|
Bioinformatics and Systems Medicine Laboratory
|
|
|
VirusFinder 2 (with VERSE algorithm)
|
|
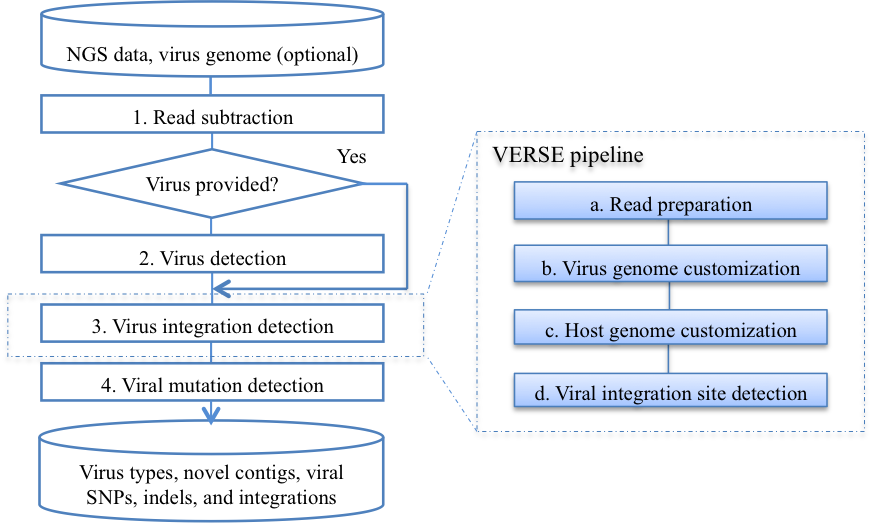
VirusFinder 2 is a new software tool for characterizing intra-host viruses through next generation sequencing (NGS) data. It implements our recently developed algorithm, Virus intEgration site detection through Reference SEquence customization (VERSE) (manuscript in consideration) as well as other new features. Specifically, it detects virus infection, co-infection with multiple viruses, mutations in the virus genomes, in addition to virus integration sites in host genomes. It also facilitates virus discovery by reporting novel contigs, long sequences assembled from short reads that map neither to the host genome nor to the genomes of known viruses. VirusFinder 2 can not only work for the human genome data, but also other organisms with available reference genome sequences (e.g. animals). VirusFinder 2 works with both paired-end and single-end data, unlike the previous 1.x versions that accepted only paired-end reads. The types of NGS data that VirusFinder 2 can deal with include whole genome sequencing (WGS), whole transcriptome sequencing (RNA-Seq), targeted sequencing data such as whole exome sequencing (WES) and ultra-deep amplicon sequencing.
|
 |
|
| Current Version |
|
|
| Documentation |
|
|
|
Release Note
|
| 12/09/2014 |
- |
The pipeline of VirusFinder 2 in the user's manual was updated.
|
| 12/01/2014 |
- |
User's manual updated to describe the implementation of VERSE in VirusFinder 2 and specify the citation of our VirusFinder 2 paper.
|
| 06/18/2014 |
- |
Code modified to improve the detection of low-confidence virus integration sites.
|
| 05/19/2014 |
- |
A bug in detect_integration.pl fixed to improve VirusFinder's portability.
|
| 04/17/2014 |
- |
A typo in Section 6.2 of User's manual corrected.
|
| 03/17/2014 |
- |
Code added to allow users to skip functions they don't need (Section 3.1 of User's manual).
|
| 02/19/2014 |
- |
VirusFinder 2 released to allow analyzing single-end NGS data, detecting virus co-infections, viral mutations, as well as novel viruses (see User's manual for detail). We thank users for the numerous feedback!
|
| 02/10/2014 |
- |
Code that deals with chromosome name without 'chr' prefix was mended.
|
| 01/08/2014 |
- |
Two bugs were fixed, one in Mosaic.pm and another in detect_integration.pl. We thank our user for helping identify and remove them.
|
| 12/26/2013 |
- |
VirusFinder 1.3 released to allow user to specify in command line all prerequisite inputs of each Perl script, e.g. detect_virus.pl, and detect_integration.pl.
|
| 12/20/2013 |
- |
User's manual updated (Sections 4.3 and FAQ).
|
| 12/18/2013 |
- |
Code added to report confidence in the virus-host integration loci (see User's manual);
A bug causing VirusFinder to fail to extract virus id from file results-virus.txt fixed.
|
| 11/11/2013 |
- |
A bug fixed. The bug caused VirusFinder not to report sites with >500 coverage.
|
| 10/25/2013 |
- |
Code changed to create file results-virus-top1.fa seperately from results-virus.txt.
|
| 10/18/2013 |
- |
VirusFinder 1.2 released to allow to detect virus integration sites in mitochondria and to address user's feedback related to software tool blat.
|
| 09/06/2013 |
- |
User's manual updated (Section FAQ).
|
| 07/31/2013 |
- |
The default value of the vairable 'flank_region_size' in configuration file changed to 4000.
|
| 07/26/2013 |
- |
VirusFinder 1.1 released. This version enables sensitive detection mode (see User's manual).
|
| 07/19/2013 |
- |
Code modified to handle virus sequence having special characters in its description line. |
| 06/25/2013 |
- |
User's manual updated to include frequently asked questions (FAQs) from users. |
| 05/03/2013 |
- |
Code for detecting virus integration events modified to better process deep sequencing data. |
| 04/16/2013 |
- | Code added to handle software installation errors. |
| 03/12/2013 |
- | Software release. |
|
| Test datasets |
|
|
| Related software |
|
|
| References |
- Wang Q, Jia P, Zhao Z (2013) VirusFinder: software for efficient and accurate detection of viruses and their integration sites in host genomes through next generation sequencing data. PLOS ONE 8(5):e64465 PubMed
- Bhaduri A, Qu K, Lee CS, Ungewickell A, Khavari PA. (2012) Rapid identification of nonhuman sequences in high throughput sequencing data sets, Bioinformatics, 28:1174-1175. PubMed
- Chen Y, Yao H, Thompson EJ, Tannir NM, Weinsten JN, Su X (2013) VirusSeq: software to identify viruses and their integration sites using next generation sequencing of human cancer tissue, Bioinformatics, 29(2):266-267. PubMed
- Kostic AD, Ojesina AI, Pedamallu CS, Jung J, Verhaak RG, Getz G, Meyerson M. (2011) PathSeq: software to identify or discover microbes by deep sequencing of human tissue, Nat Biotechnol, 29:393-396. PubMed
- Li JW, Wan R, Yu CS, Co NN, Wong N, Chan TF. (2013) ViralFusionSeq: accurately discover viral integration events and reconstruct fusion transcripts at single-base resolution, Bioinformatics, 29(5):649-651. PubMed
|
| Citation of VirusFinder 2 |
- Wang Q, Jia P, Zhao Z (2015) VERSE: a novel approach to detect virus integration in host genomes through reference genome customization. Genome Medicine 7:2 Paper Download
|
|
FAQ
|
|
Please read FAQ (Section 6 of the User's manual) in order to run VirusFinder correctly.
|
| Contact |
|
|
|
|
|
|

