| Basic gene Info. | Gene symbol | CXCR4 |
| Gene name | chemokine (C-X-C motif) receptor 4 |
| Synonyms | CD184|D2S201E|FB22|HM89|HSY3RR|LAP-3|LAP3|LCR1|LESTR|NPY3R|NPYR|NPYRL|NPYY3R|WHIM |
| Cytomap | UCSC genome browser: 2q21 |
| Type of gene | protein-coding |
| RefGenes | NM_001008540.1,
NM_003467.2, |
| Description | C-X-C chemokine receptor type 4CD184 antigenLPS-associated protein 3SDF-1 receptorfusinleukocyte-derived seven transmembrane domain receptorlipopolysaccharide-associated protein 3neuropeptide Y receptor Y3seven transmembrane helix receptorseven-t |
| Modification date | 20141221 |
| dbXrefs | MIM : 162643 |
| HGNC : HGNC |
| Ensembl : ENSG00000121966 |
| HPRD : 01217 |
| Vega : OTTHUMG00000153583 |
| Protein | UniProt: P61073
go to UniProt's Cross Reference DB Table |
| Expression | CleanEX: HS_CXCR4 |
| BioGPS: 7852 |
| Pathway | NCI Pathway Interaction Database: CXCR4 |
| KEGG: CXCR4 |
| REACTOME: CXCR4 |
| Pathway Commons: CXCR4 |
| Context | iHOP: CXCR4 |
| ligand binding site mutation search in PubMed: CXCR4 |
| UCL Cancer Institute: CXCR4 |
| Assigned class in mutLBSgeneDB | A: This gene has a literature evidence and it belongs to targetable_mutLBSgenes. |
| References showing study about ligand binding site mutation for CXCR4. | 1. "Wong RS, Bodart V, Metz M, Labrecque J, Bridger G, Fricker SP. Comparison of the potential multiple binding modes of bicyclam, monocylam, and noncyclam small-molecule CXC chemokine receptor 4 inhibitors. Mol Pharmacol. 2008 Dec;74(6):1485-95. doi: 10.1124/mol.108.049775. Epub 2008 Sep 2. PubMed PMID: 18768385. " 18768385
2. "Thiele S, Mungalpara J, Steen A, Rosenkilde MM, Våbenø J. Determination of the binding mode for the cyclopentapeptide CXCR4 antagonist FC131 using a dual approach of ligand modifications and receptor mutagenesis. Br J Pharmacol. 2014 Dec;171(23):5313-29. doi: 10.1111/bph.12842. PubMed PMID: 25039237; PubMed Central PMCID: PMC4294042. " 25039237
|
| Ligand ID | Ligand short name | Ligand long name | PDB ID | PDB name | mutLBS |
ITD | | (6,6-DIMETHYL-5,6-DIHYDROIMIDAZO[2,1-B][1,3]THIAZOL-3-YL)METHYL N,N'-DICYCLOHEXYLIMIDOTHIOCARBAMATE | 3oe8 | A | W94 | ITD | | (6,6-DIMETHYL-5,6-DIHYDROIMIDAZO[2,1-B][1,3]THIAZOL-3-YL)METHYL N,N'-DICYCLOHEXYLIMIDOTHIOCARBAMATE | 3oe9 | A | W94 H113 R183 | ITD | | (6,6-DIMETHYL-5,6-DIHYDROIMIDAZO[2,1-B][1,3]THIAZOL-3-YL)METHYL N,N'-DICYCLOHEXYLIMIDOTHIOCARBAMATE | 3oe9 | B | W94 H113 R183 | ITD | | (6,6-DIMETHYL-5,6-DIHYDROIMIDAZO[2,1-B][1,3]THIAZOL-3-YL)METHYL N,N'-DICYCLOHEXYLIMIDOTHIOCARBAMATE | 3odu | A | W94 R183 | ITD | | (6,6-DIMETHYL-5,6-DIHYDROIMIDAZO[2,1-B][1,3]THIAZOL-3-YL)METHYL N,N'-DICYCLOHEXYLIMIDOTHIOCARBAMATE | 3odu | B | W94 R183 | ITD | | (6,6-DIMETHYL-5,6-DIHYDROIMIDAZO[2,1-B][1,3]THIAZOL-3-YL)METHYL N,N'-DICYCLOHEXYLIMIDOTHIOCARBAMATE | 3oe6 | A | W94 R183 | ITD | | (6,6-DIMETHYL-5,6-DIHYDROIMIDAZO[2,1-B][1,3]THIAZOL-3-YL)METHYL N,N'-DICYCLOHEXYLIMIDOTHIOCARBAMATE | 3oe8 | B | W94 R183 | ITD | | (6,6-DIMETHYL-5,6-DIHYDROIMIDAZO[2,1-B][1,3]THIAZOL-3-YL)METHYL N,N'-DICYCLOHEXYLIMIDOTHIOCARBAMATE | 3oe8 | C | W94 R183 |
| LBS | AA sequence | # species | Species |
C186 | DDRYICDRFYP | 4 | Homo sapiens, Canis lupus familiaris, Pan troglodytes, Macaca mulatta | C186 | NGQFVCDRIYP | 1 | Xenopus tropicalis | C186 | DDRYICDRLYP | 1 | Mus musculus | C186 | DERYICDRFYP | 1 | Bos taurus | D187 | DRYICDRFYP- | 4 | Homo sapiens, Canis lupus familiaris, Pan troglodytes, Macaca mulatta | D187 | GQFVCDRIYPI | 1 | Xenopus tropicalis | D187 | DRYICDRLYP- | 1 | Mus musculus | D187 | ERYICDRFYP- | 1 | Bos taurus | D97 | PFWAVDAVANW | 4 | Homo sapiens, Bos taurus, Pan troglodytes, Macaca mulatta | D97 | PFWSVDAAIGW | 1 | Xenopus tropicalis | D97 | PFWAVDAMADW | 1 | Mus musculus | D97 | PFWAVEAVANW | 1 | Canis lupus familiaris | E288 | WISITEALAFF | 6 | Homo sapiens, Mus musculus, Bos taurus, Canis lupus familiaris, Pan troglodytes, Macaca mulatta | E288 | AISITEALAFF | 1 | Xenopus tropicalis | E32 | PCFREENAHFN | 3 | Bos taurus, Canis lupus familiaris, Macaca mulatta | E32 | PCFREENANFN | 2 | Homo sapiens, Pan troglodytes | E32 | PCFMHDNSDFN | 1 | Xenopus tropicalis | E32 | PCFRDENVHFN | 1 | Mus musculus | H113 | LCKAVHVIYTV | 6 | Homo sapiens, Xenopus tropicalis, Bos taurus, Canis lupus familiaris, Pan troglodytes, Macaca mulatta | H113 | LCKAVHIIYTV | 1 | Mus musculus | I185 | ADDRYICDRFY | 4 | Homo sapiens, Canis lupus familiaris, Pan troglodytes, Macaca mulatta | I185 | ENGQFVCDRIY | 1 | Xenopus tropicalis | I185 | GDDRYICDRLY | 1 | Mus musculus | I185 | VDERYICDRFY | 1 | Bos taurus | K38 | NAHFNRIFLPT | 3 | Bos taurus, Canis lupus familiaris, Macaca mulatta | K38 | NANFNKIFLPT | 2 | Homo sapiens, Pan troglodytes | K38 | NSDFNRIFLPT | 1 | Xenopus tropicalis | K38 | NVHFNRIFLPT | 1 | Mus musculus | R183 | SEADDRYICDR | 3 | Homo sapiens, Pan troglodytes, Macaca mulatta | R183 | SDENGQFVCDR | 1 | Xenopus tropicalis | R183 | SQGDDRYICDR | 1 | Mus musculus | R183 | KEVDERYICDR | 1 | Bos taurus | R183 | READDRYICDR | 1 | Canis lupus familiaris | R188 | RYICDRFYP-- | 5 | Homo sapiens, Bos taurus, Canis lupus familiaris, Pan troglodytes, Macaca mulatta | R188 | QFVCDRIYPID | 1 | Xenopus tropicalis | R188 | RYICDRLYP-- | 1 | Mus musculus | V112 | FLCKAVHVIYT | 6 | Homo sapiens, Xenopus tropicalis, Bos taurus, Canis lupus familiaris, Pan troglodytes, Macaca mulatta | V112 | FLCKAVHIIYT | 1 | Mus musculus | W102 | DAVANWYFGNF | 3 | Homo sapiens, Pan troglodytes, Macaca mulatta | W102 | DAAIGWYFKEF | 1 | Xenopus tropicalis | W102 | DAMADWYFGKF | 1 | Mus musculus | W102 | DAVANWYFGKF | 1 | Bos taurus | W102 | EAVANWYFGNF | 1 | Canis lupus familiaris | W94 | ITLPFWAVDAV | 3 | Homo sapiens, Pan troglodytes, Macaca mulatta | W94 | FTLPFWSVDAA | 1 | Xenopus tropicalis | W94 | ITLPFWAVDAM | 1 | Mus musculus | W94 | LTLPFWAVDAV | 1 | Bos taurus | W94 | LTLPFWAVEAV | 1 | Canis lupus familiaris | Y116 | AVHVIYTVNLY | 6 | Homo sapiens, Xenopus tropicalis, Bos taurus, Canis lupus familiaris, Pan troglodytes, Macaca mulatta | Y116 | AVHIIYTVNLY | 1 | Mus musculus |
 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Lollipop-style diagram of mutations at LBS in amino-acid sequence.
Lollipop-style diagram of mutations at LBS in amino-acid sequence. 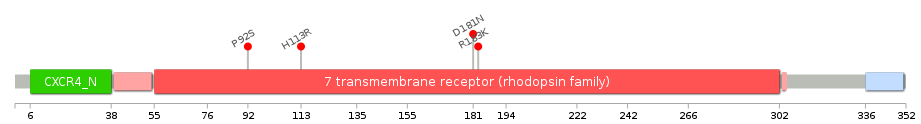
 Cancer type specific mutLBS sorted by frequency
Cancer type specific mutLBS sorted by frequency Relative protein structure stability change (ΔΔE) using Mupro 1.1
Relative protein structure stability change (ΔΔE) using Mupro 1.1  : nsSNV at non-LBS
: nsSNV at non-LBS : nsSNV at LBS
: nsSNV at LBS
 nsSNVs sorted by the relative stability change of protein structure by each mutation
nsSNVs sorted by the relative stability change of protein structure by each mutation  Structure image for CXCR4 from PDB
Structure image for CXCR4 from PDB Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types
Differential gene expression between mutated and non-mutated LBS samples in all 16 major cancer types Differential co-expressed gene network based on protein-protein interaction data (CePIN)
Differential co-expressed gene network based on protein-protein interaction data (CePIN) Gene level disease information (DisGeNet)
Gene level disease information (DisGeNet)  Mutation level pathogenic information (ClinVar annotation)
Mutation level pathogenic information (ClinVar annotation)  Gene expression profile of anticancer drug treated cell-lines (CCLE)
Gene expression profile of anticancer drug treated cell-lines (CCLE)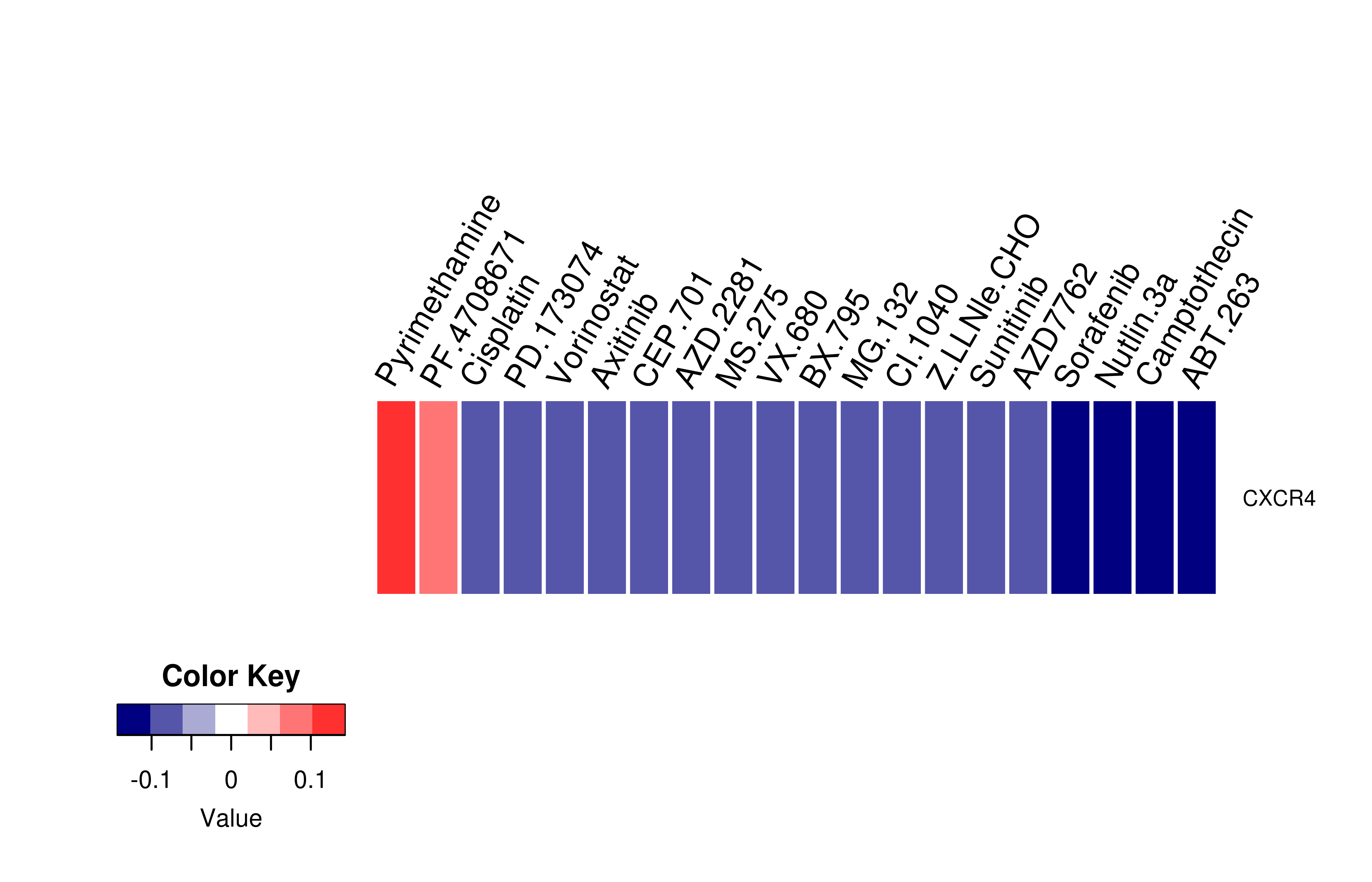
 Gene-centered drug-gene interaction network
Gene-centered drug-gene interaction network 
 Drug information targeting mutLBSgene (Approved drugs only)
Drug information targeting mutLBSgene (Approved drugs only)

 Gene-centered ligand-gene interaction network
Gene-centered ligand-gene interaction network 
 Ligands binding to mutated ligand binding site of CXCR4 go to BioLip
Ligands binding to mutated ligand binding site of CXCR4 go to BioLip Multiple alignments for P61073 in multiple species
Multiple alignments for P61073 in multiple species 
