|
Bioinformatics and Systems Medicine Laboratory
|
|
|
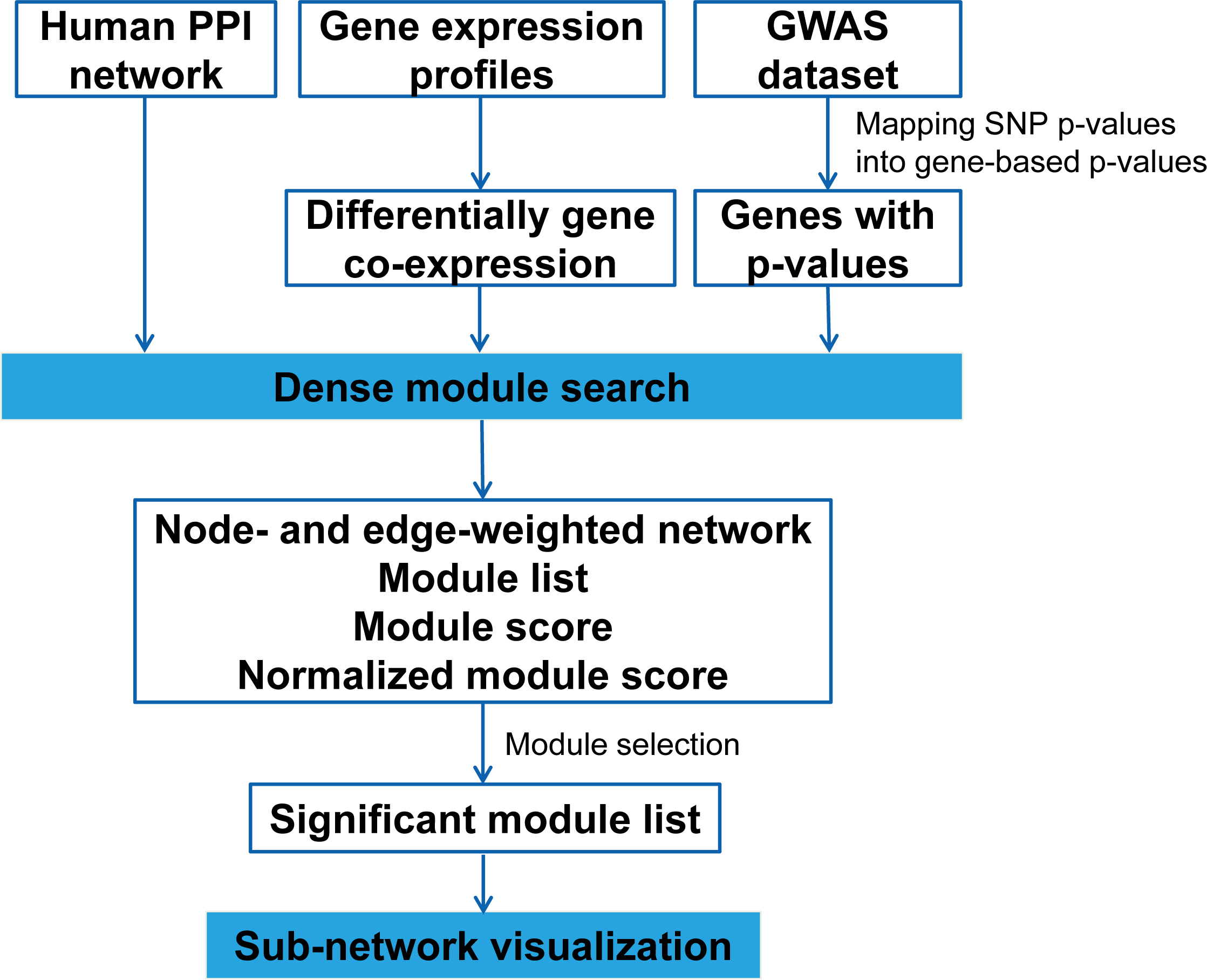
dmGWAS 3.0
|
|
On October 4, 2014, we released an upgraded version, dmGWAS 3.0, which implements a new algorithm EW_dmGWAS: Edge-weighted dense module search for genome-wide association studies and gene expression profiles.
In dmGWAS 3.0, the algorithm EW_dmGWAS introduces the following features:
|
- The new algorithm, EW_dmGWAS, combines node weight, which are computed based on GWAS signals, and edge weight, which are computed based on gene expression data.
- Allow tissue-specific gene expression profile. For example, for breast cancer GWAS analysis, gene expression data from breast cancer patients can be used; for schizophrenia, gene expression data in brain tissues can be used.
- Edge weights can be computed based on differential gene co-expression (DGCE) patterns, i.e., by comparing tumor to normal samples.
- In dmGWAS 3.0, users can still apply the "old" models that use only node weight from GWAS signal (see the user guide below).
- dmGWAS 3.0 is compatible to igraph 0.6.x.
All old versions, i.e., <3.0, are available from here.
|  |
|
| Current Version |
|
|
| Documentation |
|
|
| Citation |
- Wang Q, Yu H, Zhao Z, and Jia P (2015) EW_dmGWAS: Edge-weighted dense module search for genome-wide association studies and gene expression profiles. Bioinformatics online access
- Jia P, Zheng S, Long J, Zheng W, and Zhao Z (2011) dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics 27(1):95-102 PubMed
|
| Contact |
|
|
|
|

Copyright 2008-Present - The University of Texas Health Science Center at Houston (UTHealth)
Web File Viewing |
How to Report, Fraud, Waste and Abuse |
State of Texas |
Statewide Search |
Texas Homeland Security |
Site Policies
|

