Kinome NetworkX
A Network Atlas of 538 Human Kinase Genes
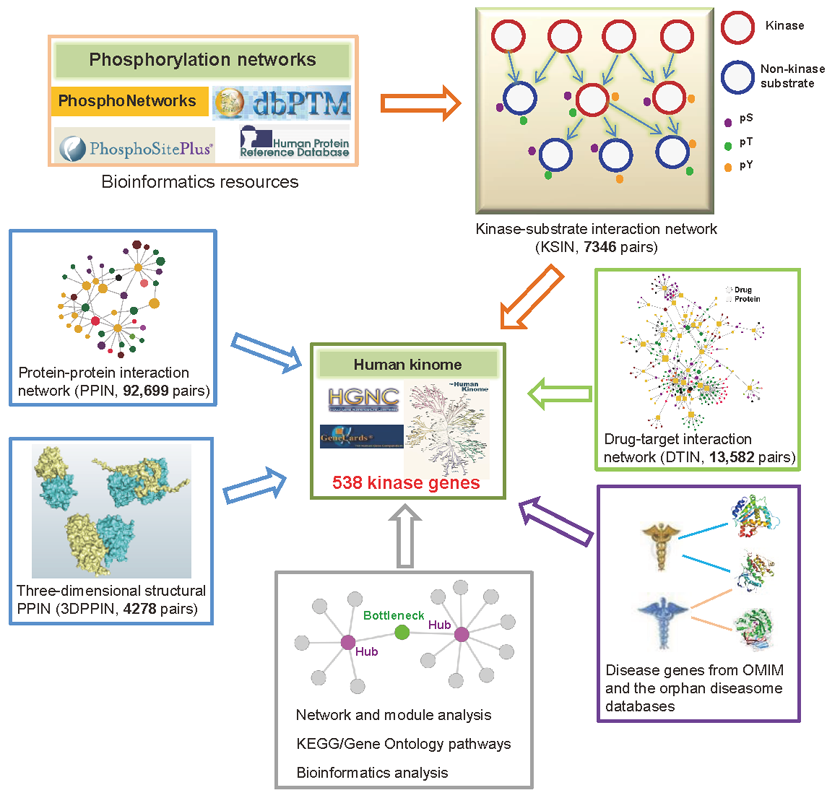
About Kinome NetworkXAlthough human protein kinases have been well studied, the currently incomplete interactome maps limit understanding of the global functional significance and network topological properties of human kinome. The Kinome NetworkX is a comprehensive functional and network atlas of 538 kinase genes built by a new integrative workflow as follow. This human kinome gene atlas included a kinase-substrate interaction network (KSIN) with 7,346 pairs collected from PhosphoNetworks, Phospho.ELM, HPRD, dbPTM3 and PhosphoSitePlus, a protein-protein interaction network (PPIN) with 92,699 pairs from PINA, and an atomic resolution three-dimensional structural PPIN with 4,278 pairs from INstruct. Among 538 kinase genes, 78.4% (422) genes were found to be cancer genes, mendelian disease genes, orphan disease mutant genes, or essential genes. We found that kinases are favorite to be functional centrality in the human protein interactome.
|
||
|
||
|
Topological analysis of the drug-kinase bipartite network quantitatively showed that the current effective kinase inhibitors were polypharmacology. Our analysis also revealed that out-of-date drug target selection strategy which used the functional hubs as drug targets link the high risk of drug side effects. Moreover, genes involved in anticancer drug sensitivity were significantly enriched to be the functional hubs, which heavily participated in a number of signaling and cancer pathways by network and bioinformatics analysis. This comprehensive catalog of human kinome map provided an overview of its network functional significance and is a useful resource for future discovery of effective kinase inhibitors in the personalized medicine.
|