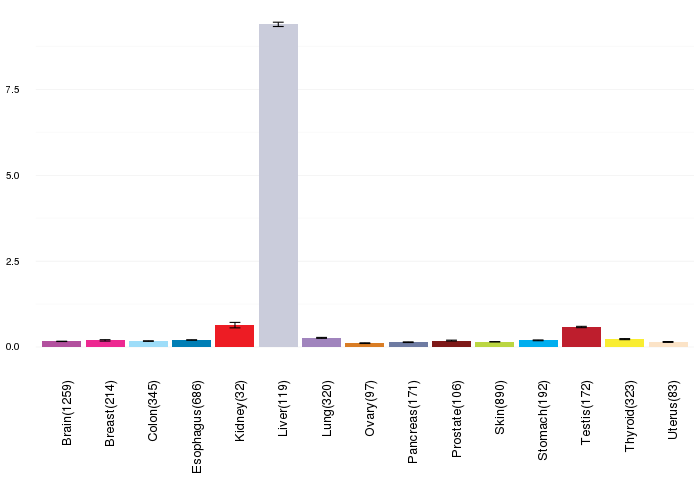
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))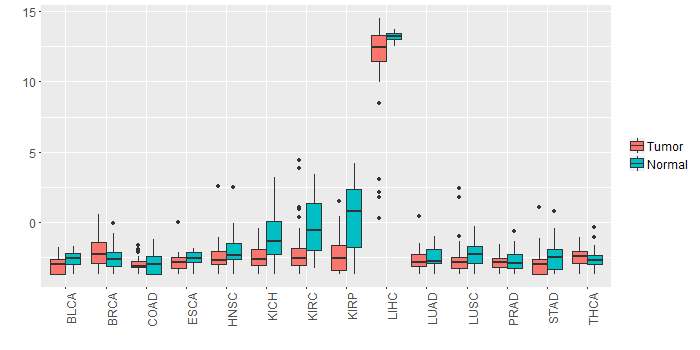
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.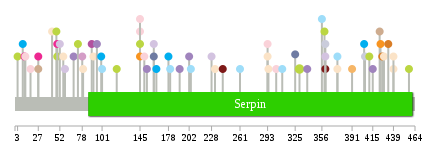

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 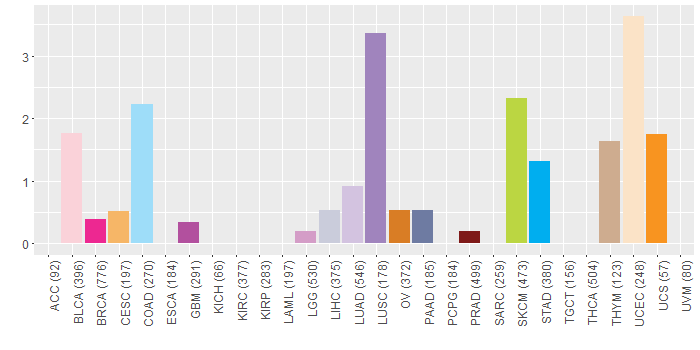
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)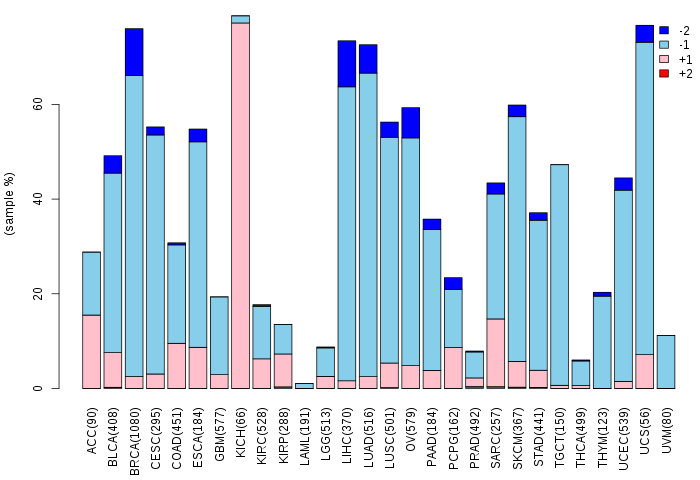
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 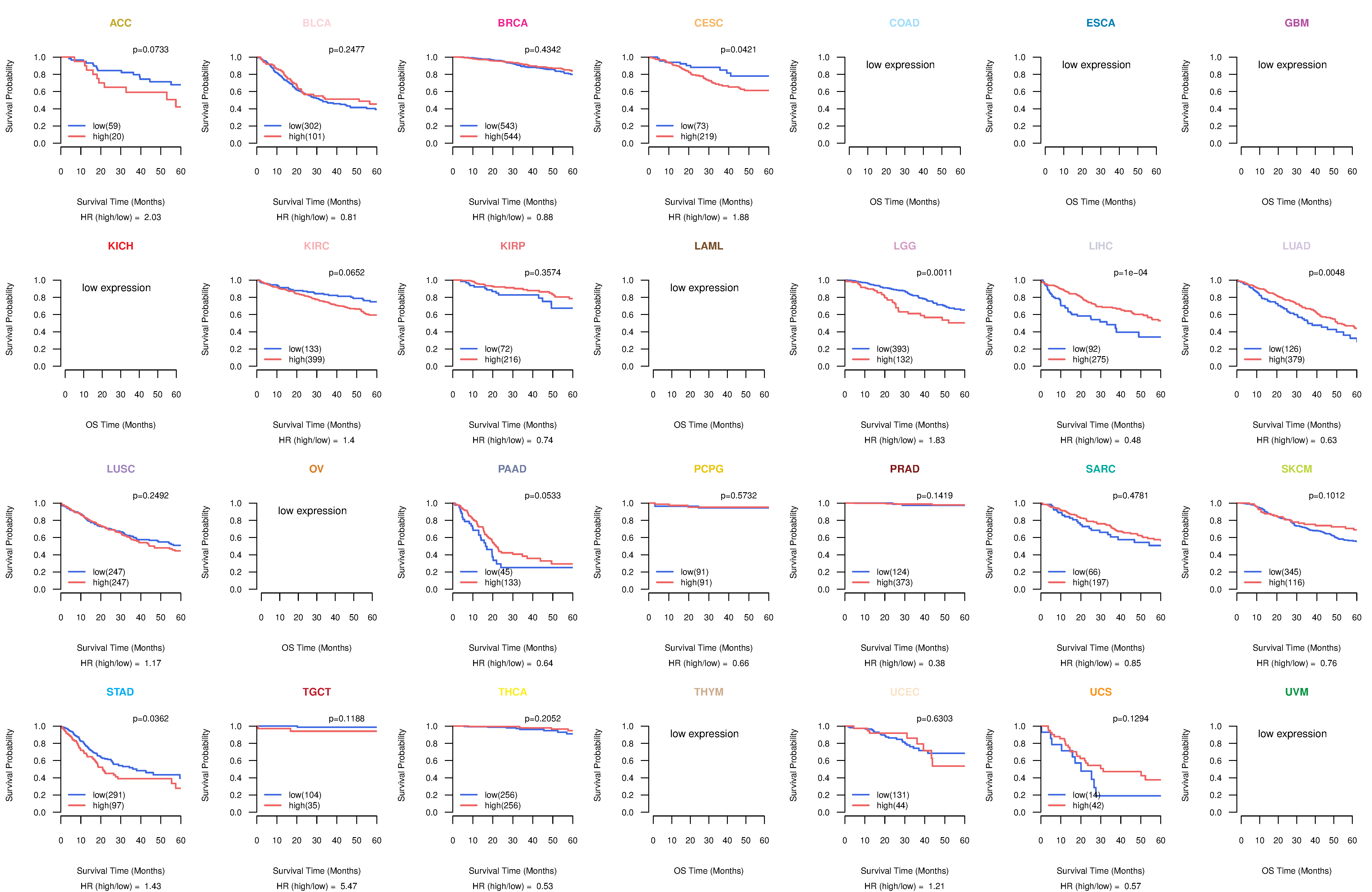
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 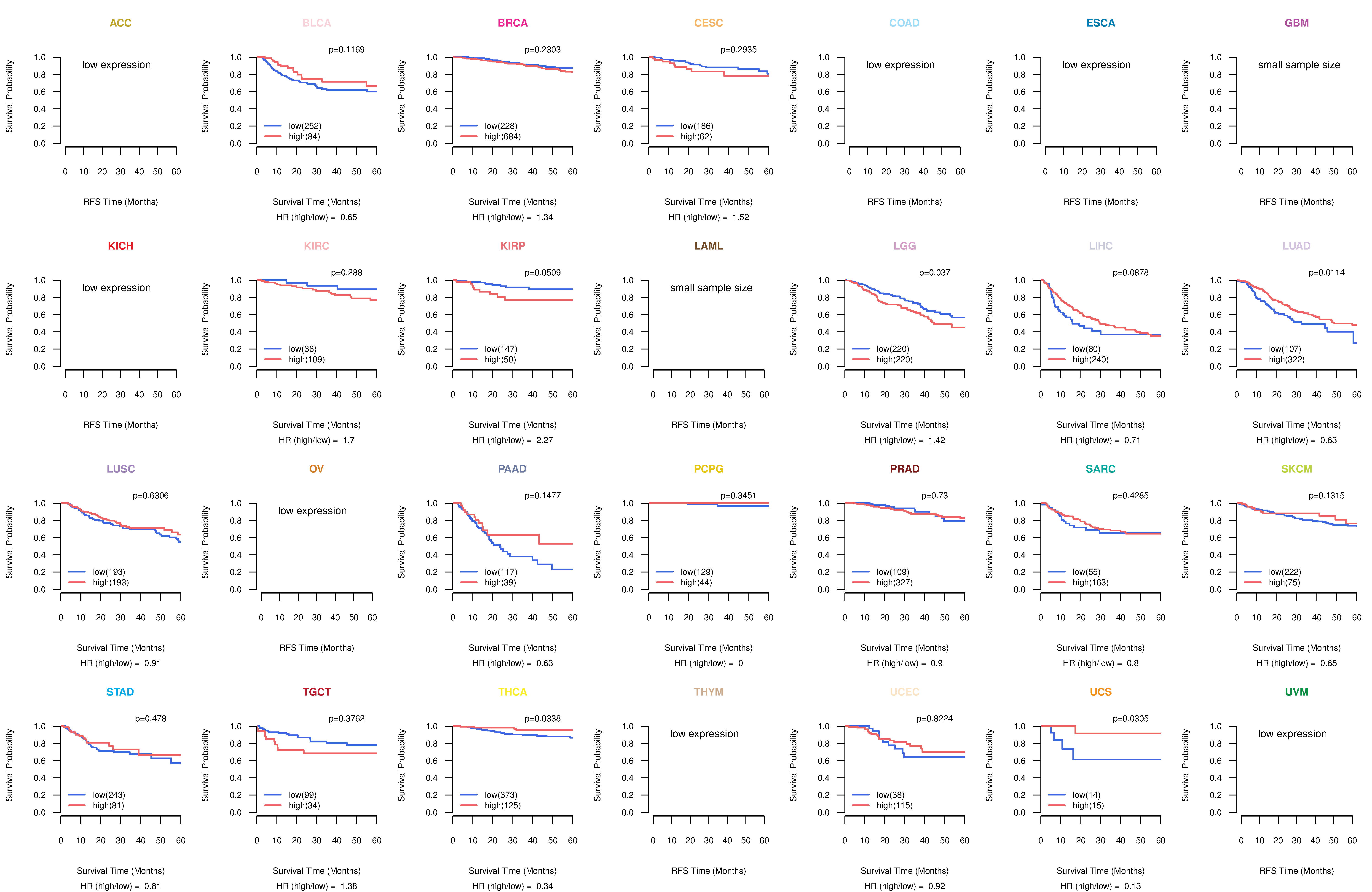
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 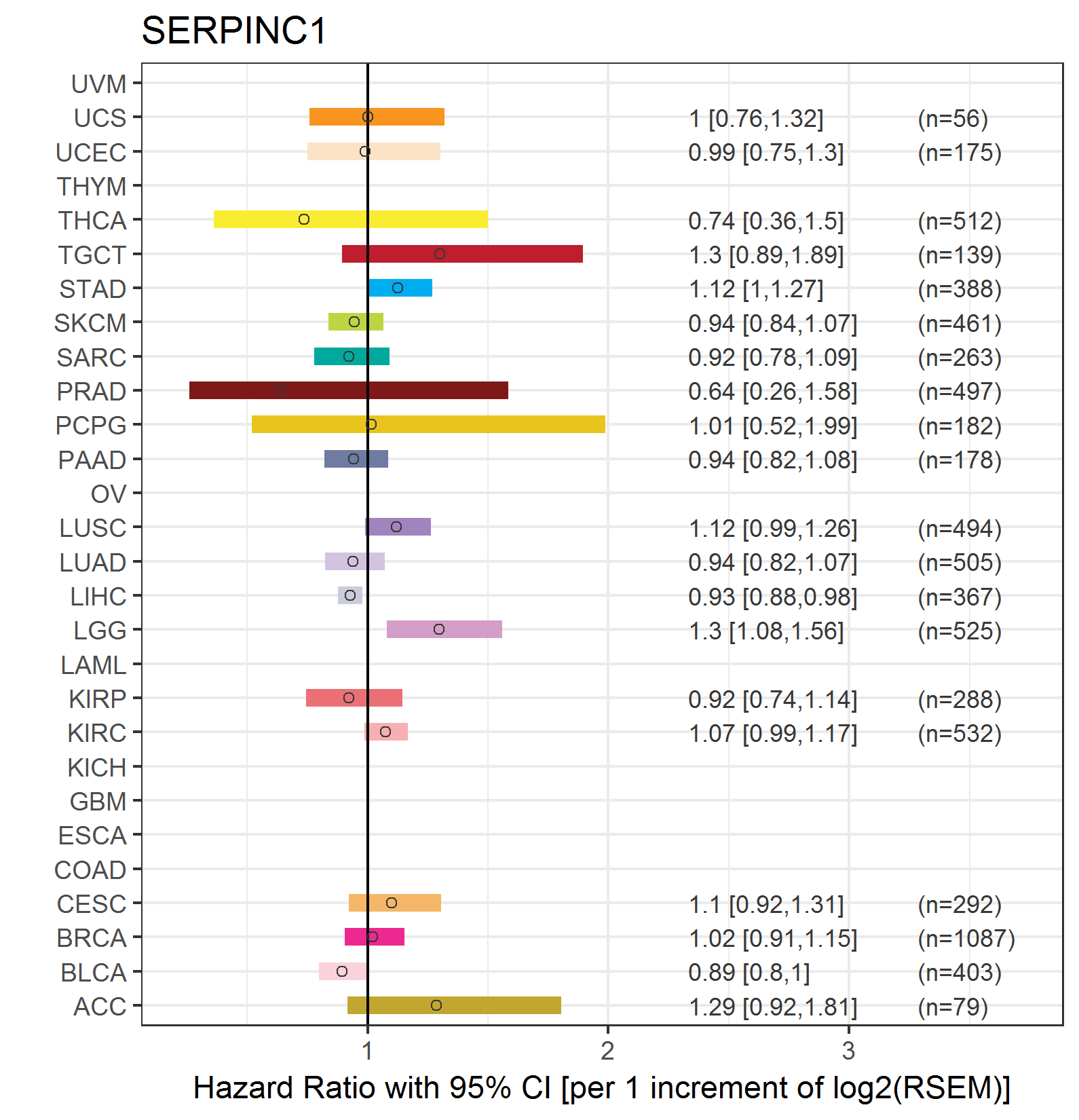
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 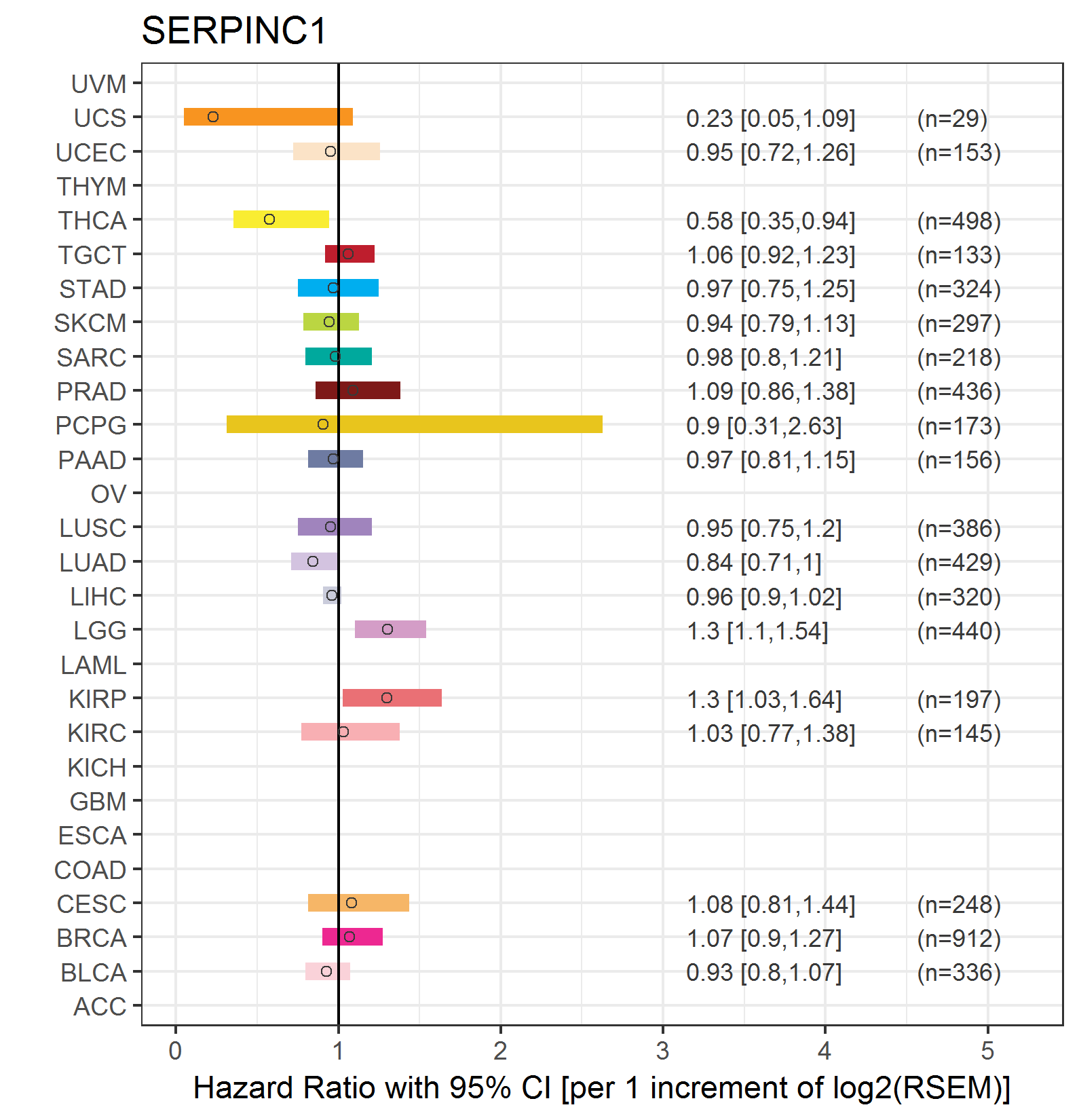
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























