1.Curation and annotations of cleft-related genes
Cleft lip with/without cleft palate (CL/P) is one of the most common birth defects with a prevalence of around 1/700 live birth worldwide. There have been many genetic, human epidemiological, mouse developmental and mutational studies conducted for CL/P, covering both cleft lip and cleft palate. The current version of CleftGeneDB (v1) hosts 560 experimentally identified genes associated with cleft lips/palate manually curated by experts in dental developmental biologists, and an expanded list of 3,261 genes related to CL/P in 11 eukaryotes (as of 4/29/2021). These genes were curated from the following resources
-
(1) Manual curation from literature. We collected cleft genes through extensive literature search and curation from Medline (Ovid), Embase (Ovid), and PubMed (NLM). We followed a guideline set forth by PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) for the systematic cleft-related gene search. The bibliographies of highly pertinent articles were further examined to avoid any errors introduced with the systematic review. To recover any missing data related to CL/P, we integrated with information in Scopus (Elsevier) and the Mouse Genome Informatics (MGI). In total, we have collected 560 experimentally identified genes associated with cleft lips and palate in two model organisms: Homo sapiens and Mus musculus. We name these genes as cleft genes. Specifically, cleft palate (CP) genes included genes associated with cleft palate only (CPO) and cleft lip and palate (CLP). Cleft lip (CL) genes included genes associated with cleft lip only (CLO) and cleft lip and palate (CLP), but excluded midline cleft and cleft palate only (CPO).
-
(2) Orthologous mapping.To annotate potential cleft genes in other species, we conducted orthologous gene search. Only those species having the reported phenotype of cleft lip or cleft palate were included. This procedure resulted in 8 other species: Bos taurus (bovine), Canis lupus familiaris (dog), Capra hircus (goat), Equus caballus (horse), Pan troglodytes (chimpanzee), Sus scrofa (pig), Oryctolagus cuniculus (rabbit), Rattus norvegicus (rat) and Danio rerio (zebrafish). Using the Orthologous Matrix (OMA) ortholog database, we identified 429 cleft related orthologous groups, each of which contained at least one experimentally reported cleft gene in humans or mice. We then integrated 2,701 homologous genes in the orthologous groups as candidate genes into the database.
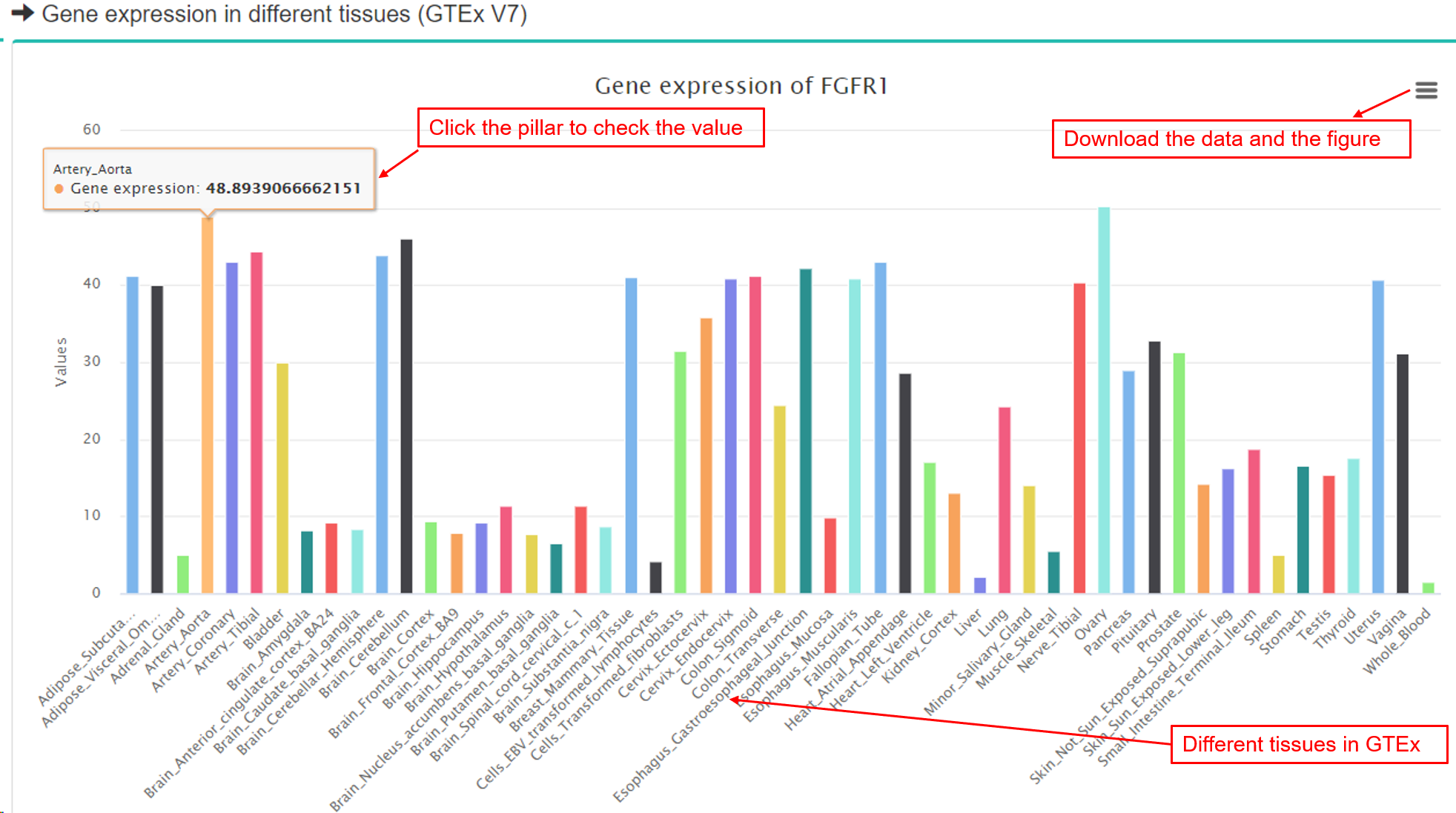
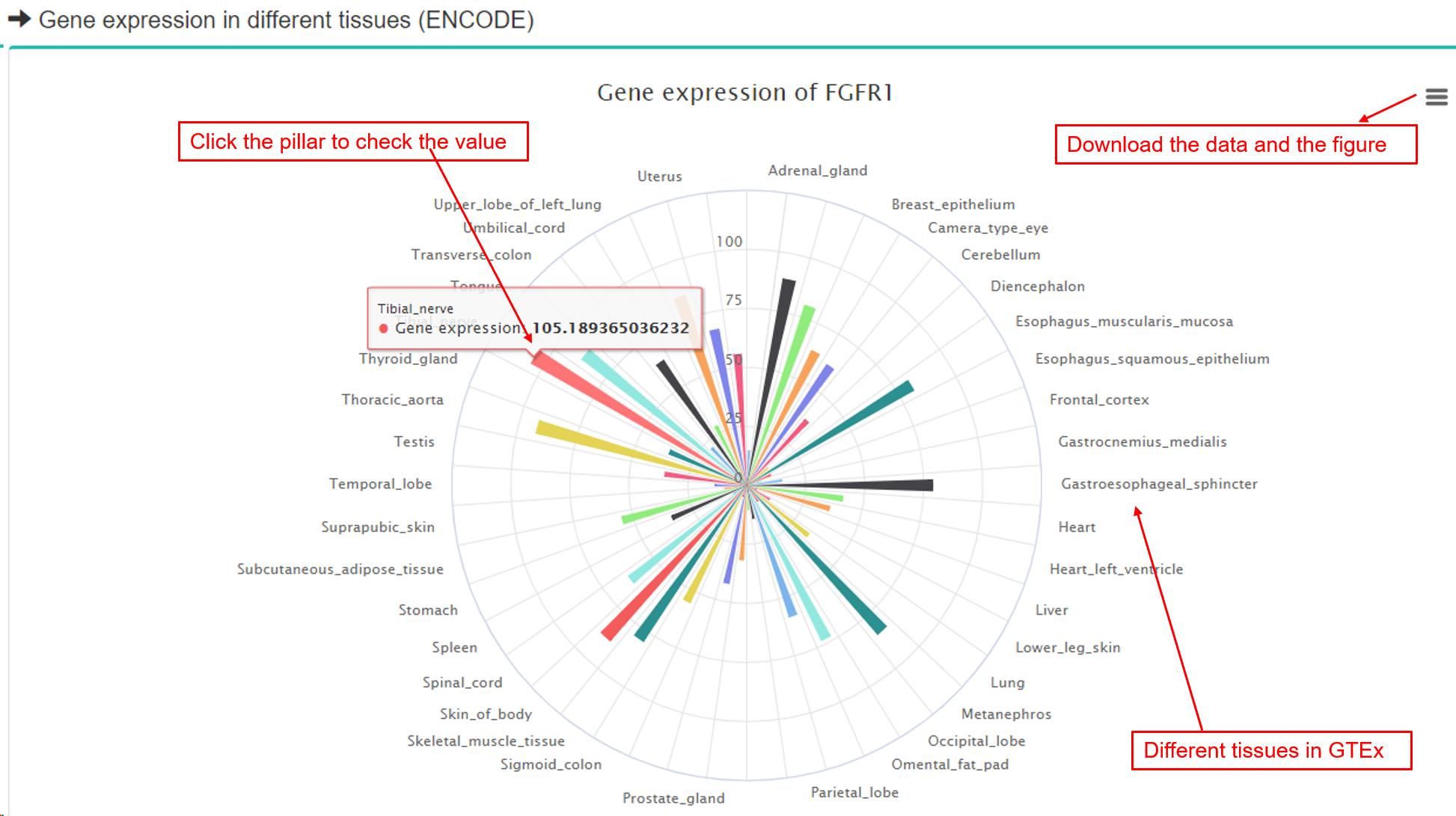
Next, we linked all the cleft genes through multiple cross-referencing. For example, gene and protein names were taken mainly from NCBI and UniProt, whereas corresponding accession numbers were integrated from UniProt, Ensembl, EMBL and NCBI GenBank. For experimentally identified cleft genes, the corresponding cleft types and developmental stages in mouse embryogenesis were added. Moreover, functional descriptions and protein sequences were derived from UniProt to provide the basic information for each cleft gene, while the PubMed references for each gene were listed via NCBI PMID number. To comprehensively annotate these cleft genes, in addition to basic information, we integrated the knowledge from more than 30 publicly available resources, including UniProt, ENCODE, GTEx, PDB, InterPro, BioGRID, STRING, Reactome, MGI, D2P2, DisGeNET, CTD, dbSNP, The 1000 Genomes project, The Cancer Genome Atlas (TCGA), ClinVar, ExAC, TOPMed, gnomAD, PROSITE, Pfam, among others. These annotations cover 9 distinct features, including (1) gene expression information, (2) protein structural annotations, (3) orthologous information, (4) genetic variants & mutations, (5) disease or phenotype associated information, (6) drugs and compounds information, (7) Gene Ontology (GO)/biological pathways, (8) functional annotations, (9) protein-protein interaction and protein-miRNA interaction.
2. Data statistics
3. Illustration of the CleftGeneDB browse function
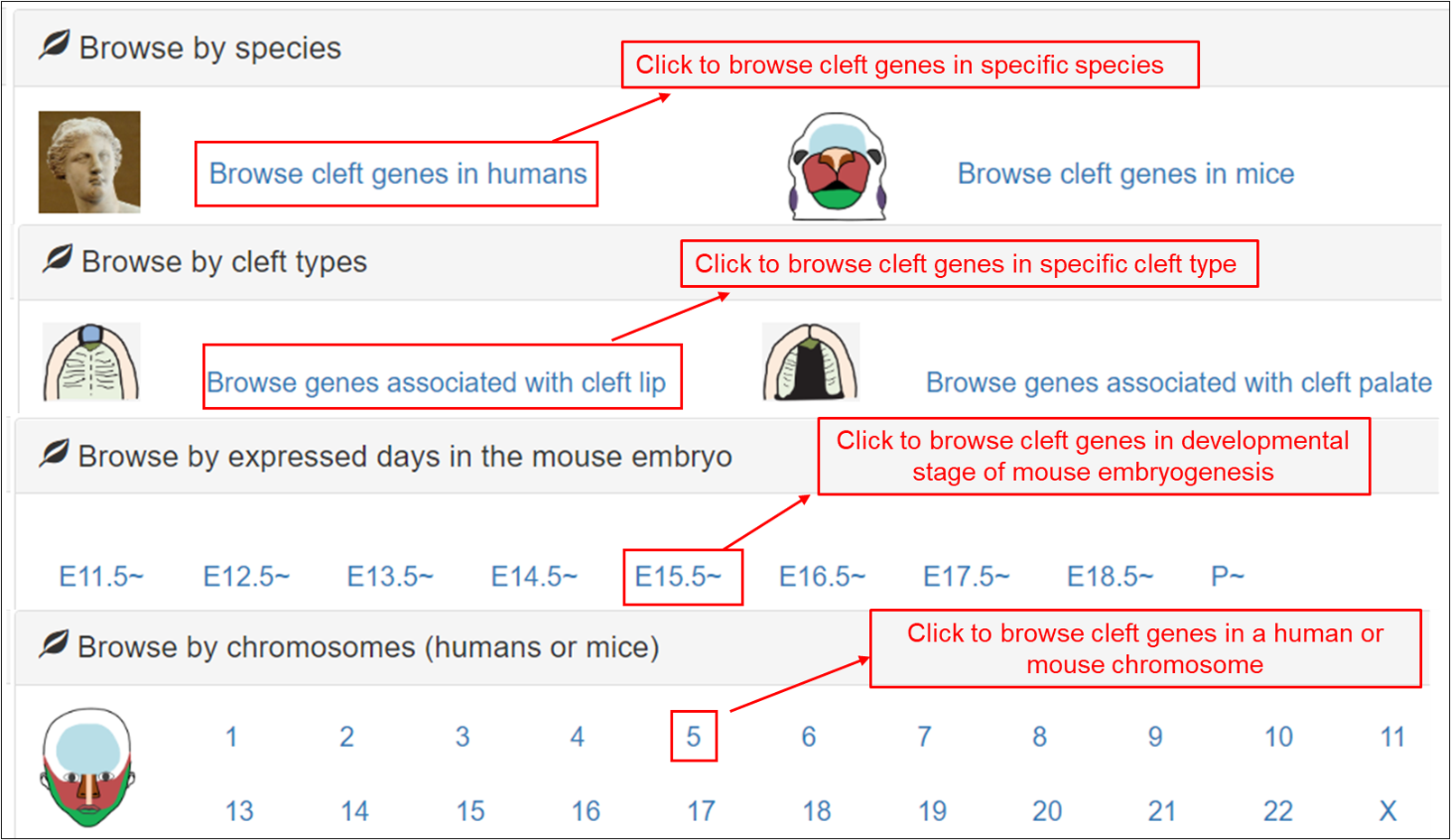
CleftGeneDB is developed with a user-friendly website interface. Multiple browse and search options are implemented to conveniently query and present the data by user’s interest. The user has 5 browse options:
-
(1) Browse by species. In the option “Browse by species”, 10 representative pictures for all species are listed. The user can click the phylum to link the taxonomic of the given phylum. For example, the user can select “Browse cleft genes in humans” to retrieve a list of human cleft genes in a tabular format.
-
(2) Browse by cleft type. In the option “Browse by cleft type”, the user can choose specific cleft types, such as cleft lip or cleft palate, to browse the curated cleft genes. For example, by clicking the diagram of “Browse genes associated with cleft lip”, all genes associated with cleft lip are listed in a tabular format with “Gene symbol”, “Gene name”, “Entrez ID”, “UniProt ID”, “Chr.”, “Cleft types”, “Species” and “Evidence”.
-
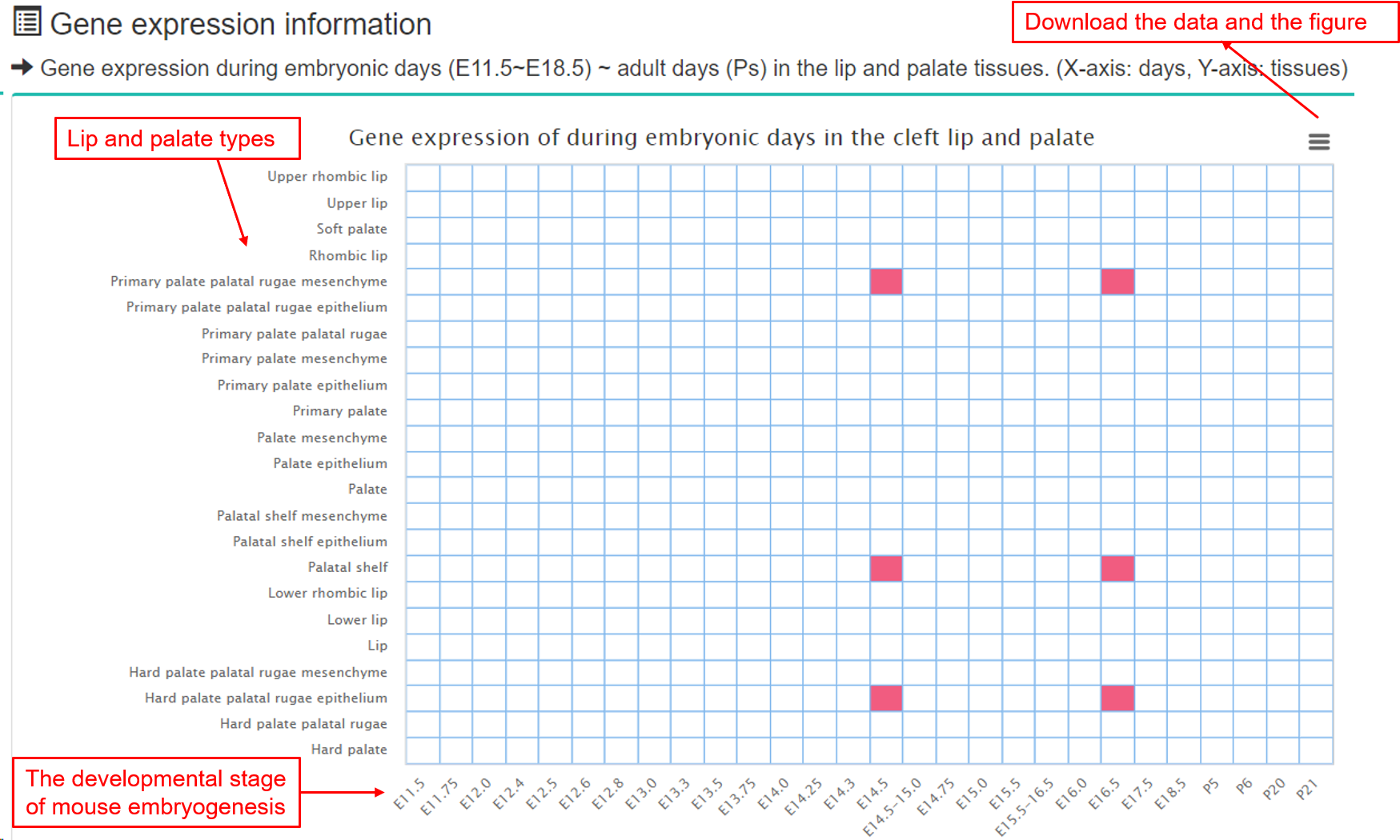
(3) Browse by developmental stage of mouse embryogenesis. In the option “Browse by developmental stage of mouse embryogenesis”, the user can choose different symbols to browse the cleft genes expressed at the given developmental days in the mouse embryo. For example, by clicking the diagram of “E11.5~”, all the cleft genes at this specific embryonic development stage are listed in a tabular format with “Gene symbol”, “Gene name”, “Entrez ID”, “UniProt ID”, “Chr.”, “Cleft types”, “Developmental stage” and “Evidence”.
-
(4) Browse by chromosome (human or mouse). The user can retrieve a gene list in each human or mouse chromosome.
-
(5) Browse all cleft genes. The user can check all cleft genes in a tabular format and conduct a basic search.
4. Description of the CleftGeneDB search functions:
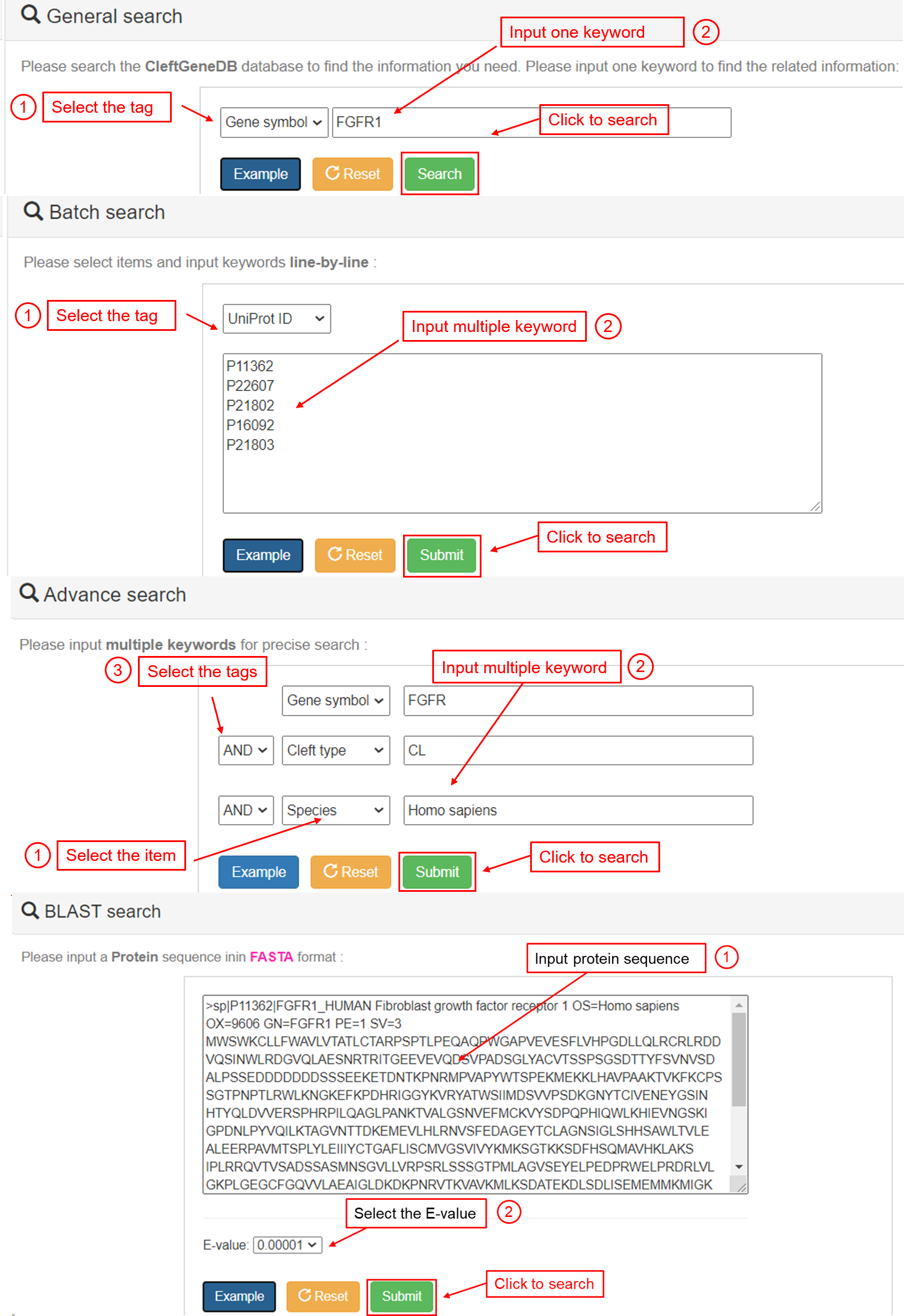
The CleftGeneDB has 4 search options. The user can search for genes using a keyword or combination of keywords. These include UniProt ID, Entrez ID, gene symbol, gene name, chromosome location, cleft type and species. We also implemented a search function based on the NCBI BLAST program to allow the user to search new sequences. The user can provide their own protein sequences in the FASTA format, and the search can output whether there are similar or homologous proteins to our cleft genes.
- The user can directly search the CleftGeneDB database by typing a keyword and the related tag. For example, if the keyword “FGFR1” under “Gene symbol” is submitted, the retrieved results are shown in a tabular format, including “Gene symbol”, “Gene name”, “Entrez ID”, “UniProt ID”, “Chr.”, “Cleft types”, “Species” and “Evidence”. The user could click the ”More>>” link to find more annotations.
-
(2) Batch search. The user can enter multiple keywords in different fields, such as entrez ID, UniProt ID, gene symbol, gene name, chromosome and cleft type, in a line-by-line format for querying. For example, if the user has the following term: “UniProt ID”, including “P11362”, “P22607”, “P21802”, “P16092” and “P21803”, the retrieved results are shown in a tabular format, including “Gene symbol”, “Gene name”, “Entrez ID”, “UniProt ID”, “Chr.”, “Cleft types”, “Species” and “Evidence”. Users could click the “More>>” link to find more annotations.
-
(3) Advanced search. The user can have complex or combinatory list of keywords to retrieve a specific set of genes with annotated information. The interface of search engine allows the querying by different database fields and linking queries through three operators based on logic “and” and “or”. For example, if the use has this combination of search term: keyword “FGFR1” in the field “Gene symbol”, keyword “CL” in the field “cleft type”, and the keyword “Homo sapiens” in the field “Species”, the corresponding results are shown in a tabular format, including “Gene symbol”, “Gene name”, “Entrez ID”, “UniProt ID”, “Chr.”, “Cleft types”, “Species” and “Evidence”. The user can click the “More>>” link to find more annotations.
-
(4) BLASTP search. This option is for querying the related information in CleftGeneDB by using a protein sequence or a batch of protein sequences. The blastp program of NCBI BLAST packages was implemented in the database. The user can enter a protein sequence in FASTA format for searching identical or homologous proteins. For example, if the protein sequence for “FGFR1” gene and specific “E-value” is submitted, the homologous proteins greater than the threshold E-value will be presented in a tabular format, including “Gene symbol”, “Gene name”, “Entrez ID”, “UniProt ID”, “Species”, “Identity”, “E-value” and “Score (bits)”. Users can click the “More>>” link to find more annotations.
5. Description of integrative analysis of CleftGeneDB:
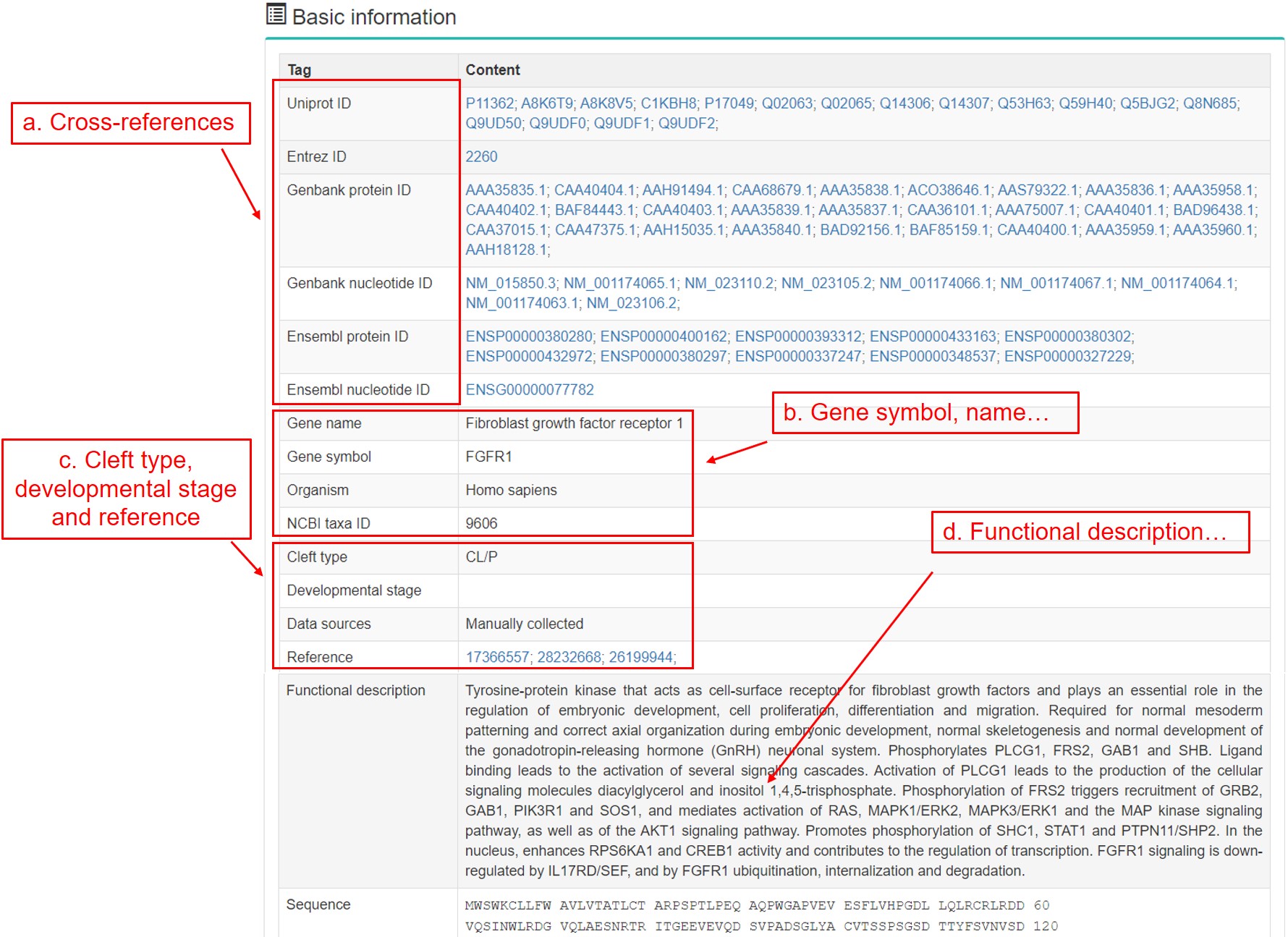
For each gene, we provide comprehensive annotations for user’s convenience. The gene page organizes these annotations as the following components. We use FGFR1, which encodes fibroblast growth factor receptor, as an example.
-
(1) Orthologous information
- a. Cross-references from UniProt, Ensembl, EMBL and NCBI GenBank.
- b. Gene symbol, gene name, organism and NCBI taxa ID.
- c. Cleft type and developmental stage.
- d. Functional description.
-
(2) Gene expression information
-
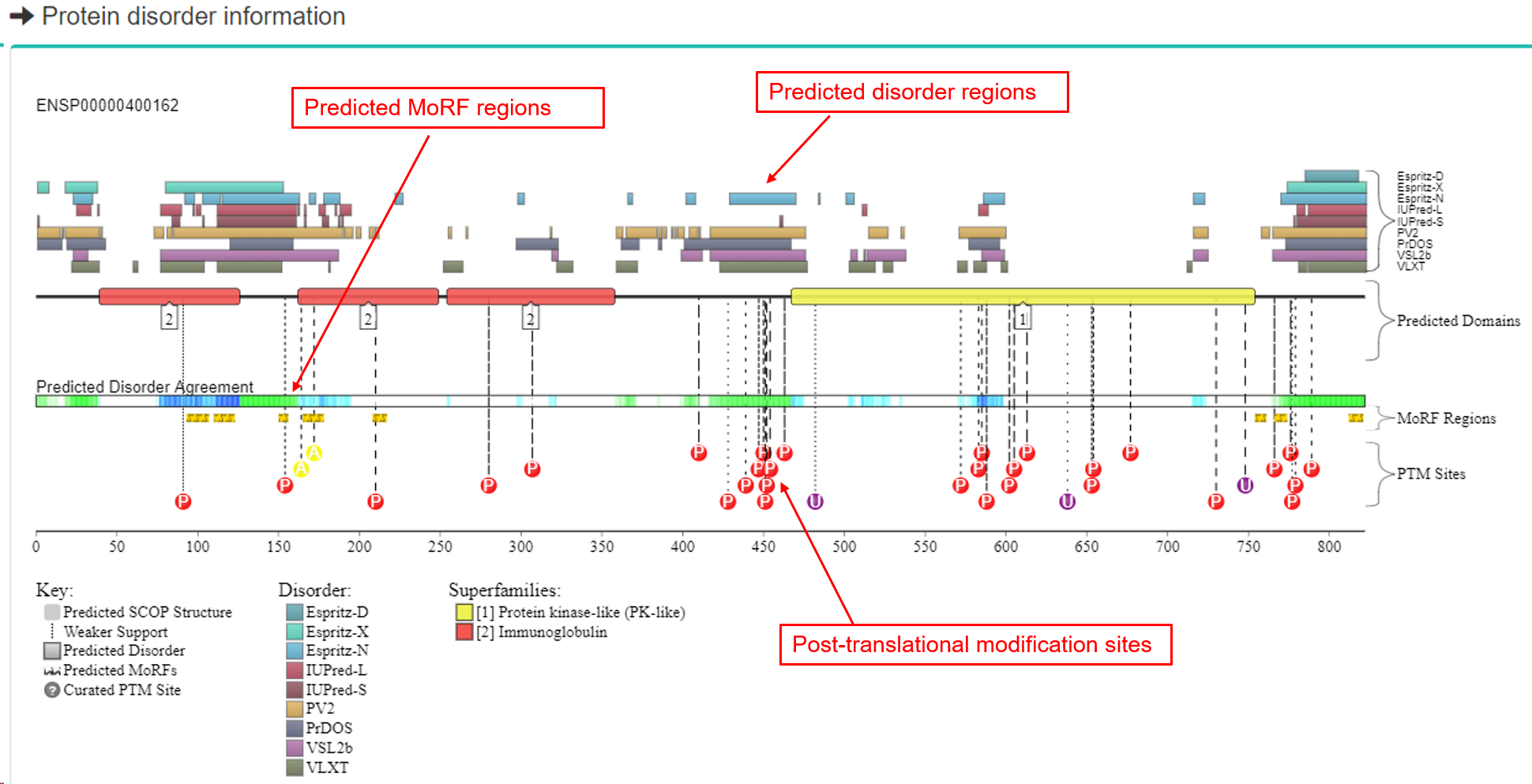
(3) Protein structural annotations
- a. 3D structure in PDB database.
- b. Protein disorder information (if applies).

-
(4) Orthologous information
- a. Cleft related orthologous groups from OMA ortholog database.

-
(5) Genetic variants & mutations
- a. All variants & mutations records were collected and integrated from dbSNP,
The 1000 Genomes project,
The Cancer Genome Atlas (TCGA), ClinVar, ExAC, TOPMed, gnomAD and UniProt.
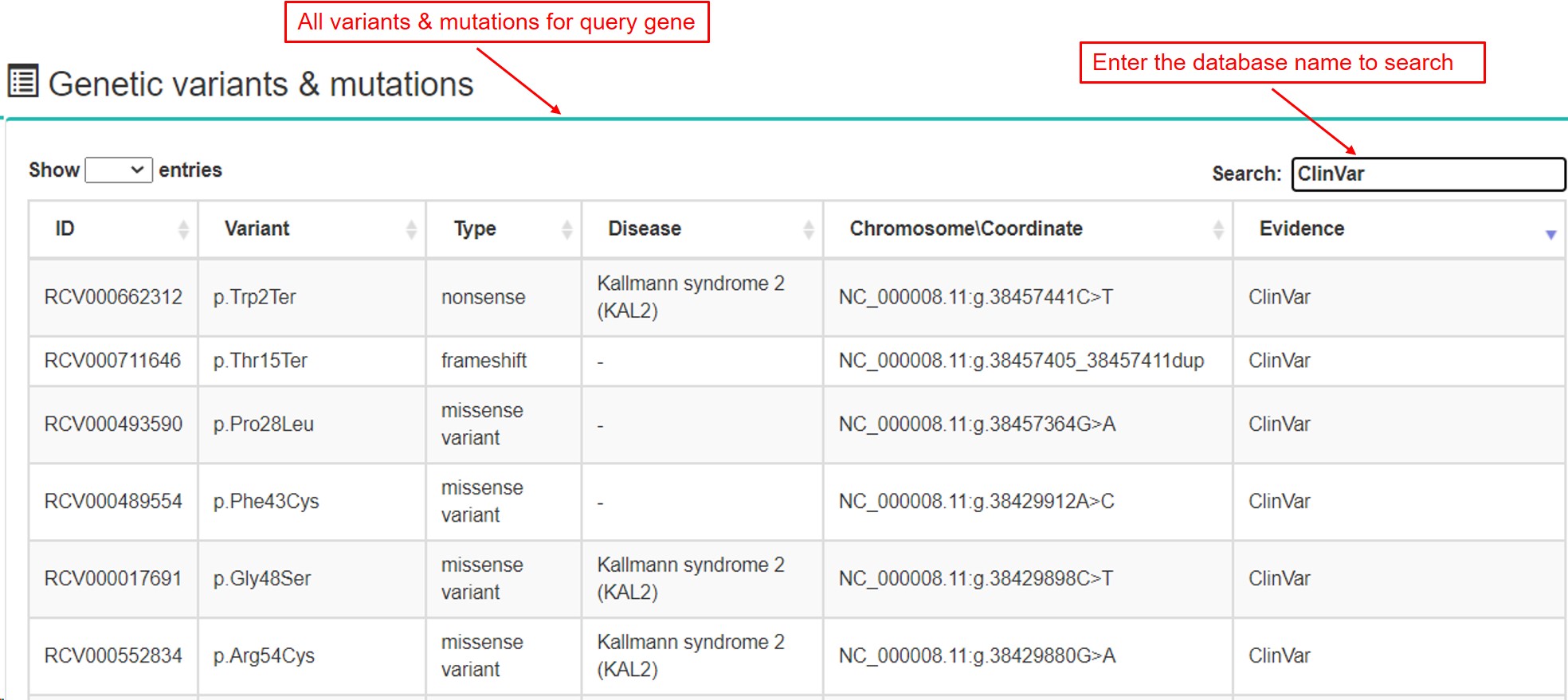
- a. All variants & mutations records were collected and integrated from dbSNP,
The 1000 Genomes project,
-
(6) Disease or phenotype associated information
- a. Disease or phenotype associated records were collected from DisGeNET database.
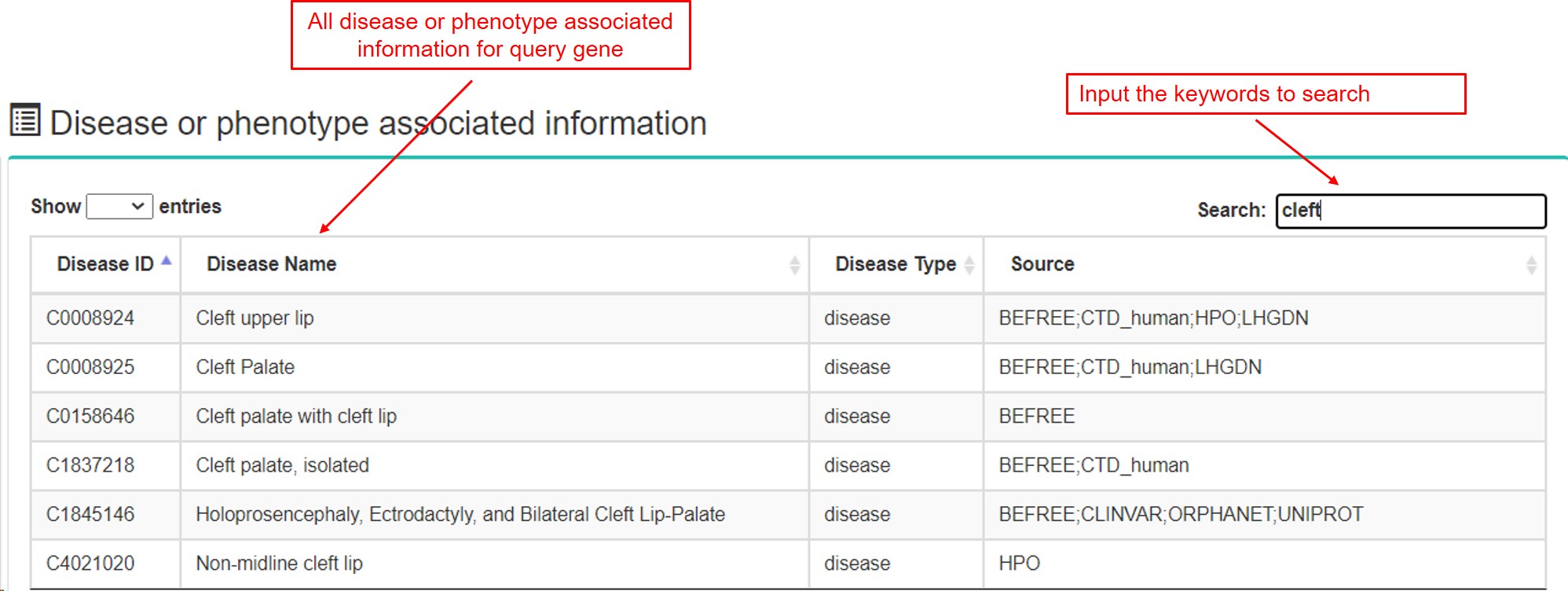
-
(7) Drugs and compounds information
- a. Drugs and compounds records were collected from The Comparative Toxicogenomics Database (CTD).

-
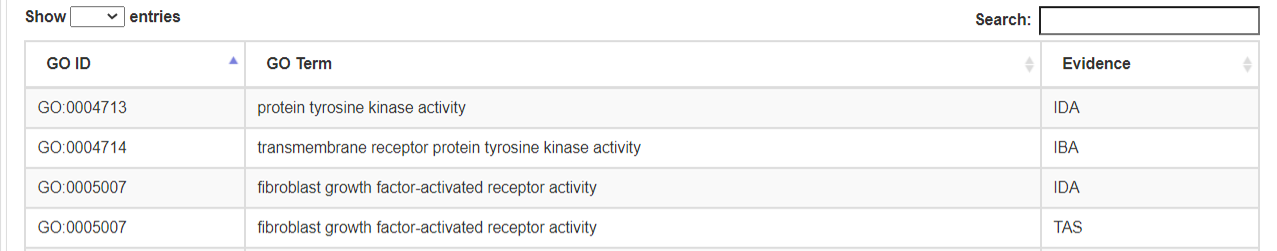
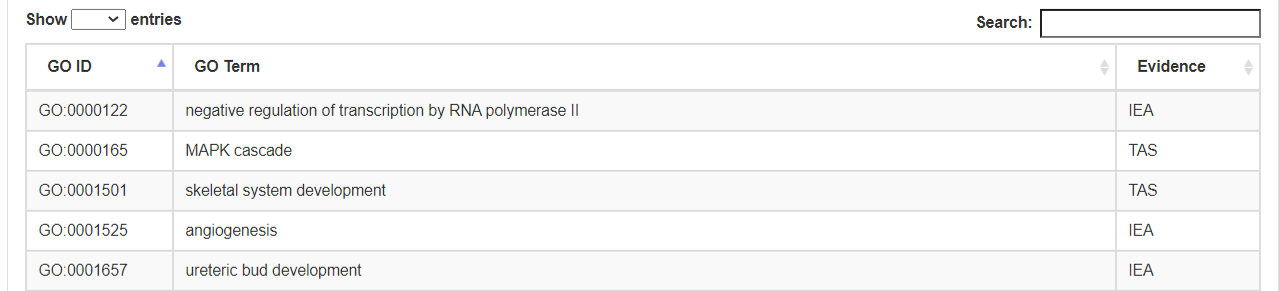
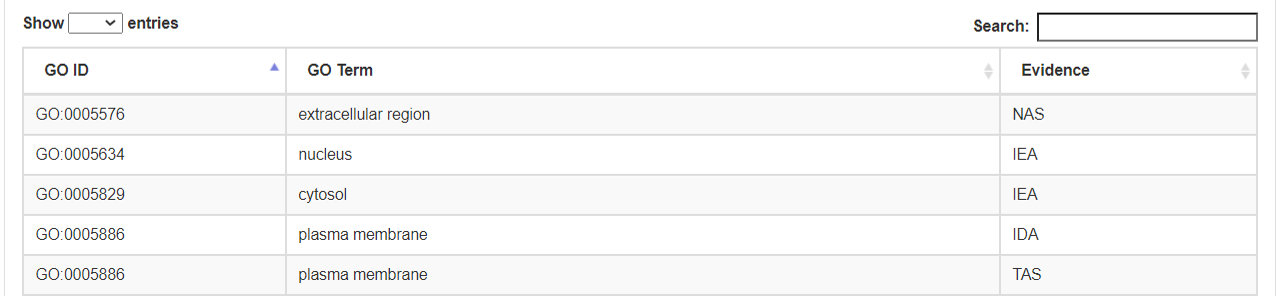
(8) Gene Ontology (GO)/biological pathways
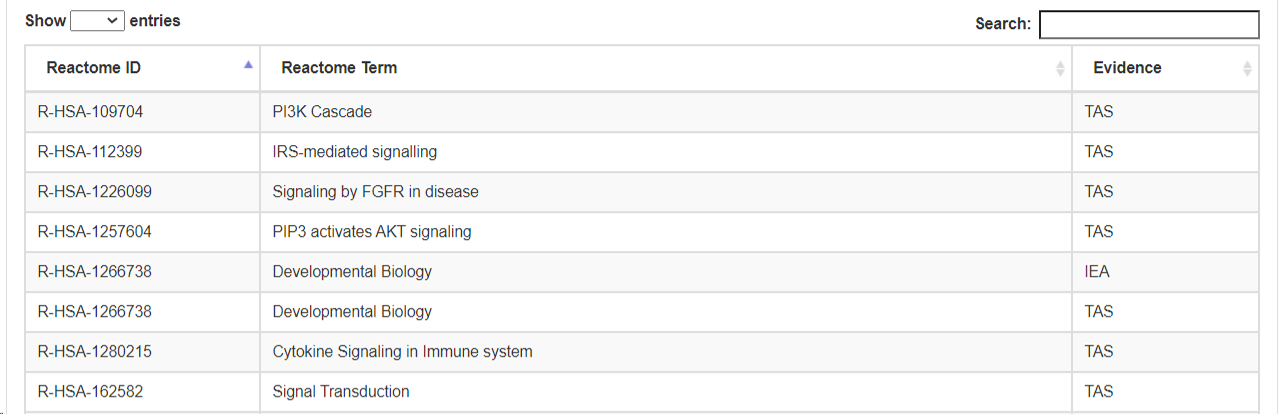
- Gene Ontology (GO) /biological pathways information were collected and integrated from the Gene Ontology Resource and Reactome Pathway Database.
- a. Molecular Function.
- b. Biological Process.
- c. Cellular Component.
- d. Reactome pathway.
-
(9) Functional annotations
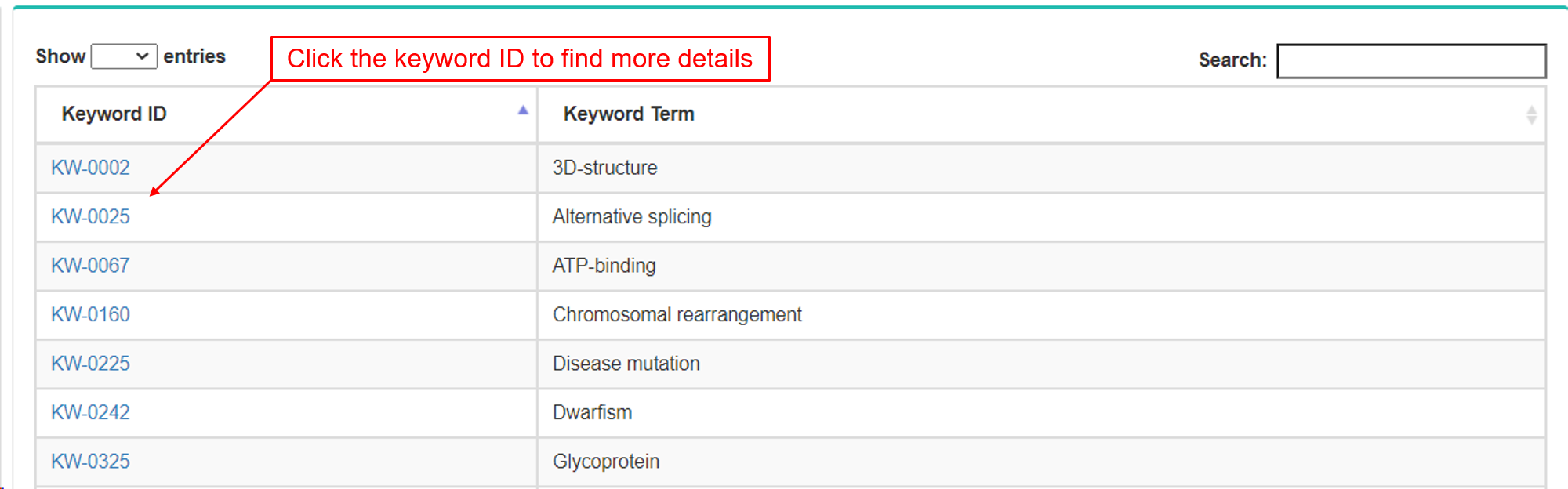
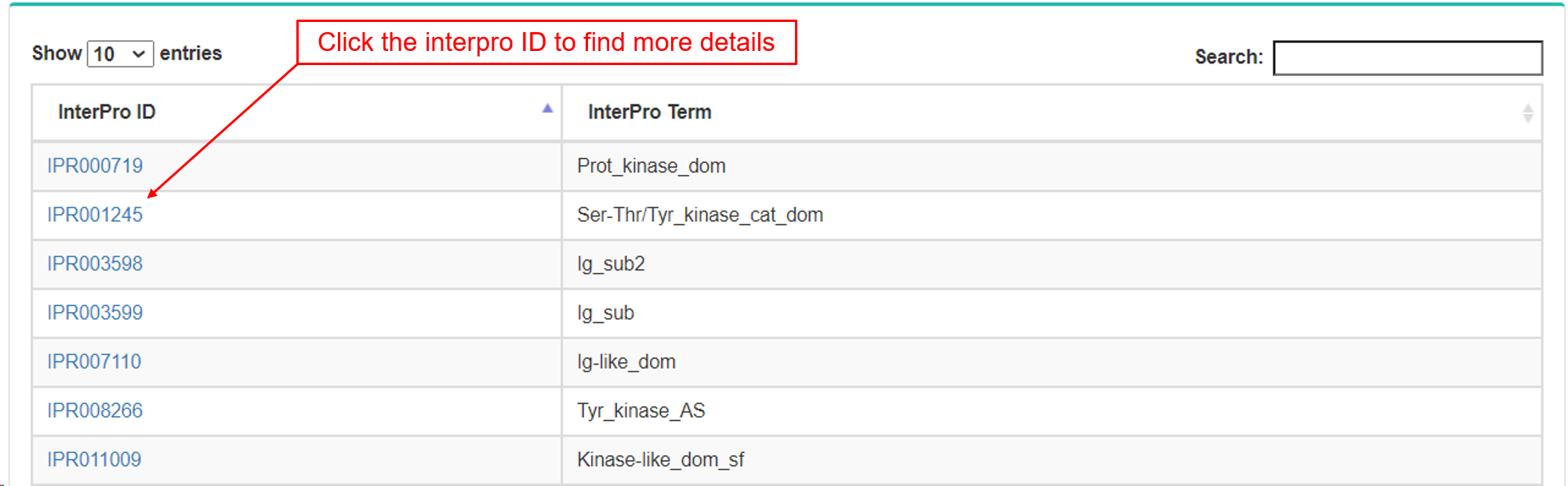
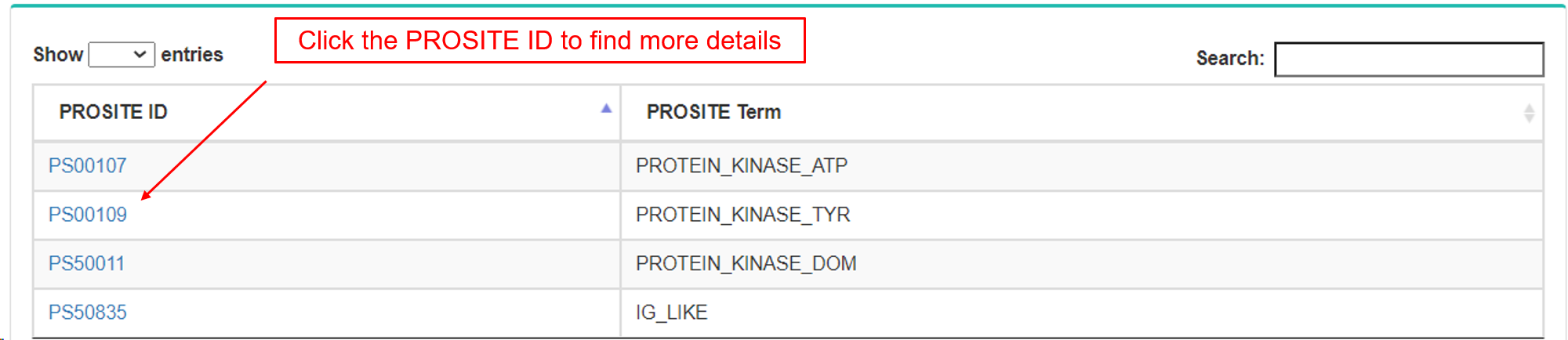
- Functional annotations were collected and integrated from the keyword (UniProt), Interpro, PROSITE and Pfam databases.
- a. Keywords.
- b. InterPro.
- c. PROSITE.
- d. Pfam.
-
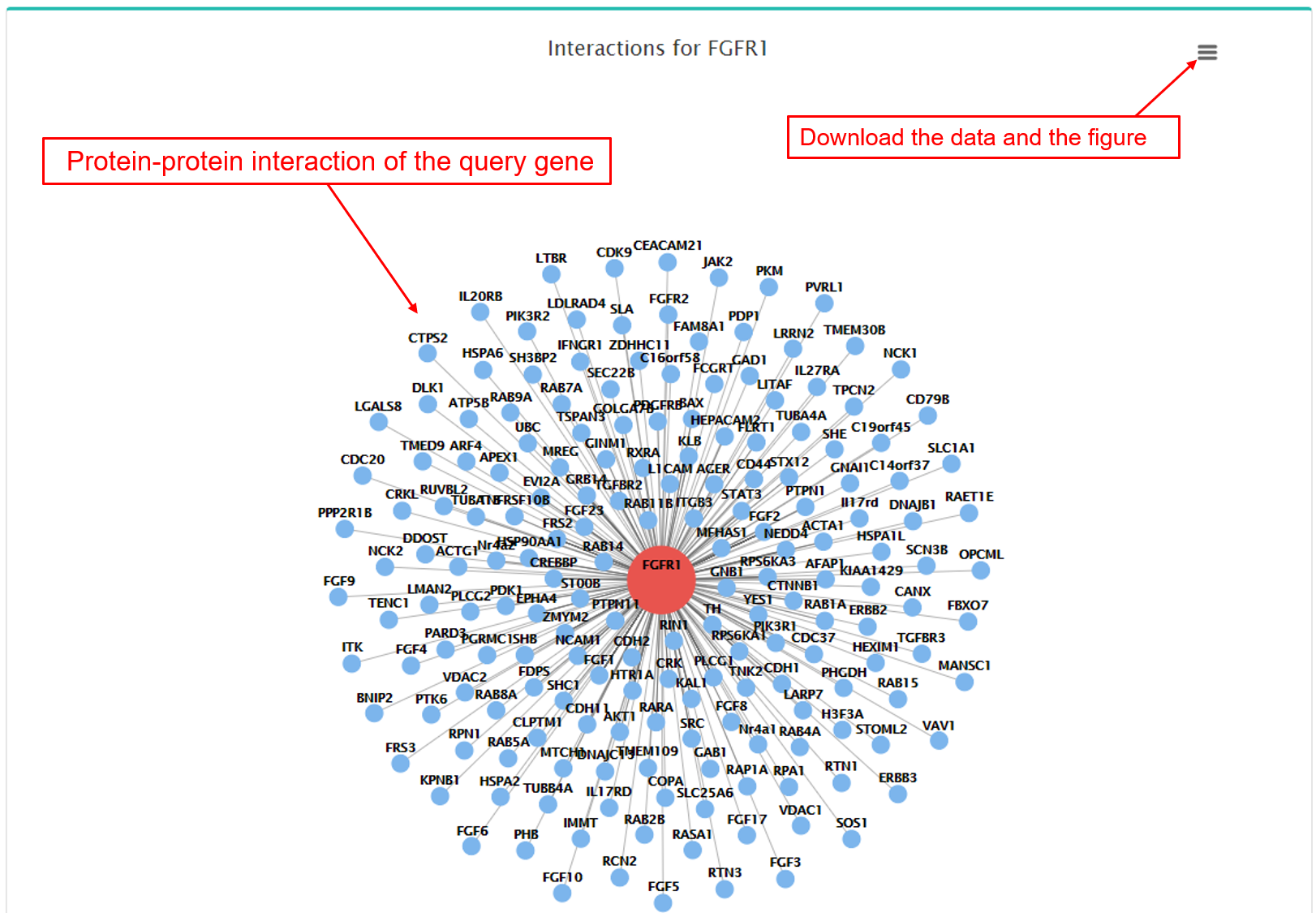
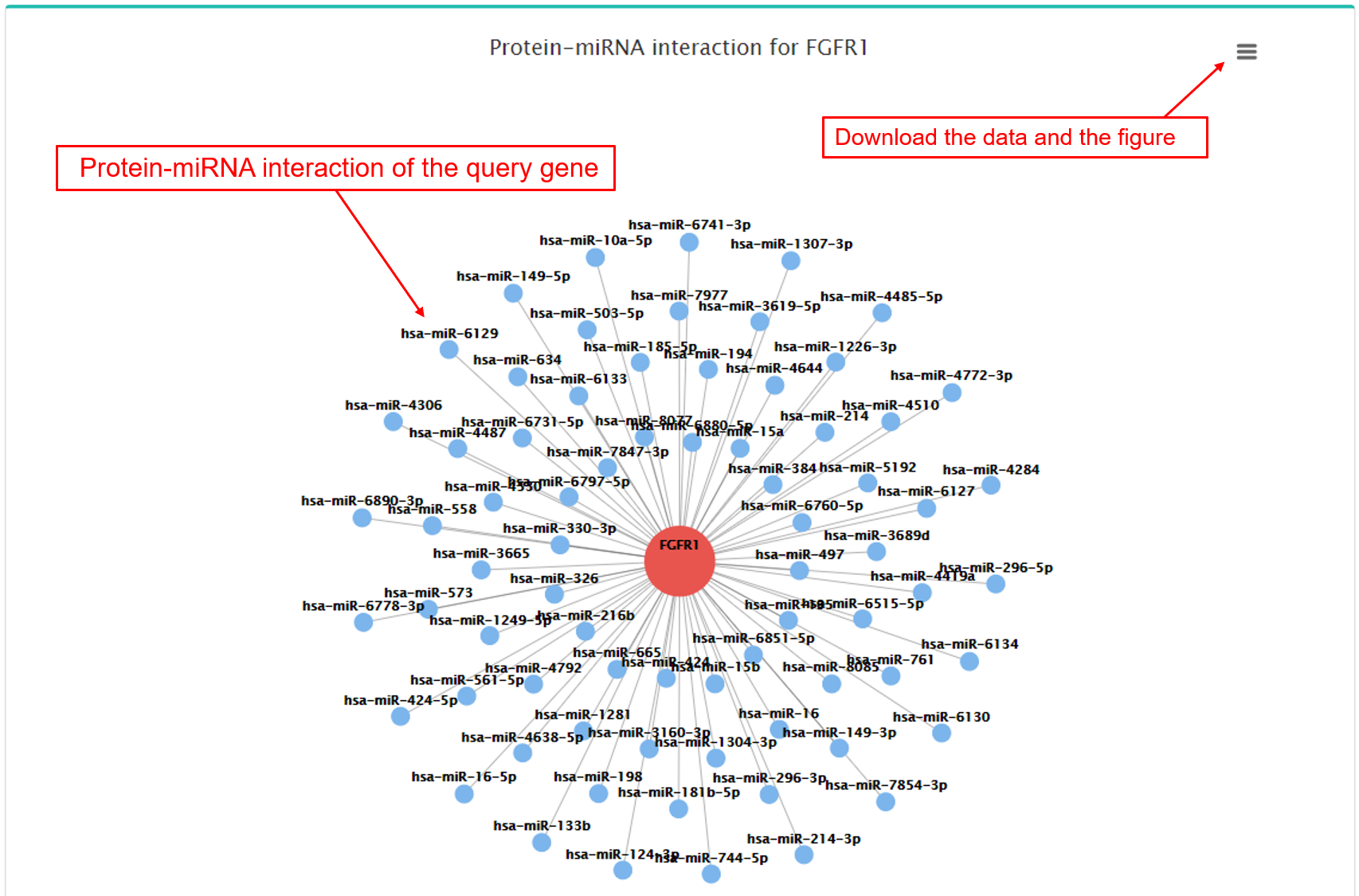
(10) Protein-protein interactions and protein-miRNA interactions
- a. Protein-protein interactions.
- b. Protein-miRNA interactions.
6. Download:
All cleft genes with rich annotations are publicly available for downloading at https://bioinfo.uth.edu/CleftGeneDB/Download.php
7. Citation:
Please cite: Xu, Haodong; Yan, Fangfang; Hu, Ruifeng; Suzuki, Akiko; Iwaya, Chihiro; Jia, Peilin; Iwata, Junichi; Zhao, Zhongming. CleftGeneDB: a resource for annotating genes associated with cleft lip and cleft palate. Science Bulletin, 2021, 66(23): 2340-2342. Web site: https://bioinfo.uth.edu/CleftGeneDB.
8. References:
Previous publications on systematic review and analyses of cleft lip and cleft palate studies in humans and mice.
- Suzuki A, Abdallah N, Gajera M, Jia P, Jun G, Zhao Z, Iwata J (2018) Genes and microRNAs associated with mouse cleft palate: A systematic review and bioinformatics analysis. Mechanisms of Development 150 (2018): 21-27. [Full Text]
- Gajera M, Desai N, Suzuki A, Li A, Zhang M, Jun G, Jia P, Zhao Z, Iwata J (2019) MicroRNA-655-3p and microRNA-497-5p inhibit cell proliferation in cultured human lip cells through the regulation of genes related to human cleft lip. BMC Medical Genomics 12(1):70. [Full Text]
- Suzuki A, Li A, Gajera M, Abdallah N, Pelikan RC, Zhao Z, Iwata J (2019) MicroRNA-374a, -4680, and -133b suppress cell proliferation through the regulation of genes associated with human cleft palate in cultured human palate cells. BMC Medical Genomics 12:93. [Full Text]
- Suzuki A, Yoshioka H, Summakia D, Desai N, Jun G, Jia P, Loose D, Ogata K, Gajera M, Zhao Z, Iwata J (2019) MicroRNA-124-3p suppresses mouse lip mesenchymal cell proliferation through the regulation of genes associated with cleft lip in the mouse. BMC Genomics 20(1):852. [Full Text]
- Li A, Qin G, Suzuki A, Gejera M, Iwata J, Jia P, Zhao Z* (2019) Network-based identification of critical regulators as putative drivers of human cleft lip. BMC Medical Genomics 12(Suppl 1): 16. [Full Text]
- Yan F, Dai Y, Iwata J, Zhao Z*, Jia P* (2020) An integrative, genomic, transcriptomic and network-assisted study to identify genes associated with human cleft lip with or without cleft palate. BMC Medical Genomics. 13(suppl 5):39. [Full Text]
- Li A, Jia P, Mallik S, Fei R, Yoshioka H, Suzuki A, Iwata J, Zhao Z* (2019) Critical microRNAs and regulatory motifs in cleft palate identified by a conserved miRNA-TF-gene network approach in humans and mice. Briefing in Bioinformatics, bbz082. [Full Text]