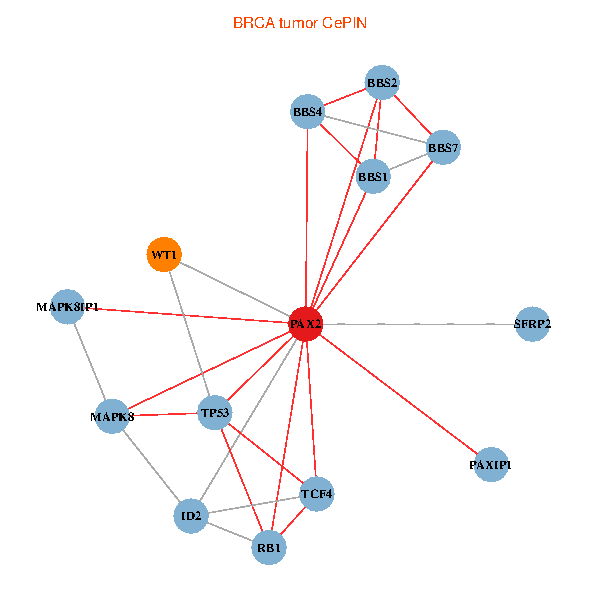
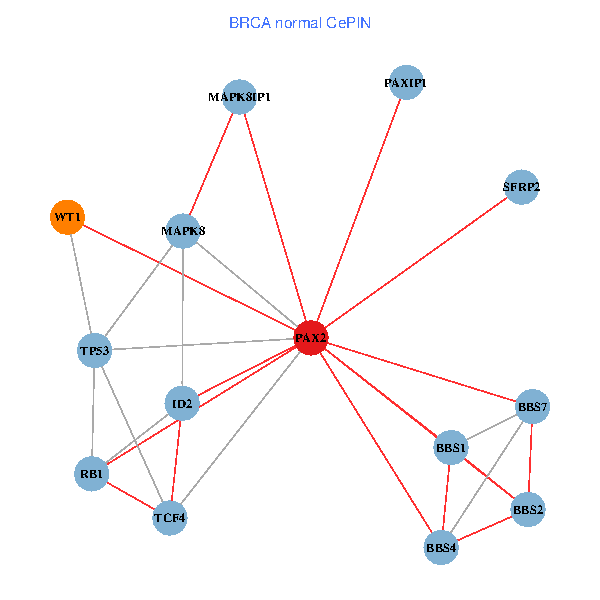
| BRCA (tumor) | BRCA (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
 |  |
|
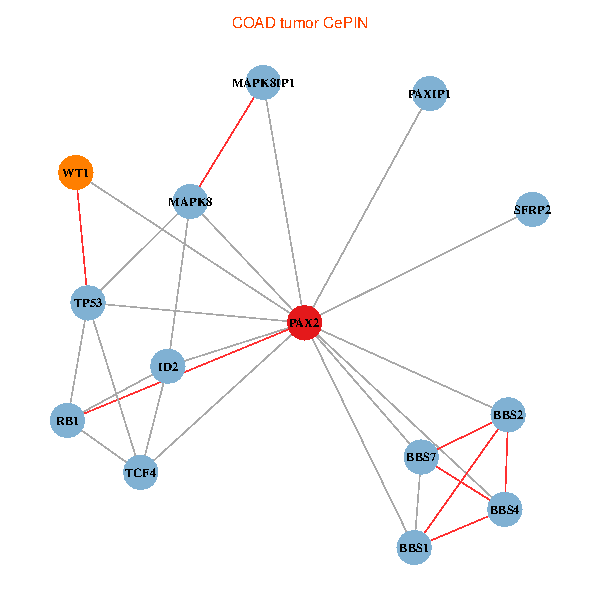
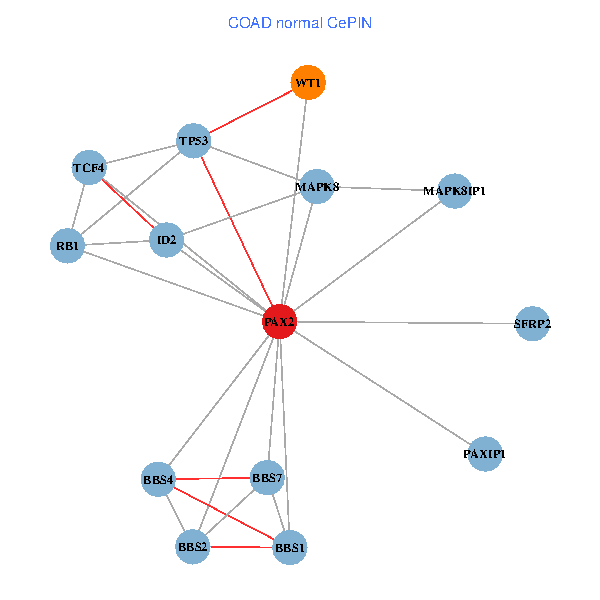
| COAD (tumor) | COAD (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
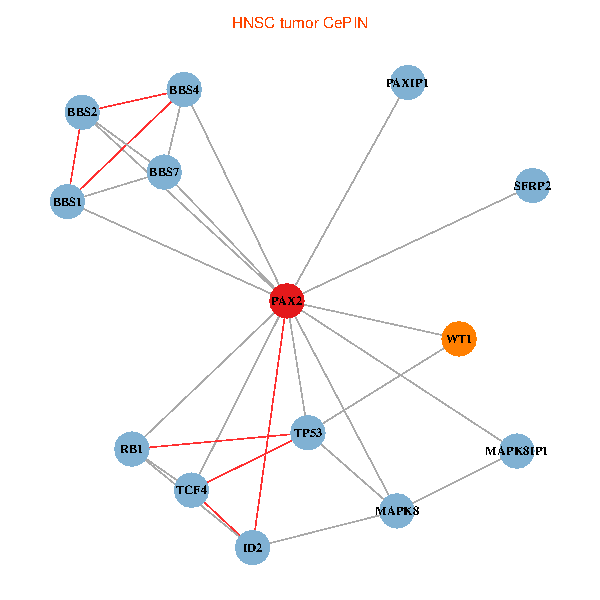 | 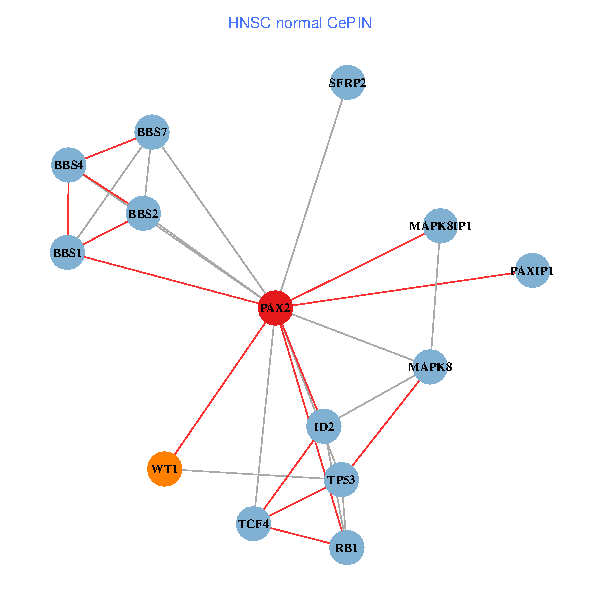 |
|
| KICH (tumor) | KICH (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
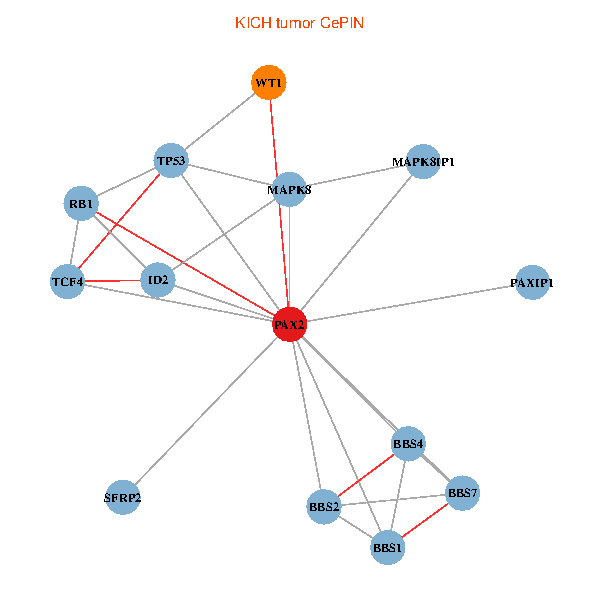 | 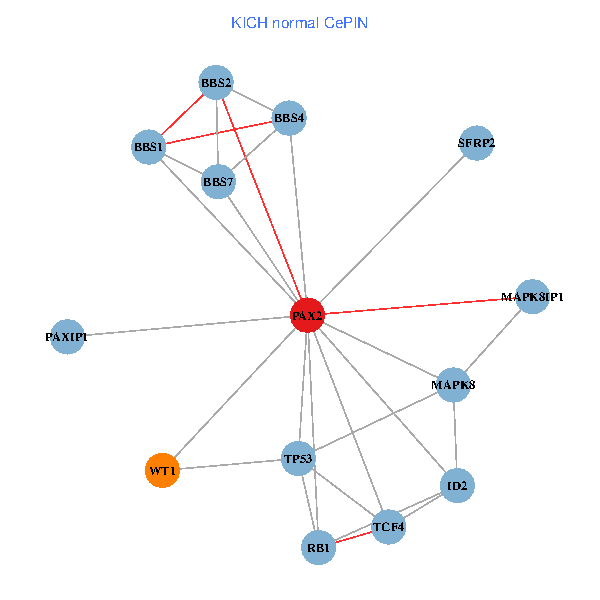 |
|
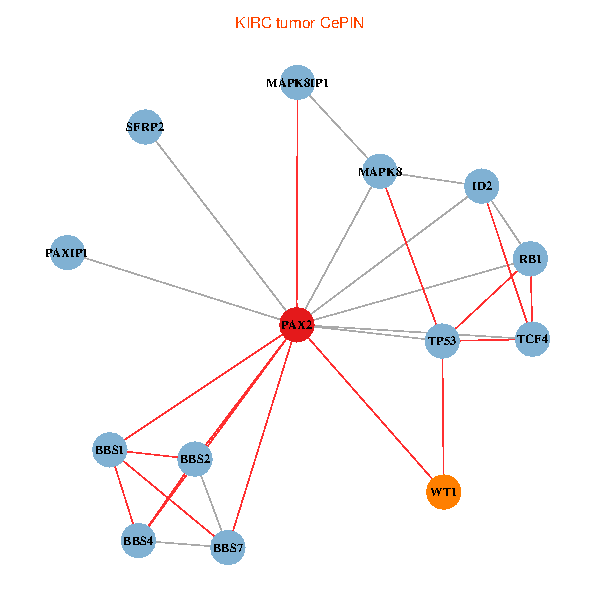
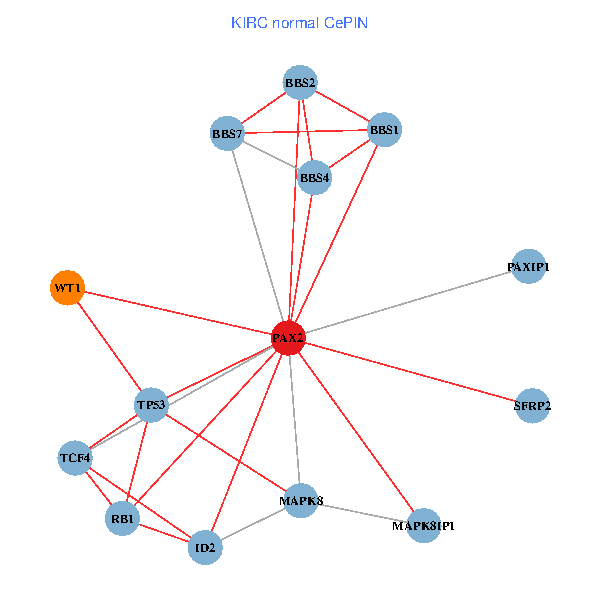
| KIRC (tumor) | KIRC (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
 |  |
|
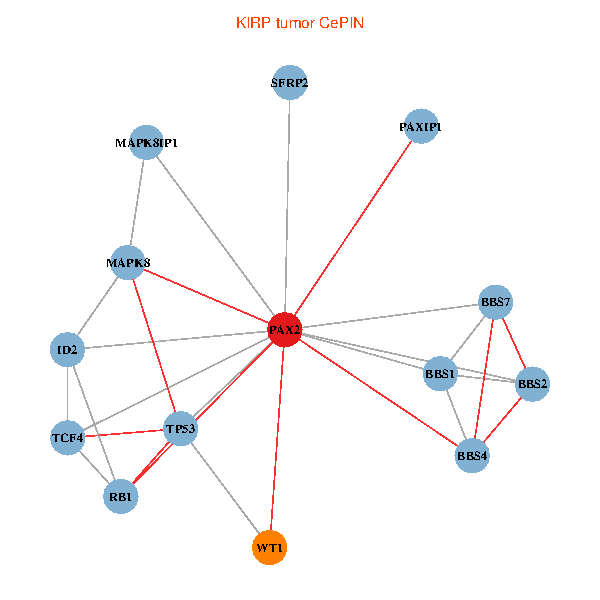
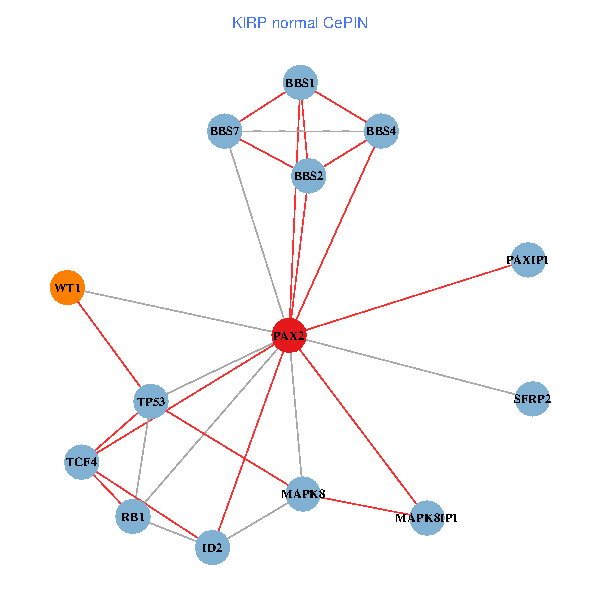
| KIRP (tumor) | KIRP (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
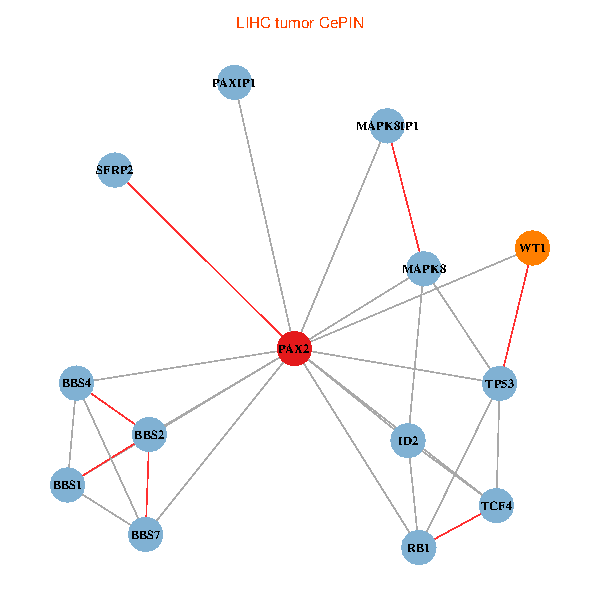 | 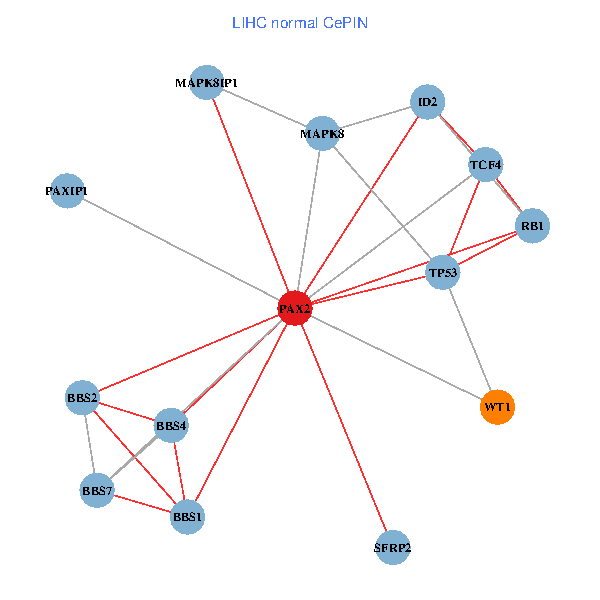 |
|
| LUAD (tumor) | LUAD (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
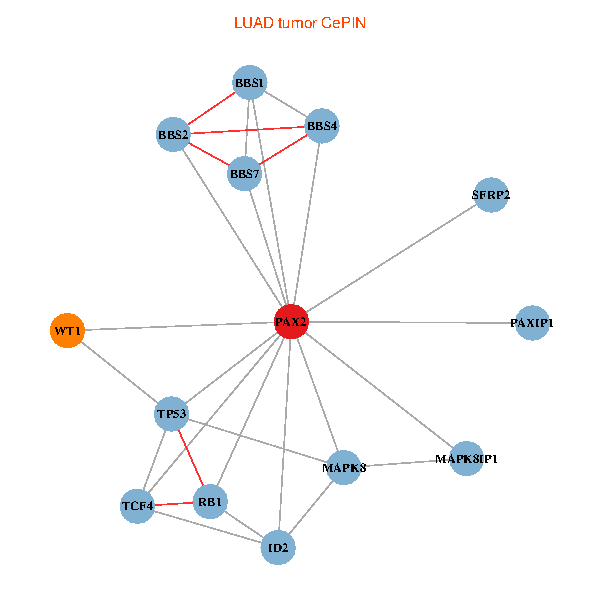 | 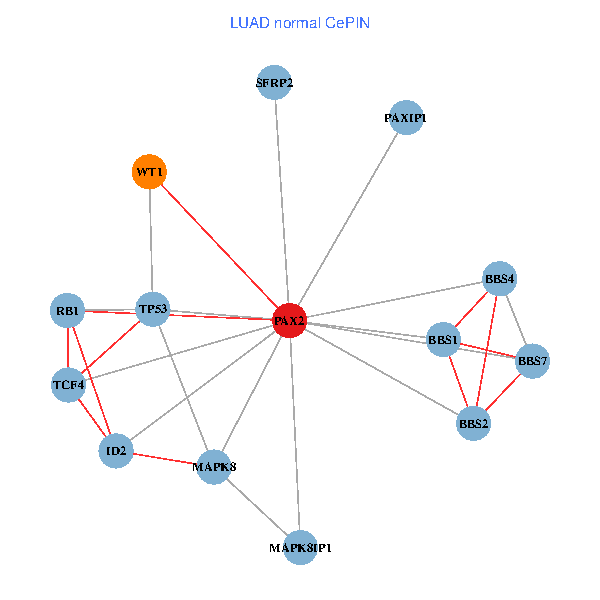 |
|
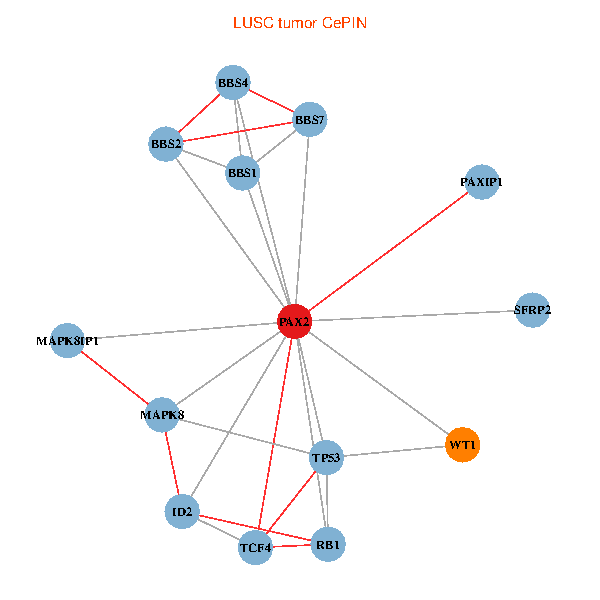
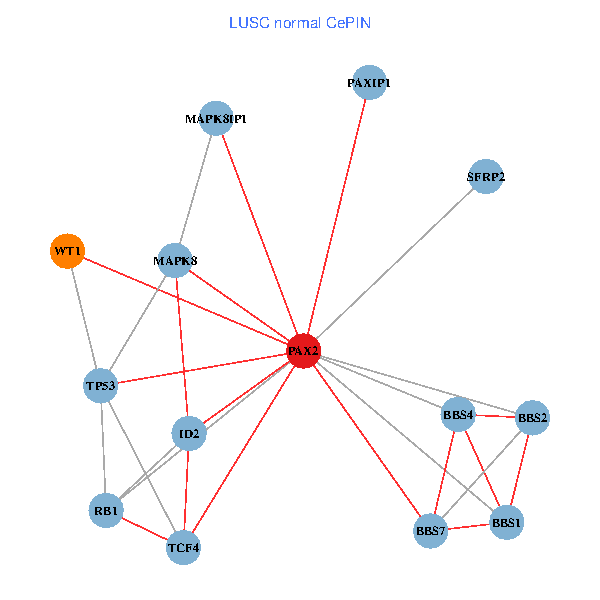
| LUSC (tumor) | LUSC (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
 |  |
|
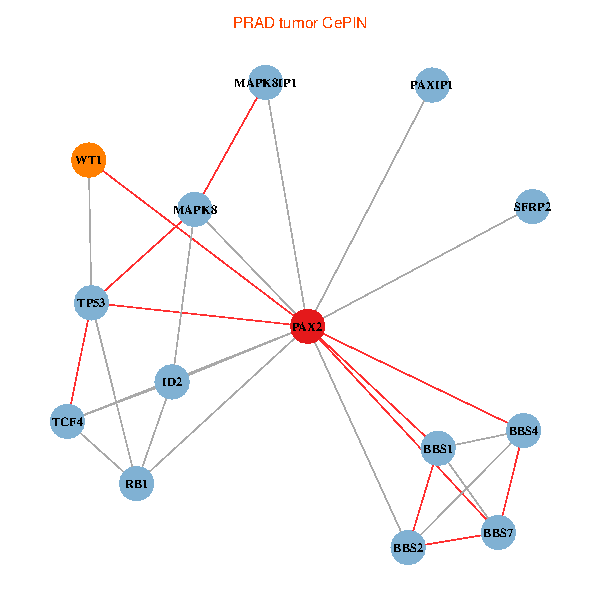
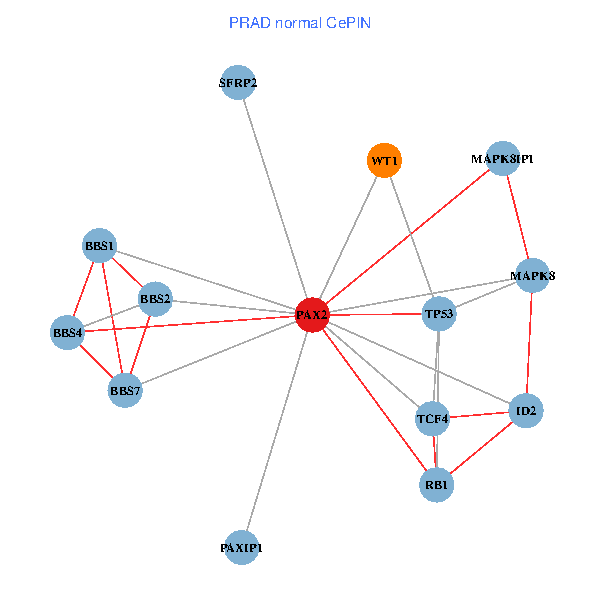
| PRAD (tumor) | PRAD (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
 |  |
|
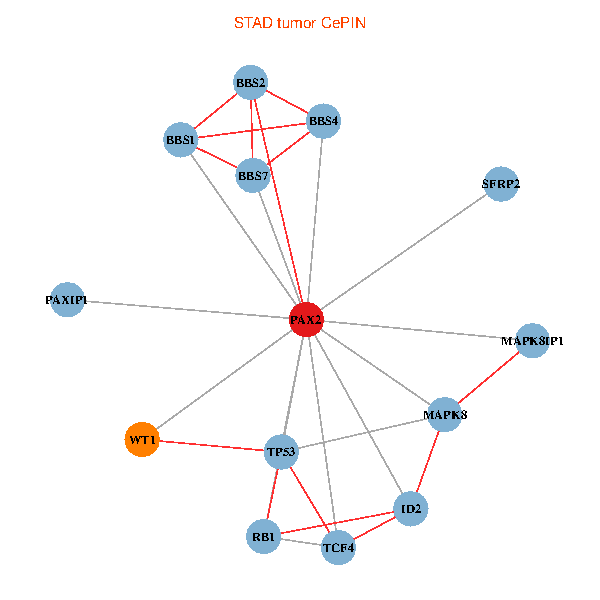
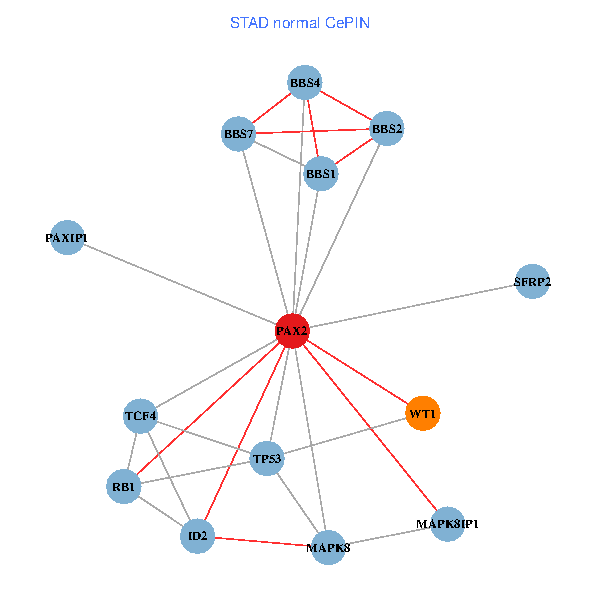
| STAD (tumor) | STAD (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
 |  |
|
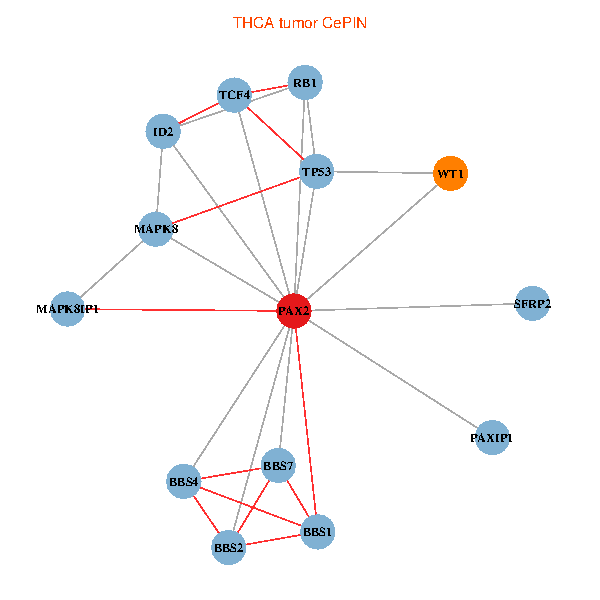
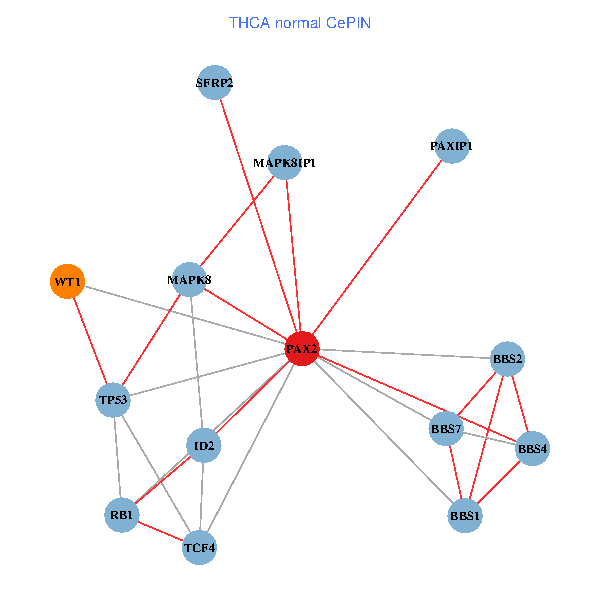
| THCA (tumor) | THCA (normal) |
| PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (tumor) | PAX2, TP53, MAPK8, BBS1, TCF4, PAXIP1, MAPK8IP1, RB1, ID2, BBS2, BBS4, WT1, BBS7, SFRP2 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C1852759 | Papillorenal syndrome | 34 | BeFree,CLINVAR,CTD_human,MGD,ORPHANET,UNIPROT |
| umls:C0027708 | Nephroblastoma | 15 | BeFree,RGD |
| umls:C0007134 | Renal Cell Carcinoma | 13 | BeFree,LHGDN |
| umls:C0000768 | Congenital Abnormality | 10 | BeFree |
| umls:C0596263 | Carcinogenesis | 10 | BeFree |
| umls:C0042580 | Vesico-Ureteral Reflux | 8 | BeFree,GAD |
| umls:C0266292 | Congenital anomaly of the kidney | 6 | BeFree |
| umls:C0266295 | Congenital hypoplasia of kidney | 6 | BeFree |
| umls:C0009363 | Congenital ocular coloboma (disorder) | 5 | BeFree |
| umls:C0022658 | Kidney Diseases | 5 | BeFree |
| umls:C0022665 | Kidney Neoplasm | 5 | BeFree,LHGDN |
| umls:C0027627 | Neoplasm Metastasis | 5 | BeFree |
| umls:C0376358 | Malignant neoplasm of prostate | 5 | BeFree |
| umls:C0600139 | Prostate carcinoma | 5 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 4 | BeFree |
| umls:C0010709 | Cyst | 4 | BeFree |
| umls:C0155299 | Coloboma of optic disc | 4 | BeFree |
| umls:C0476089 | Endometrial Carcinoma | 4 | BeFree |
| umls:C0678222 | Breast Carcinoma | 4 | BeFree |
| umls:C0007103 | Malignant neoplasm of endometrium | 3 | BeFree |
| umls:C0035078 | Kidney Failure | 3 | BeFree |
| umls:C1378703 | Renal carcinoma | 3 | BeFree |
| umls:C1883486 | Uterine Corpus Cancer | 3 | BeFree |
| umls:C3536714 | Renal dysplasia | 3 | BeFree |
| umls:C0001418 | Adenocarcinoma | 2 | BeFree |
| umls:C0022661 | Kidney Failure, Chronic | 2 | BeFree,CTD_human |
| umls:C0022680 | Polycystic Kidney Diseases | 2 | BeFree |
| umls:C0027697 | Nephritis | 2 | BeFree |
| umls:C0034152 | Henoch-Schoenlein Purpura | 2 | BeFree |
| umls:C0041956 | Ureteral obstruction | 2 | BeFree |
| umls:C0085413 | Polycystic Kidney, Autosomal Dominant | 2 | BeFree |
| umls:C0235989 | Renal interstitial fibrosis | 2 | BeFree |
| umls:C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 2 | BeFree |
| umls:C0333497 | Segmental glomerulosclerosis | 2 | BeFree |
| umls:C0740457 | Malignant neoplasm of kidney | 2 | BeFree |
| umls:C1968949 | Cakut | 2 | BeFree |
| umls:C4014925 | FOCAL SEGMENTAL GLOMERULOSCLEROSIS 7 | 2 | UNIPROT |
| umls:C0001430 | Adenoma | 1 | LHGDN |
| umls:C0005684 | Malignant neoplasm of urinary bladder | 1 | BeFree |
| umls:C0007102 | Malignant tumor of colon | 1 | BeFree |
| umls:C0009404 | Colorectal Neoplasms | 1 | BeFree |
| umls:C0011881 | Diabetic Nephropathy | 1 | RGD |
| umls:C0014175 | Endometriosis | 1 | BeFree,CTD_human |
| umls:C0014511 | Epithelial cyst | 1 | BeFree |
| umls:C0014859 | Esophageal Neoplasms | 1 | BeFree |
| umls:C0019562 | Von Hippel-Lindau Syndrome | 1 | BeFree |
| umls:C0020255 | Hydrocephalus | 1 | BeFree |
| umls:C0022679 | Cystic kidney | 1 | BeFree |
| umls:C0022681 | Medullary sponge kidney | 1 | BeFree |
| umls:C0023418 | leukemia | 1 | BeFree |
| umls:C0023440 | Acute Erythroblastic Leukemia | 1 | BeFree |
| umls:C0025202 | melanoma | 1 | BeFree |
| umls:C0026499 | Monosomy | 1 | BeFree |
| umls:C0027341 | Nail-Patella Syndrome | 1 | BeFree |
| umls:C0033578 | Prostatic Neoplasms | 1 | BeFree |
| umls:C0035126 | Reperfusion Injury | 1 | RGD |
| umls:C0036220 | Kaposi Sarcoma | 1 | LHGDN |
| umls:C0039101 | synovial sarcoma | 1 | BeFree |
| umls:C0152018 | Esophageal carcinoma | 1 | BeFree |
| umls:C0154823 | Retinal defect | 1 | BeFree |
| umls:C0158698 | Congenital malformation of the urinary system | 1 | BeFree |
| umls:C0162809 | Kallmann Syndrome | 1 | BeFree |
| umls:C0178879 | Urinary tract obstruction | 1 | BeFree |
| umls:C0206701 | Cystadenocarcinoma, Serous | 1 | BeFree |
| umls:C0235831 | Renal Cell Dysplasia | 1 | BeFree |
| umls:C0265234 | Branchio-Oto-Renal Syndrome | 1 | BeFree |
| umls:C0266449 | Congenital anomaly of brain | 1 | BeFree |
| umls:C0268164 | Primary hyperoxaluria, type I | 1 | BeFree |
| umls:C0268747 | Diffuse mesangial sclerosis (disorder) | 1 | BeFree |
| umls:C0268800 | Simple renal cyst | 1 | BeFree |
| umls:C0279626 | Squamous cell carcinoma of esophagus | 1 | BeFree |
| umls:C0332910 | bilateral agenesis | 1 | BeFree |
| umls:C0376545 | Hematologic Neoplasms | 1 | BeFree |
| umls:C0546837 | Malignant neoplasm of esophagus | 1 | BeFree |
| umls:C0686619 | Secondary malignant neoplasm of lymph node | 1 | BeFree |
| umls:C0699790 | Colon Carcinoma | 1 | BeFree |
| umls:C0699885 | Carcinoma of bladder | 1 | BeFree |
| umls:C0879615 | Stromal Neoplasm | 1 | BeFree |
| umls:C0919267 | ovarian neoplasm | 1 | BeFree |
| umls:C0920646 | Renal ischaemia | 1 | BeFree |
| umls:C0949541 | Hurthle Cell Tumor | 1 | BeFree |
| umls:C0950121 | Denys-Drash Syndrome | 1 | BeFree |
| umls:C1153706 | Endometrial adenocarcinoma | 1 | BeFree |
| umls:C1261473 | Sarcoma | 1 | BeFree |
| umls:C1335177 | Ovarian Serous Adenocarcinoma | 1 | BeFree |
| umls:C1378050 | Oncocytic Neoplasm | 1 | BeFree |
| umls:C1458155 | Mammary Neoplasms | 1 | BeFree |
| umls:C1510502 | Oxyphilic Adenoma | 1 | BeFree |
| umls:C1513269 | Microcysts | 1 | BeFree |
| umls:C1516170 | Cancer Cell Growth | 1 | BeFree |
| umls:C1565489 | Renal Insufficiency | 1 | BeFree,LHGDN |
| umls:C1567742 | Alport Syndrome, X-Linked | 1 | BeFree |
| umls:C1569637 | Adenocarcinoma, Endometrioid | 1 | BeFree |
| umls:C1619700 | RENAL ADYSPLASIA | 1 | BeFree |
| umls:C1654637 | androgen independent prostate cancer | 1 | BeFree |
| umls:C1704272 | Benign Prostatic Hyperplasia | 1 | BeFree |
| umls:C1709661 | Primary Focal Segmental Glomerulosclerosis | 1 | BeFree |
| umls:C1720887 | Female Urogenital Diseases | 1 | CTD_human |
| umls:C2721603 | Henoch-Schonlein purpura nephritis | 1 | GAD |
| umls:C3665489 | Ovarian low malignant potential tumour | 1 | BeFree |
| umls:C3714581 | Multicystic Dysplastic Kidney | 1 | BeFree |
| umls:C3839280 | High grade serous carcinoma | 1 | BeFree |
| umls:C0431692 | Bilateral renal hypoplasia | 0 | ORPHANET |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))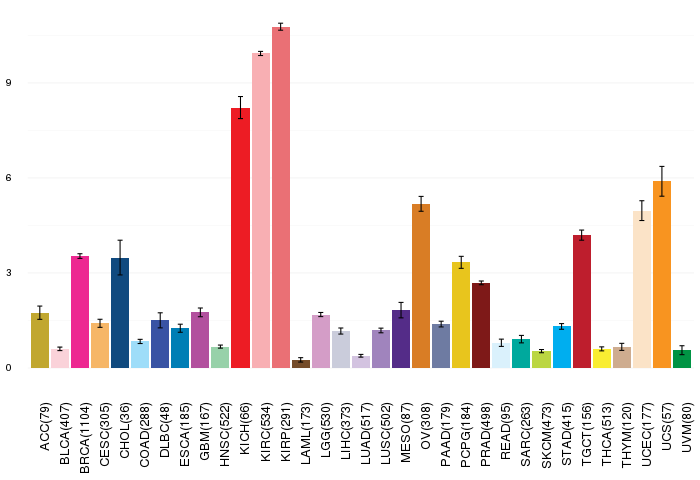
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))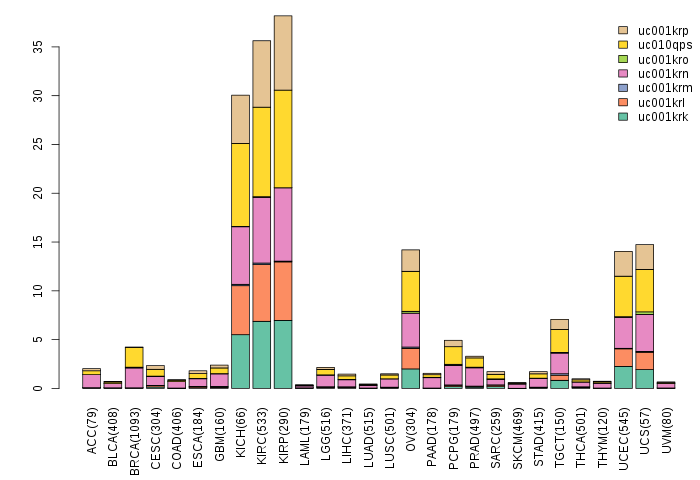
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data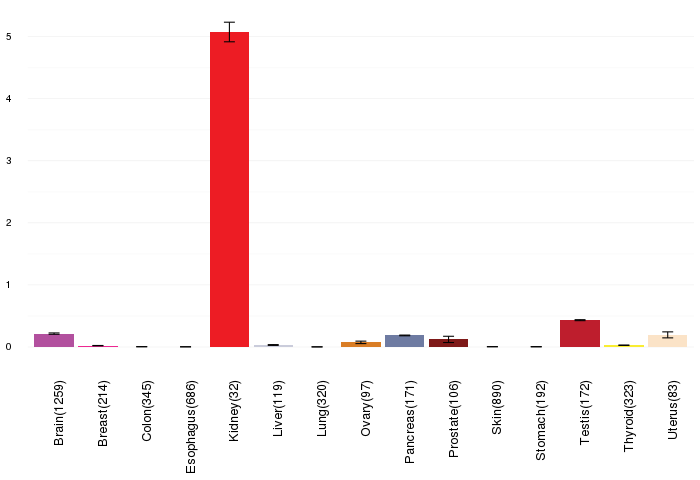
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))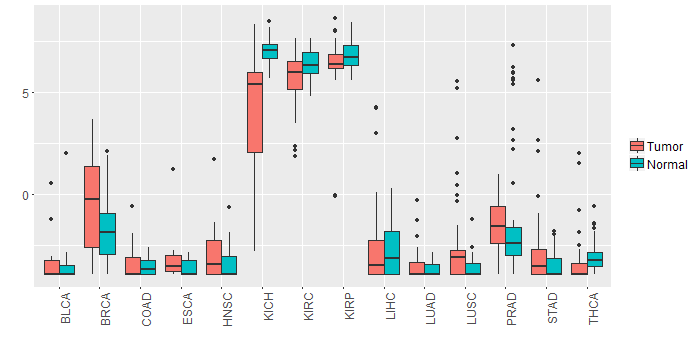
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.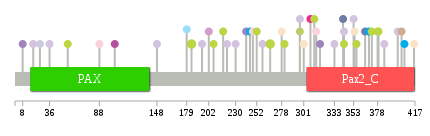

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 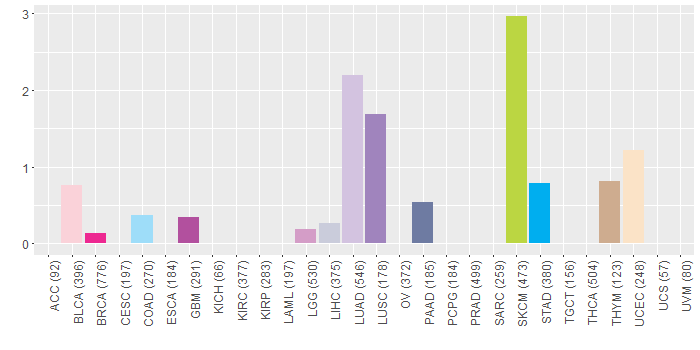
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)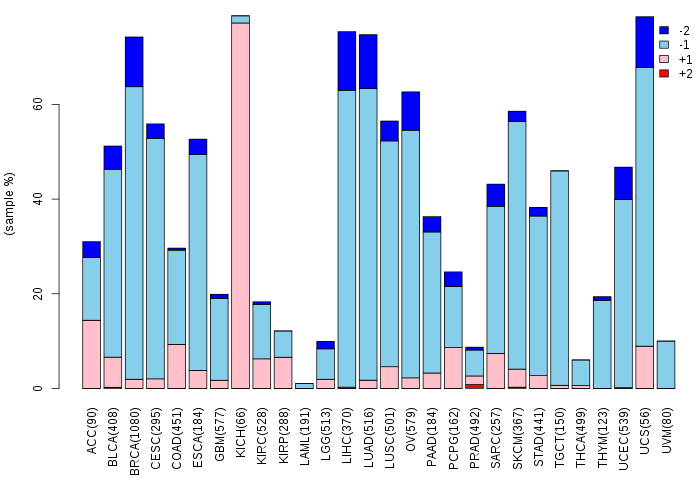
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 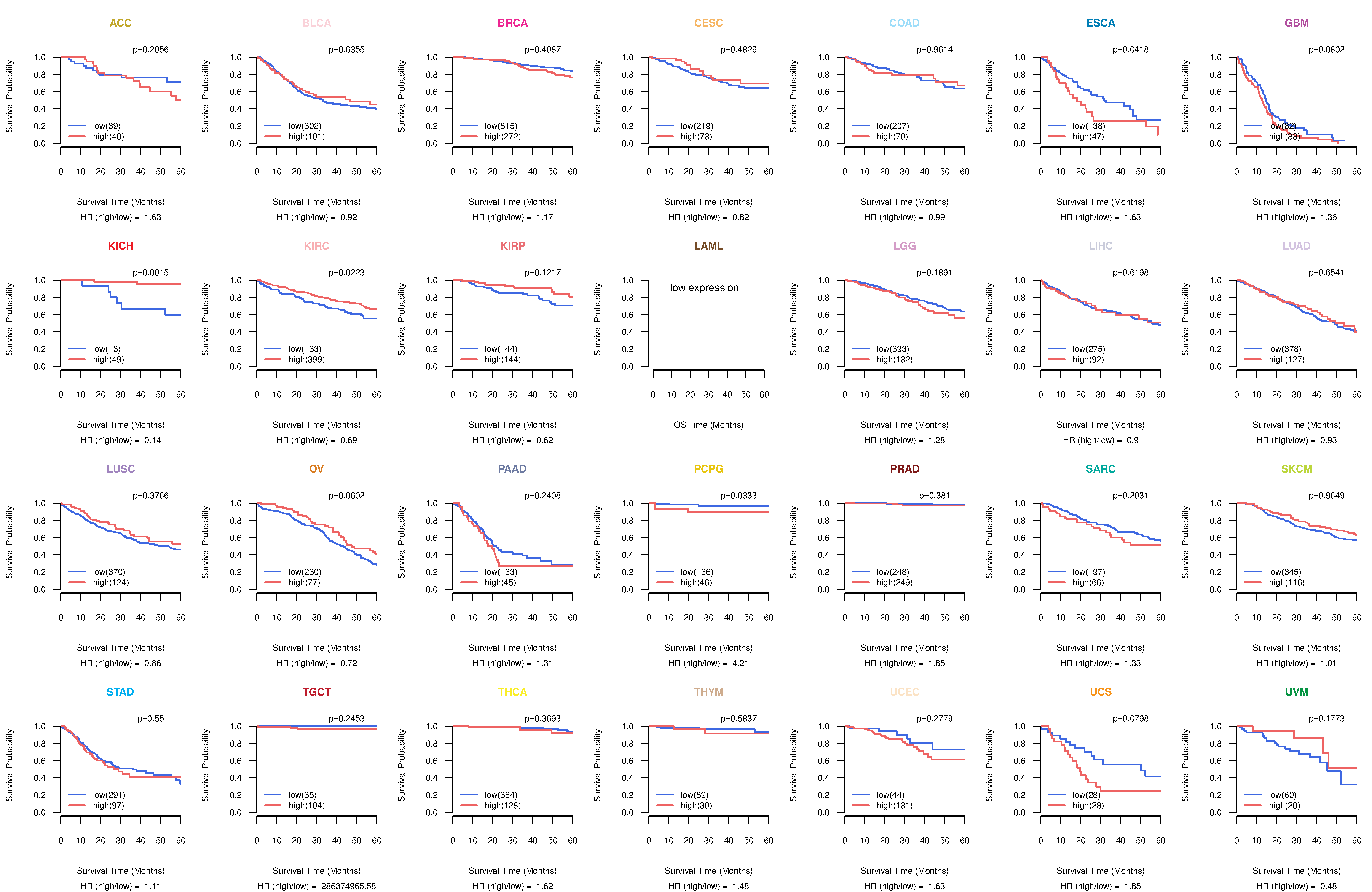
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 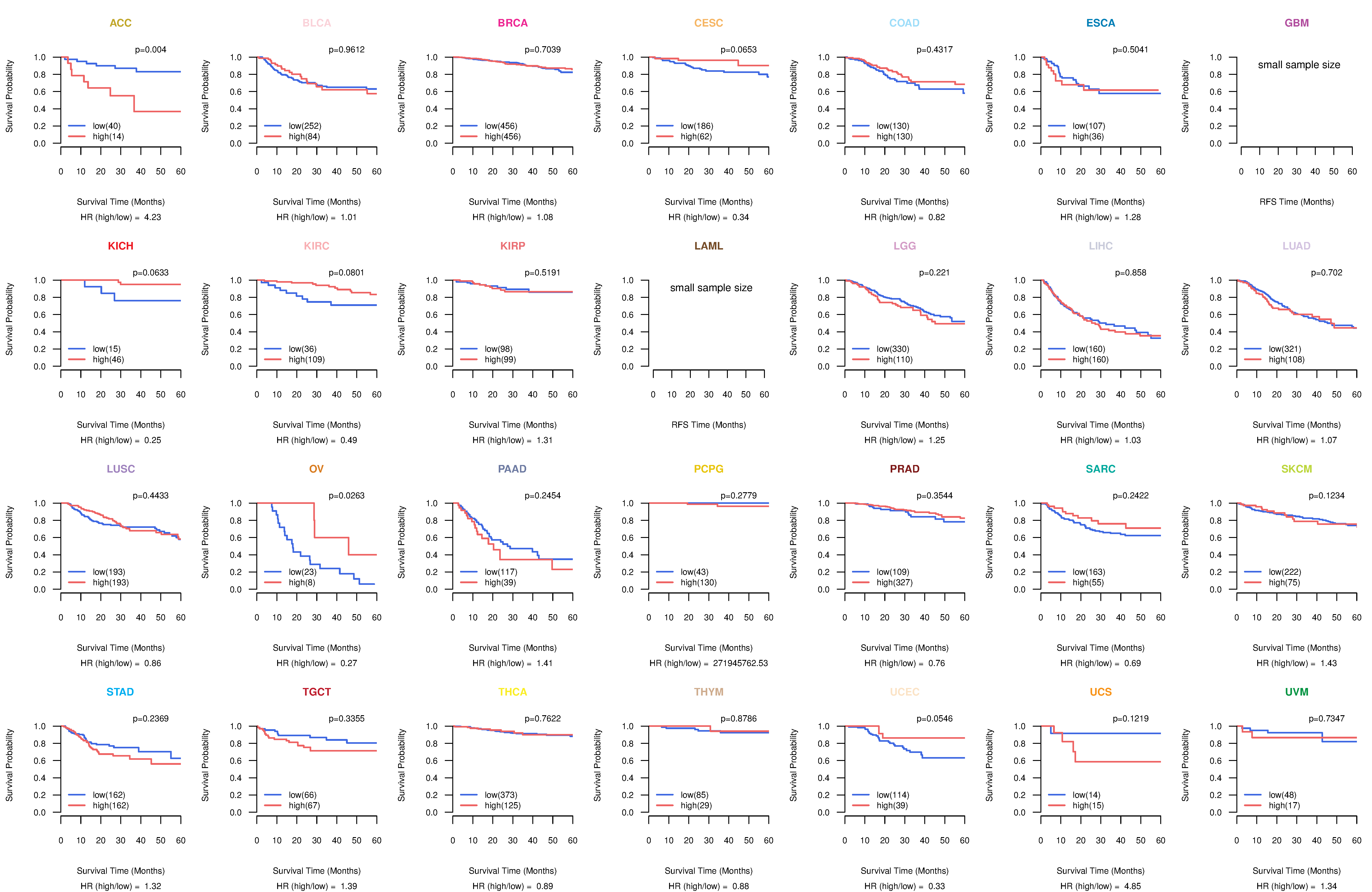
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 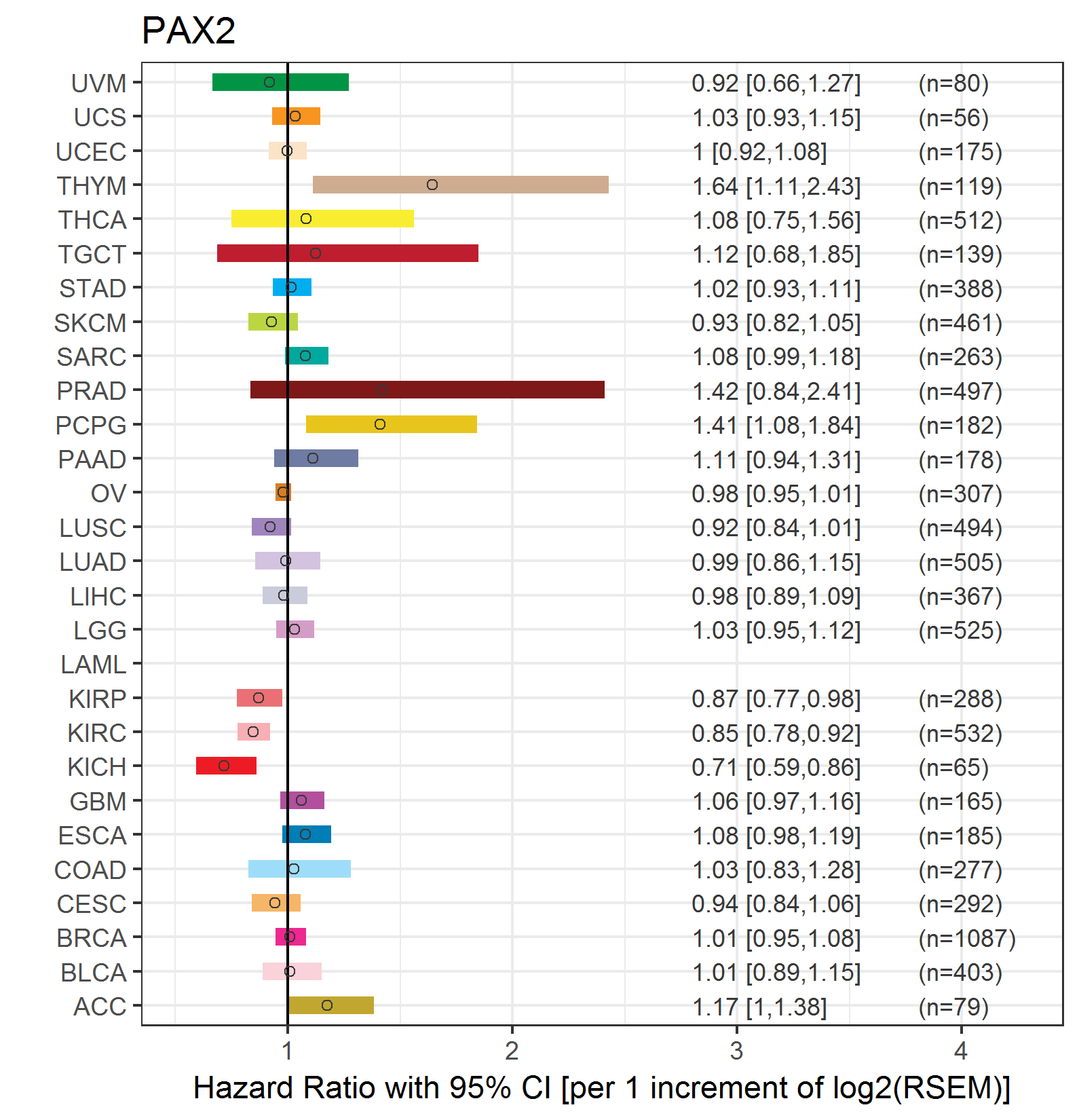
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 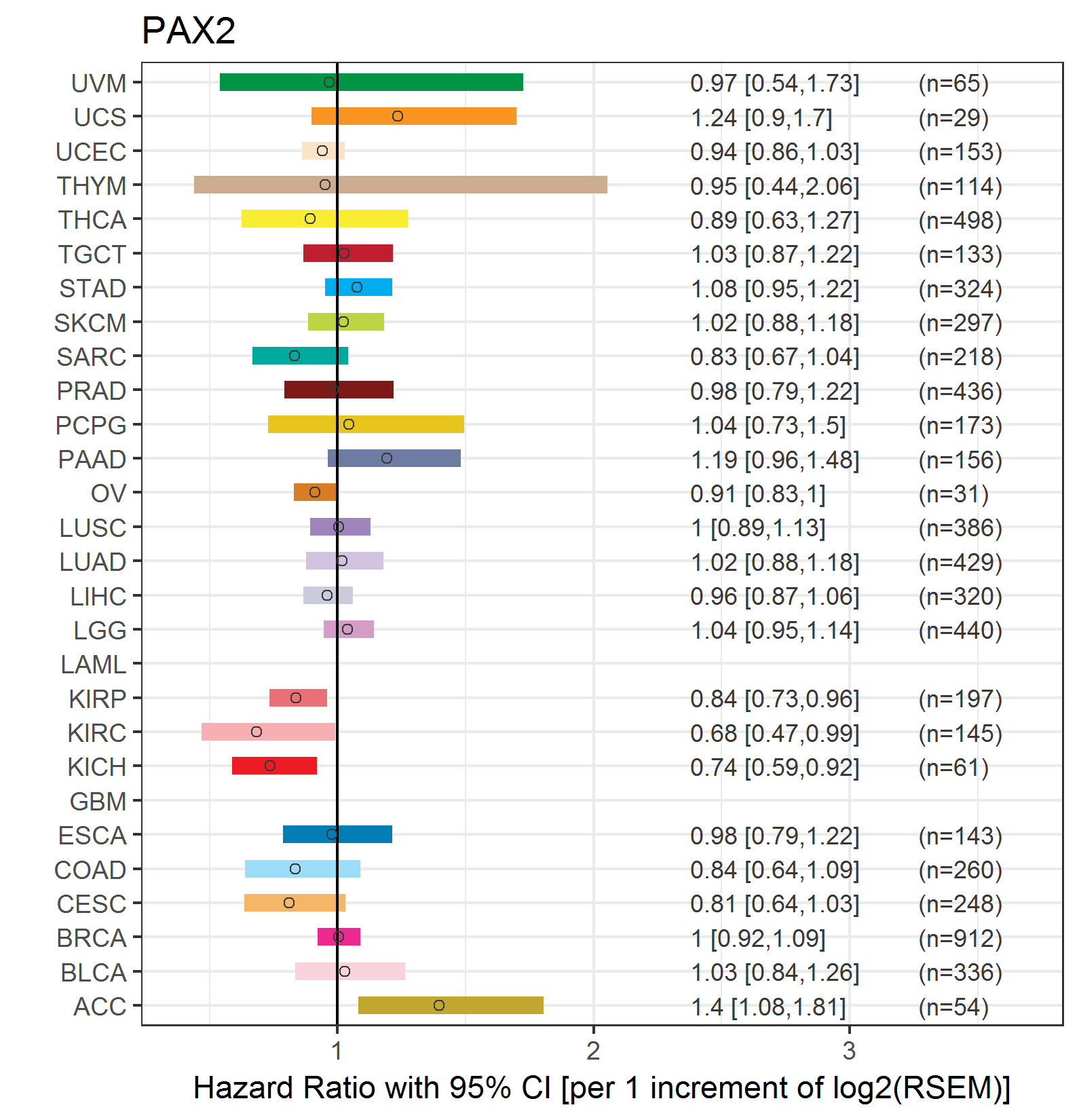
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























