| BRCA (tumor) | BRCA (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| COAD (tumor) | COAD (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| KICH (tumor) | KICH (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| KIRC (tumor) | KIRC (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| KIRP (tumor) | KIRP (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| LUAD (tumor) | LUAD (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| LUSC (tumor) | LUSC (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| PRAD (tumor) | PRAD (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| STAD (tumor) | STAD (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| THCA (tumor) | THCA (normal) |
| ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (tumor) | ABCC2, HNF4A, ONECUT1, NFKB1, PDZK1, SLC9A3R1, RDX, UBQLN1, AZIN1, HNF1A, CANX, PDZD3, MCPH1, IRF3, SHANK3 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0022350 | Jaundice, Chronic Idiopathic | 32 | BeFree,CLINVAR,CTD_human,LHGDN,ORPHANET,RGD,UNIPROT |
| umls:C0014544 | Epilepsy | 17 | BeFree,GAD |
| umls:C0008370 | Cholestasis | 11 | BeFree,CTD_human,RGD |
| umls:C0268307 | Conjugated hyperbilirubinemia | 11 | BeFree |
| umls:C0020433 | Hyperbilirubinemia | 10 | BeFree,CTD_human,RGD |
| umls:C0029925 | Ovarian Carcinoma | 9 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 8 | BeFree,GAD |
| umls:C0019693 | HIV Infections | 8 | BeFree,GAD |
| umls:C0677886 | Epithelial ovarian cancer | 8 | BeFree |
| umls:C2239176 | Liver carcinoma | 8 | BeFree |
| umls:C1527249 | Colorectal Cancer | 7 | BeFree,GAD |
| umls:C0242379 | Malignant neoplasm of lung | 6 | BeFree,GAD |
| umls:C0678222 | Breast Carcinoma | 6 | BeFree |
| umls:C0009402 | Colorectal Carcinoma | 5 | BeFree |
| umls:C0023530 | Leukopenia | 5 | BeFree,GAD |
| umls:C0027947 | Neutropenia | 5 | BeFree,GAD,LHGDN |
| umls:C0001418 | Adenocarcinoma | 4 | BeFree,GAD,LHGDN |
| umls:C0007131 | Non-Small Cell Lung Carcinoma | 4 | BeFree |
| umls:C0011991 | Diarrhea | 4 | BeFree,GAD |
| umls:C0023452 | Leukemia, Lymphocytic, Acute, L1 | 4 | BeFree |
| umls:C0268318 | Cholestasis of pregnancy | 4 | BeFree |
| umls:C0007134 | Renal Cell Carcinoma | 3 | BeFree |
| umls:C0009404 | Colorectal Neoplasms | 3 | CTD_human,GAD,LHGDN |
| umls:C0019158 | Hepatitis | 3 | BeFree |
| umls:C0019159 | Hepatitis A | 3 | BeFree |
| umls:C0041755 | Adverse reaction to drug | 3 | CTD_human |
| umls:C1140680 | Malignant neoplasm of ovary | 3 | BeFree,GAD |
| umls:C0003873 | Rheumatoid Arthritis | 2 | CTD_human,GAD |
| umls:C0008312 | Primary biliary cirrhosis | 2 | BeFree |
| umls:C0008372 | Intrahepatic Cholestasis | 2 | GAD,LHGDN |
| umls:C0009375 | Colonic Neoplasms | 2 | GAD,LHGDN |
| umls:C0011853 | Diabetes Mellitus, Experimental | 2 | RGD |
| umls:C0014547 | Epilepsies, Partial | 2 | BeFree |
| umls:C0015695 | Fatty Liver | 2 | GAD,RGD |
| umls:C0023895 | Liver diseases | 2 | BeFree,LHGDN |
| umls:C0025202 | melanoma | 2 | BeFree |
| umls:C0030297 | Pancreatic Neoplasm | 2 | GAD,LHGDN |
| umls:C0031117 | Peripheral Neuropathy | 2 | CTD_human,GAD |
| umls:C0032962 | Pregnancy Complications | 2 | GAD |
| umls:C0036572 | Seizures | 2 | BeFree,CTD_human |
| umls:C0079731 | B-Cell Lymphomas | 2 | BeFree |
| umls:C0149925 | Small cell carcinoma of lung | 2 | BeFree |
| umls:C0206698 | Cholangiocarcinoma | 2 | BeFree,GAD |
| umls:C0235974 | Pancreatic carcinoma | 2 | BeFree |
| umls:C0279626 | Squamous cell carcinoma of esophagus | 2 | BeFree,CTD_human |
| umls:C0346647 | Malignant neoplasm of pancreas | 2 | BeFree |
| umls:C0684249 | Carcinoma of lung | 2 | BeFree |
| umls:C0948380 | Colorectal cancer metastatic | 2 | BeFree |
| umls:C1168401 | Squamous cell carcinoma of the head and neck | 2 | BeFree,CTD_human |
| umls:C1378703 | Renal carcinoma | 2 | BeFree |
| umls:C0001430 | Adenoma | 1 | BeFree |
| umls:C0002170 | Alopecia | 1 | CTD_human |
| umls:C0005396 | Bile Duct Neoplasms | 1 | GAD |
| umls:C0005398 | Cholestasis, Extrahepatic | 1 | BeFree |
| umls:C0005684 | Malignant neoplasm of urinary bladder | 1 | GAD |
| umls:C0007137 | Squamous cell carcinoma | 1 | BeFree |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | RGD |
| umls:C0013182 | Drug Allergy | 1 | CTD_human |
| umls:C0013221 | Drug toxicity | 1 | GAD |
| umls:C0014548 | Epilepsy, Generalized | 1 | BeFree |
| umls:C0016057 | Fibrosarcoma | 1 | BeFree |
| umls:C0017178 | Gastrointestinal Diseases | 1 | CTD_human |
| umls:C0018799 | Heart Diseases | 1 | CTD_human |
| umls:C0019193 | Hepatitis, Toxic | 1 | GAD |
| umls:C0019196 | Hepatitis C | 1 | BeFree |
| umls:C0022354 | Jaundice, Obstructive | 1 | BeFree |
| umls:C0022658 | Kidney Diseases | 1 | GAD |
| umls:C0022661 | Kidney Failure, Chronic | 1 | RGD |
| umls:C0023467 | Leukemia, Myelocytic, Acute | 1 | BeFree |
| umls:C0023473 | Myeloid Leukemia, Chronic | 1 | BeFree |
| umls:C0023474 | Leukemia, Myeloid, Chronic-Phase | 1 | GAD |
| umls:C0023892 | Biliary cirrhosis | 1 | CTD_human |
| umls:C0023903 | Liver neoplasms | 1 | BeFree |
| umls:C0024117 | Chronic Obstructive Airway Disease | 1 | GAD |
| umls:C0024121 | Lung Neoplasms | 1 | LHGDN |
| umls:C0024141 | Lupus Erythematosus, Systemic | 1 | LHGDN |
| umls:C0024299 | Lymphoma | 1 | BeFree |
| umls:C0025500 | Mesothelioma | 1 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 1 | BeFree |
| umls:C0027765 | nervous system disorder | 1 | CTD_human |
| umls:C0028754 | Obesity | 1 | BeFree |
| umls:C0028860 | Oculocerebrorenal Syndrome | 1 | BeFree |
| umls:C0030286 | Pancreatic Diseases | 1 | BeFree |
| umls:C0031048 | Pericarditis, Constrictive | 1 | BeFree |
| umls:C0033774 | Pruritus | 1 | GAD |
| umls:C0035078 | Kidney Failure | 1 | GAD |
| umls:C0036117 | Salmonella infections | 1 | BeFree |
| umls:C0038220 | Status Epilepticus | 1 | CTD_human,RGD |
| umls:C0042769 | Virus Diseases | 1 | BeFree |
| umls:C0080178 | Spina Bifida | 1 | BeFree,LHGDN |
| umls:C0086692 | Benign Neoplasm | 1 | BeFree |
| umls:C0149521 | Pancreatitis, Chronic | 1 | BeFree |
| umls:C0149958 | Complex partial seizures | 1 | BeFree |
| umls:C0151747 | Renal tubular disorder | 1 | BeFree |
| umls:C0153467 | Malignant tumor of peritoneum | 1 | BeFree |
| umls:C0206624 | Hepatoblastoma | 1 | BeFree |
| umls:C0206681 | Adenocarcinoma, Clear Cell | 1 | BeFree |
| umls:C0280220 | stage, ovarian epithelial cancer | 1 | BeFree |
| umls:C0345967 | Malignant mesothelioma | 1 | BeFree |
| umls:C0376544 | Hematopoietic Neoplasms | 1 | BeFree |
| umls:C0376618 | Endotoxemia | 1 | RGD |
| umls:C0400966 | Non-alcoholic Fatty Liver Disease | 1 | BeFree |
| umls:C0400979 | Obstruction of biliary tree | 1 | BeFree |
| umls:C0524910 | Hepatitis C, Chronic | 1 | BeFree |
| umls:C0596263 | Carcinogenesis | 1 | BeFree |
| umls:C0699791 | Stomach Carcinoma | 1 | BeFree |
| umls:C0740457 | Malignant neoplasm of kidney | 1 | BeFree |
| umls:C0860207 | Drug-Induced Liver Injury | 1 | CTD_human |
| umls:C0877382 | Delayed renal graft function | 1 | GAD |
| umls:C0948168 | Bone marrow toxicity | 1 | BeFree |
| umls:C1096063 | Drug Resistant Epilepsy | 1 | BeFree |
| umls:C1112160 | Gastrooesophageal cancer | 1 | BeFree |
| umls:C1302401 | Adenoma of large intestine | 1 | BeFree |
| umls:C1443924 | Severe diarrhea | 1 | BeFree |
| umls:C1458155 | Mammary Neoplasms | 1 | LHGDN |
| umls:C2931822 | Nasopharyngeal carcinoma | 1 | BeFree |
| umls:C2931852 | Clear-cell metastatic renal cell carcinoma | 1 | BeFree |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))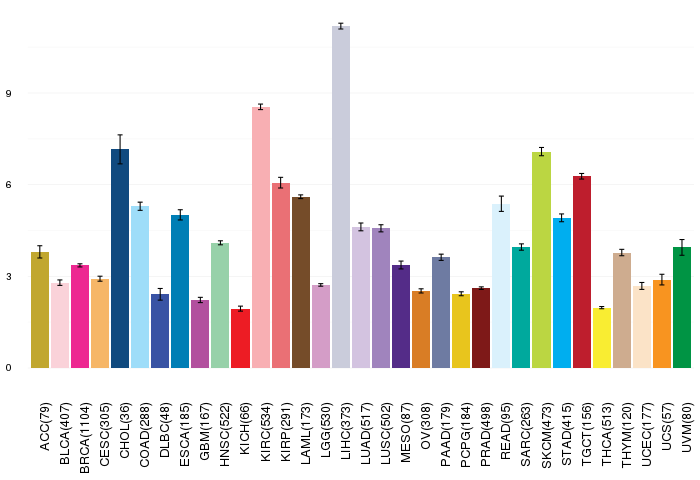
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))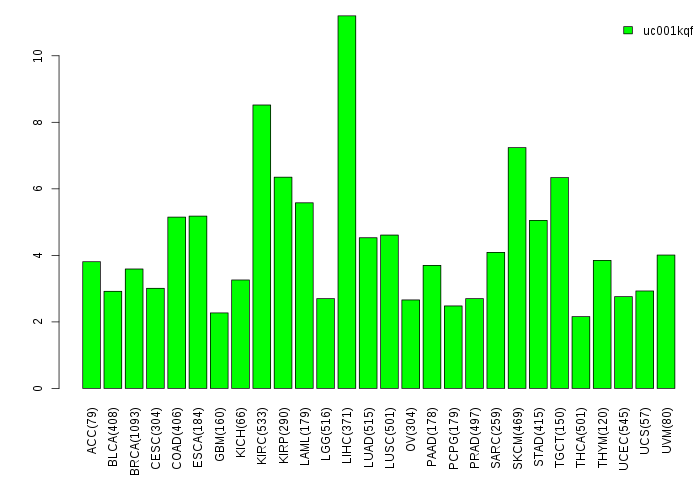
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data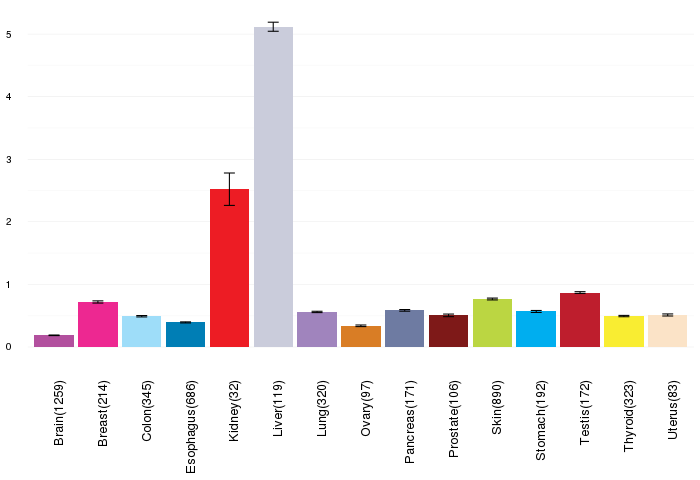
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))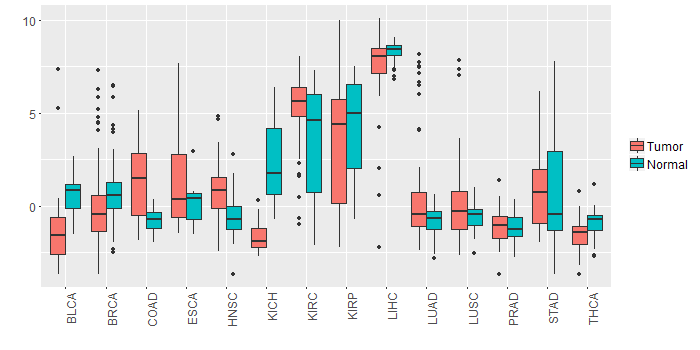
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 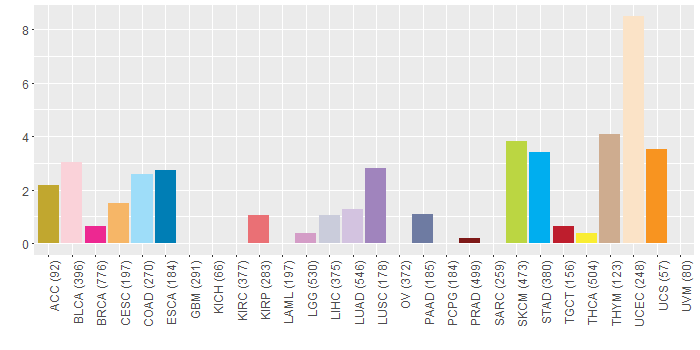
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)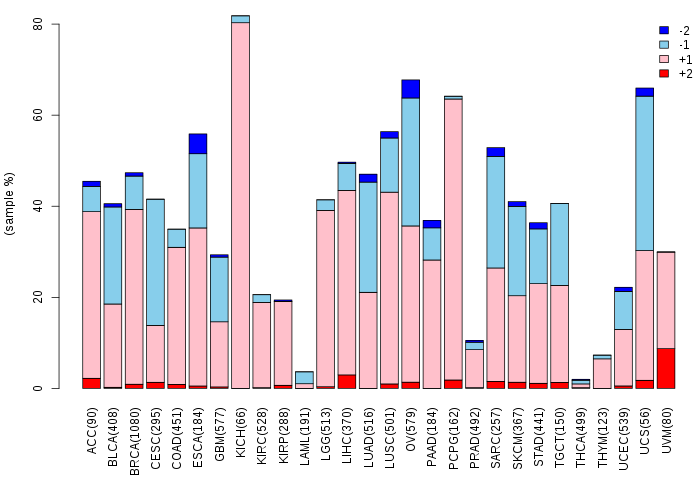
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























