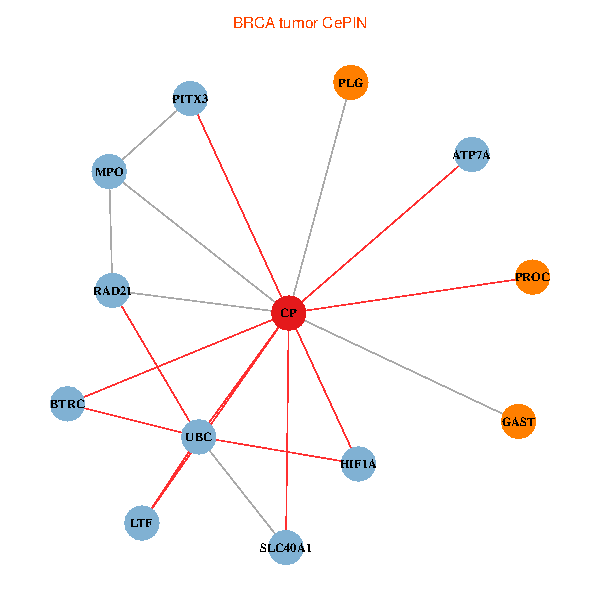
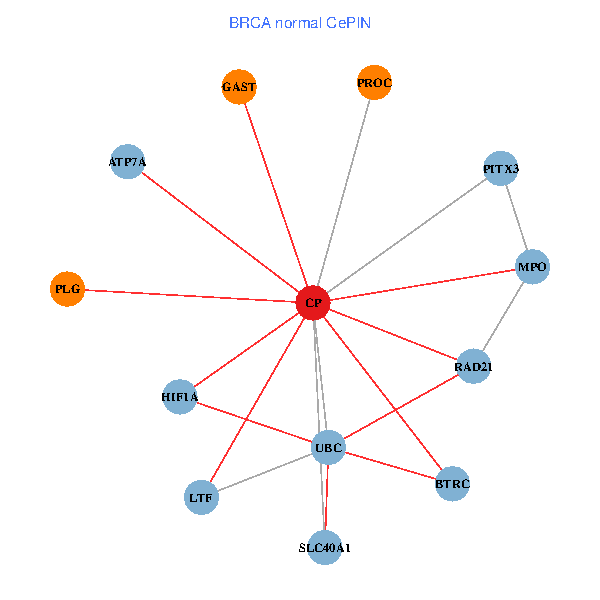
| BRCA (tumor) | BRCA (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
 |  |
|
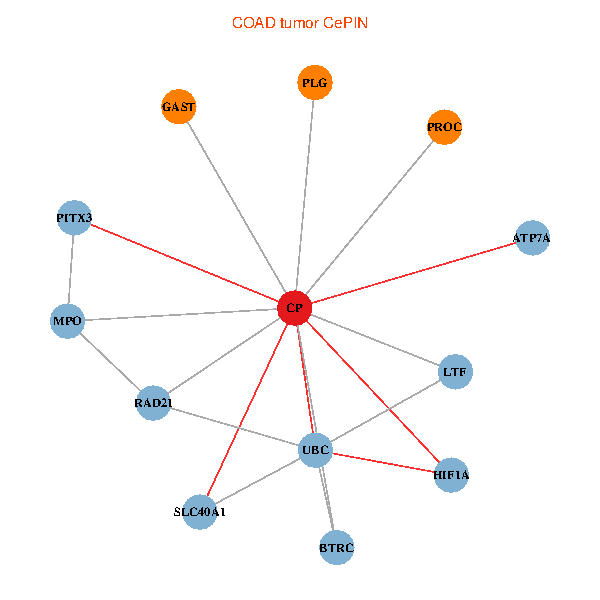
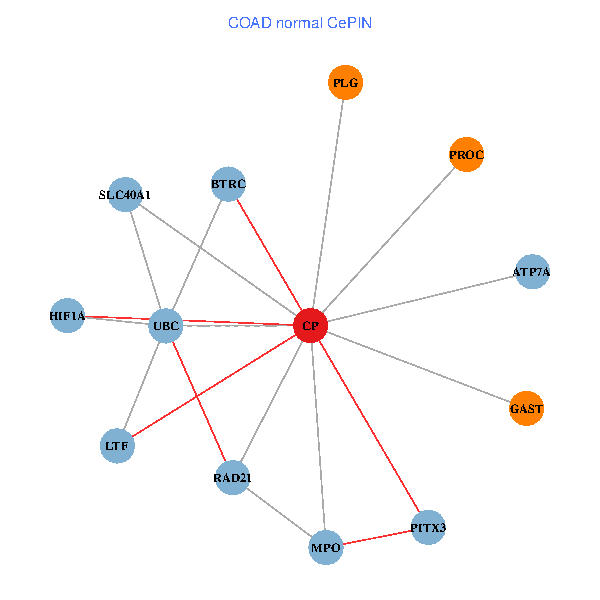
| COAD (tumor) | COAD (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
 |  |
|
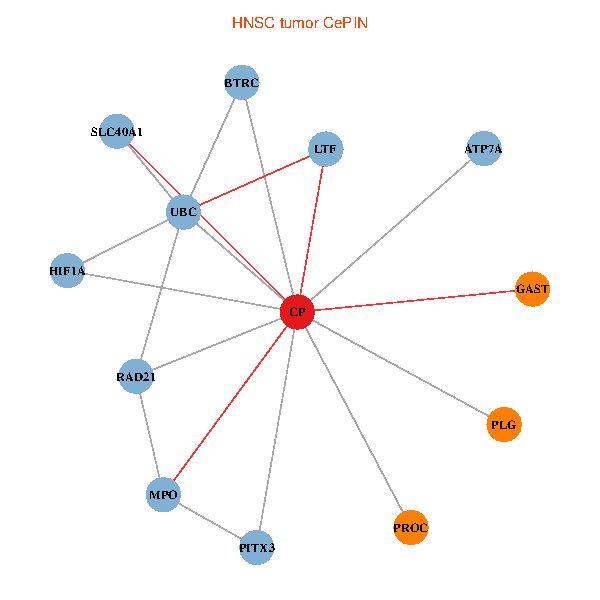
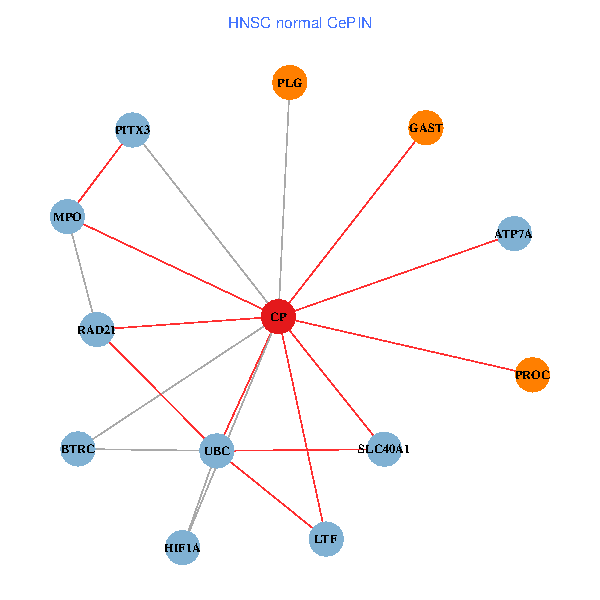
| HNSC (tumor) | HNSC (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
 |  |
|
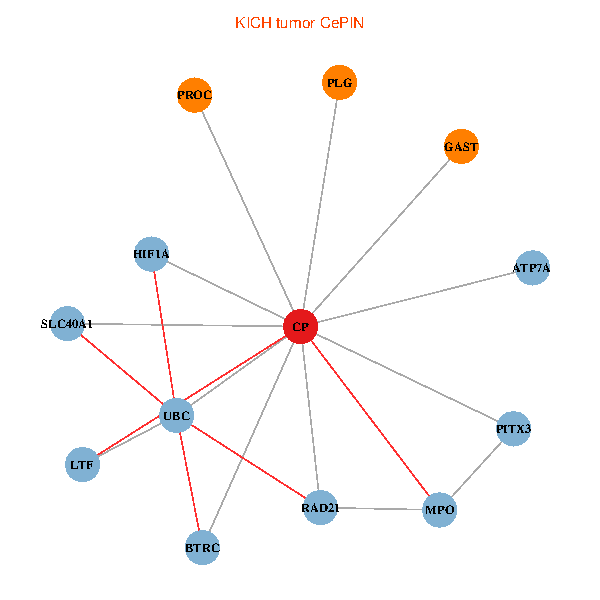
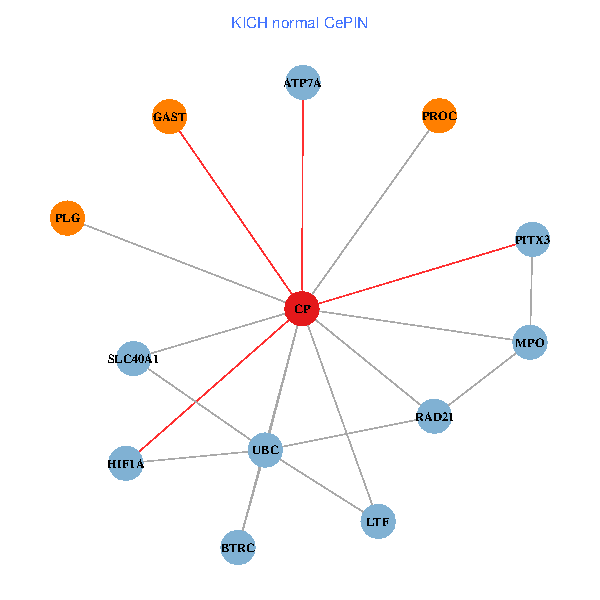
| KICH (tumor) | KICH (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
 |  |
|
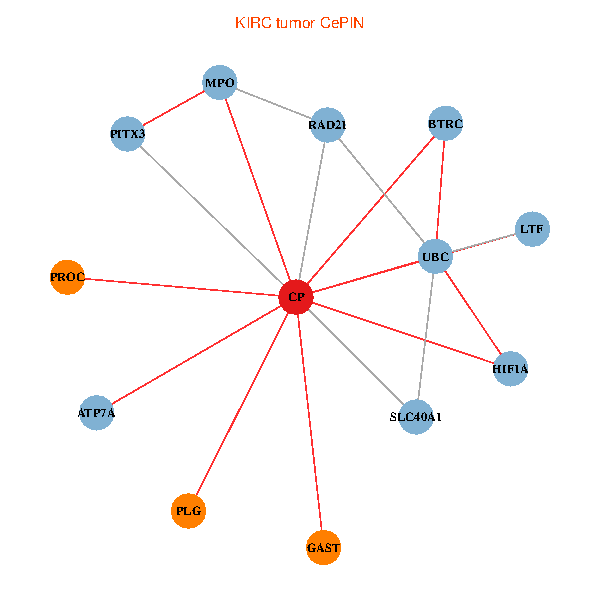
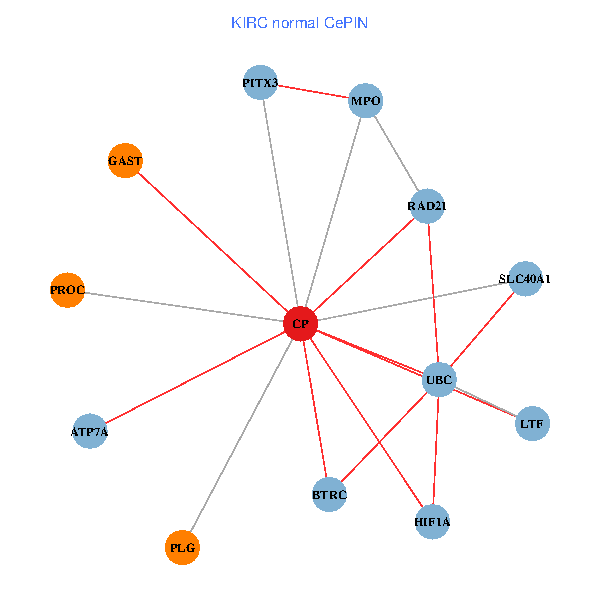
| KIRC (tumor) | KIRC (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
 |  |
|
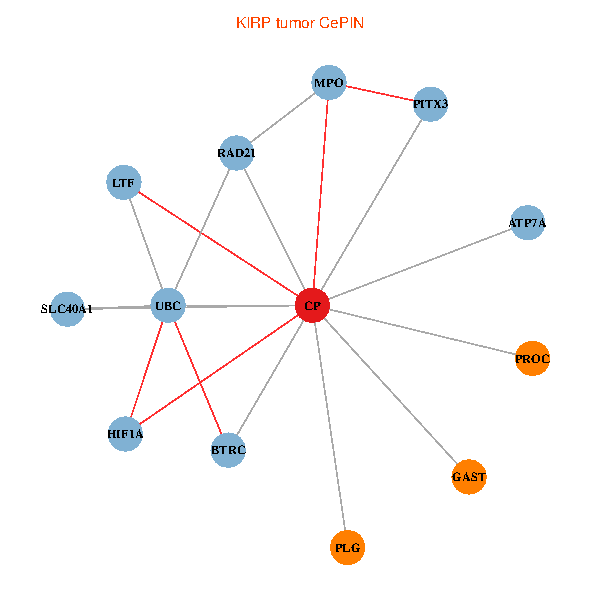
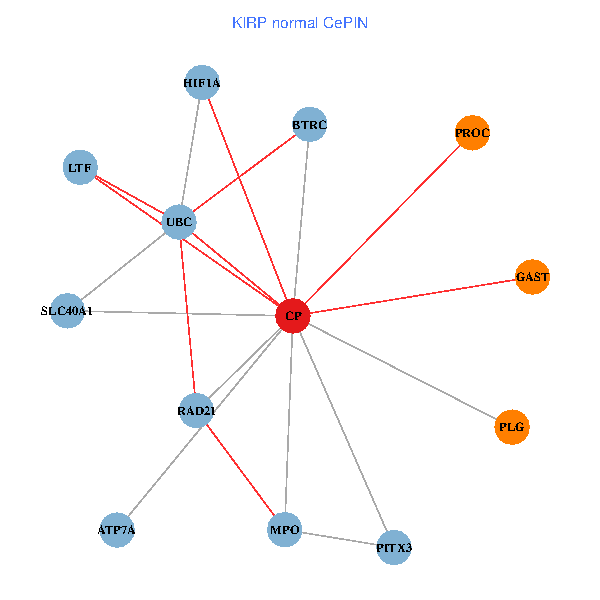
| KIRP (tumor) | KIRP (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
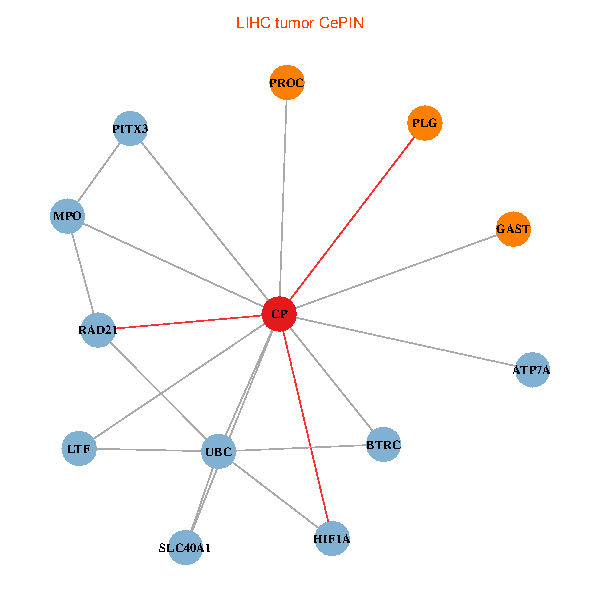 | 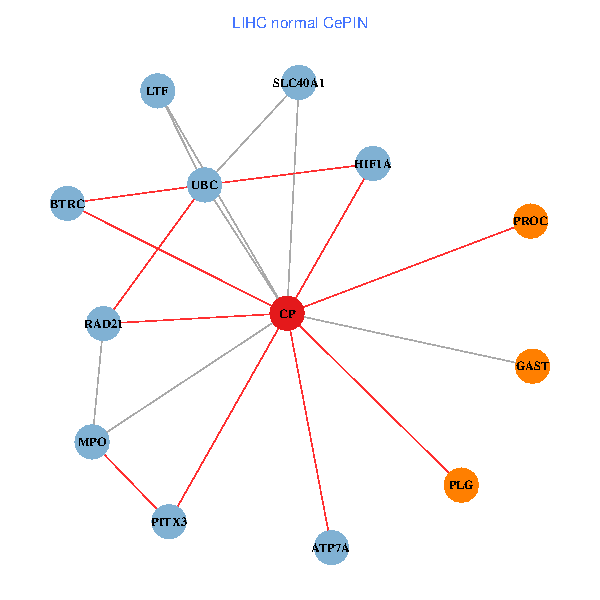 |
|
| LUAD (tumor) | LUAD (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
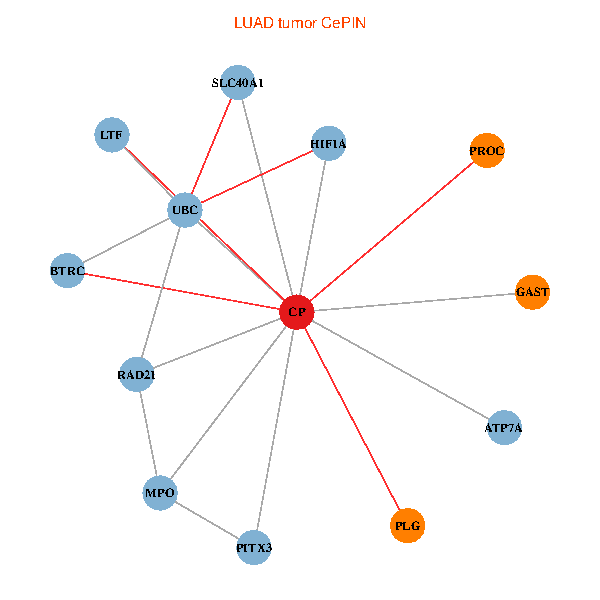 | 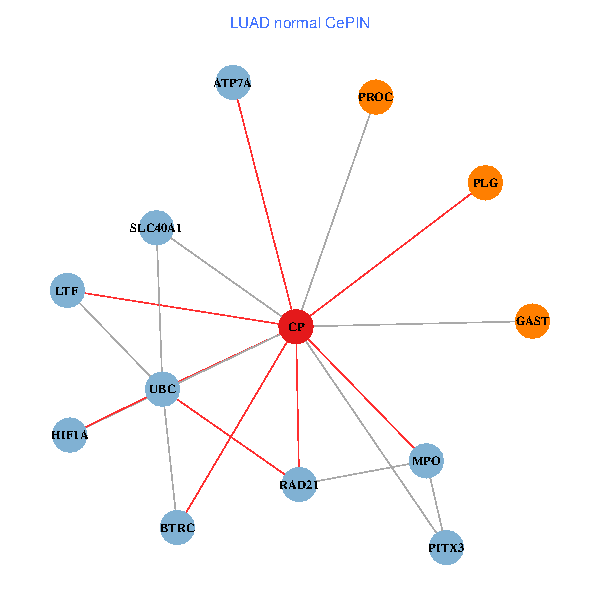 |
|
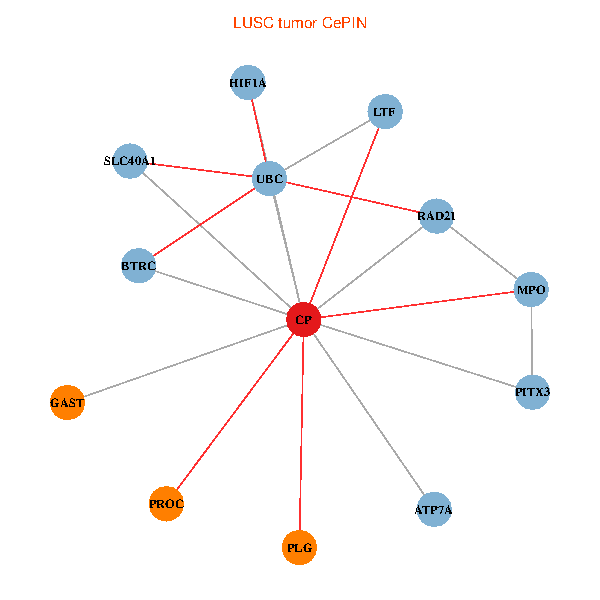
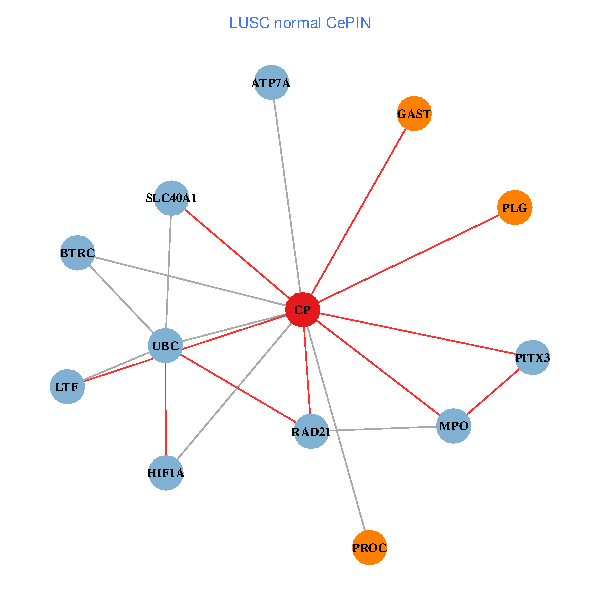
| LUSC (tumor) | LUSC (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
 |  |
|
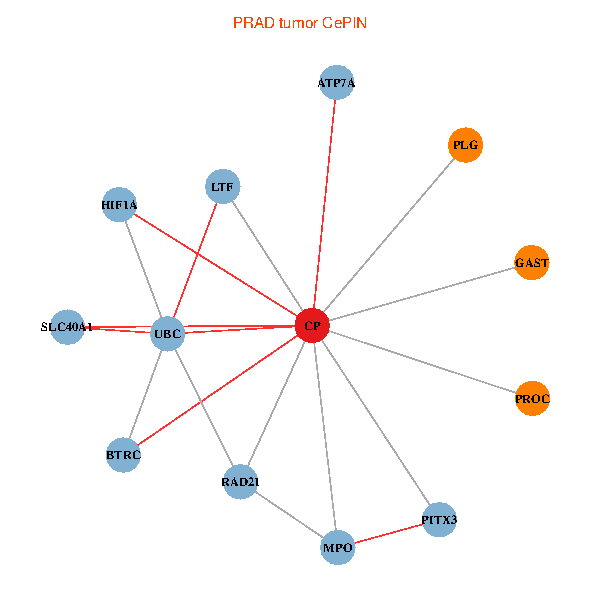
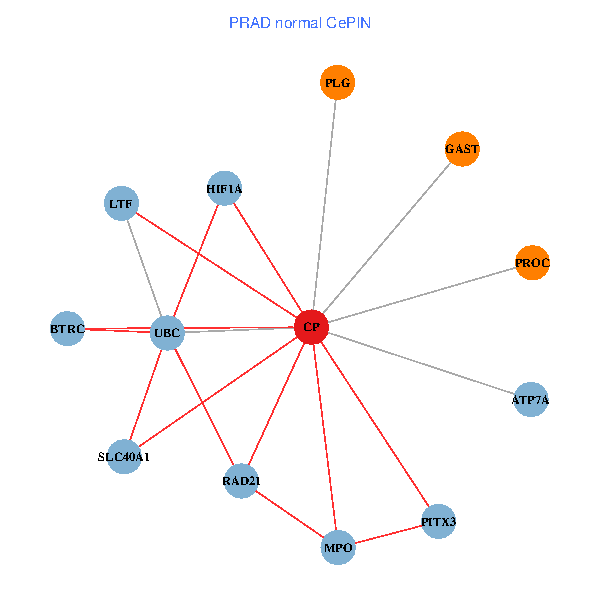
| PRAD (tumor) | PRAD (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
 |  |
|
| STAD (tumor) | STAD (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
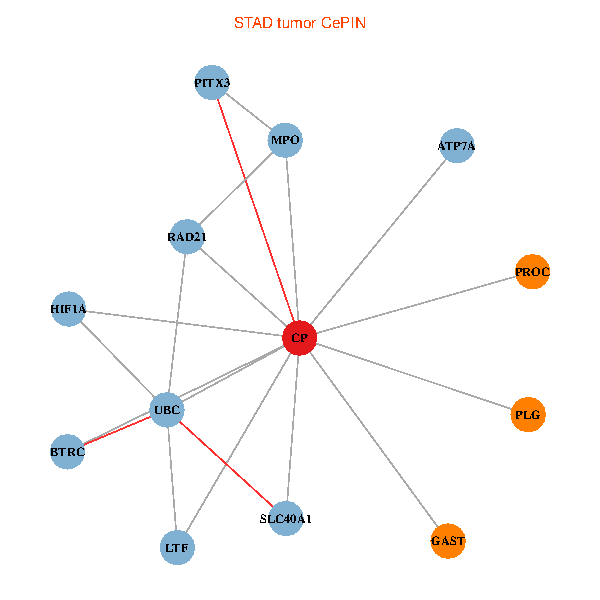 | 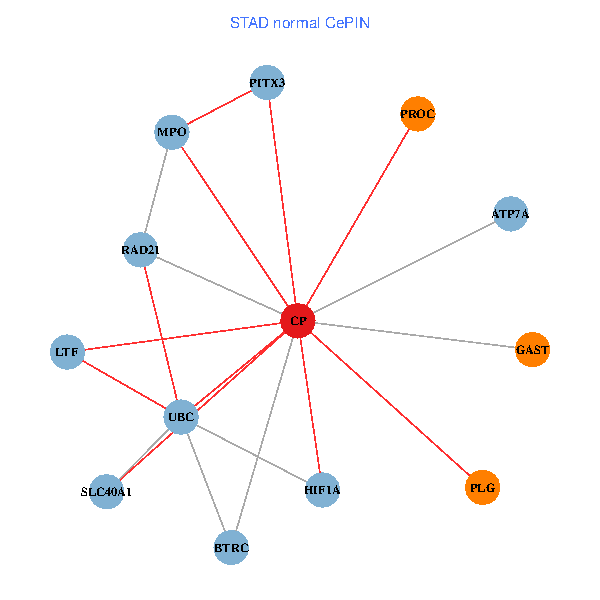 |
|
| THCA (tumor) | THCA (normal) |
| CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (tumor) | CP, UBC, HIF1A, BTRC, PITX3, PLG, RAD21, LTF, MPO, SLC40A1, PROC, ATP7A, GAST (normal) |
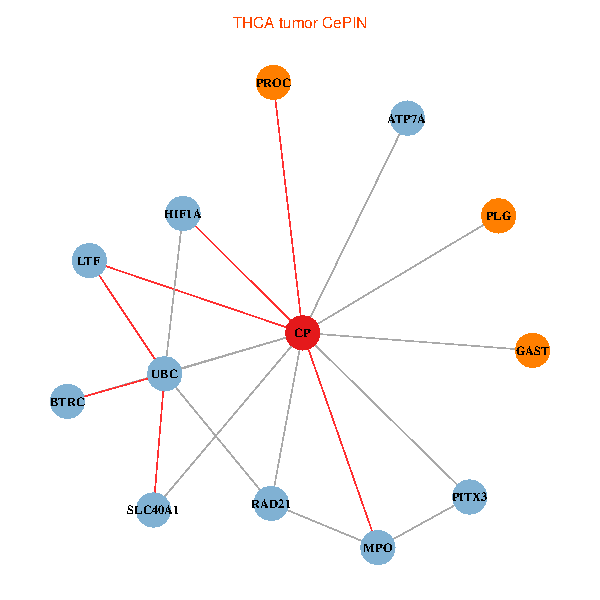 | 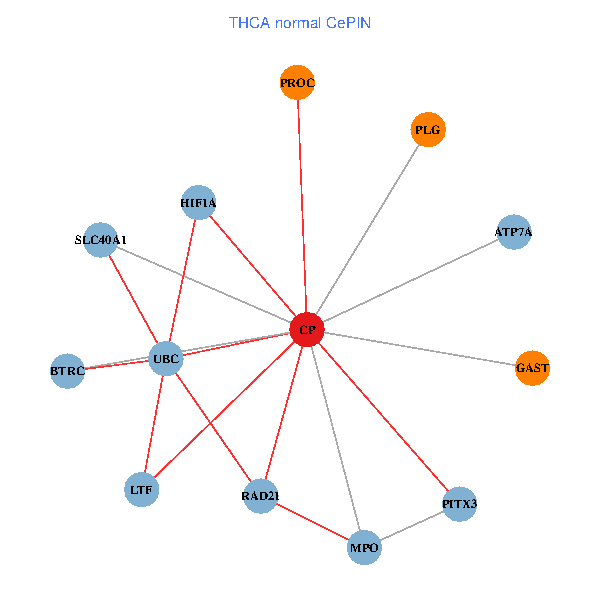 |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0878682 | Ceruloplasmin deficiency | 28 | BeFree,CLINVAR,MGD,ORPHANET |
| umls:C0019202 | Hepatolenticular Degeneration | 26 | BeFree,CTD_human |
| umls:C0282193 | Iron Overload | 12 | BeFree,CTD_human |
| umls:C0012715 | Iron Metabolism Disorders | 8 | BeFree,CTD_human,RGD |
| umls:C0152457 | Kayser-Fleischer ring | 8 | BeFree |
| umls:C0524851 | Neurodegenerative Disorders | 8 | BeFree |
| umls:C0030567 | Parkinson Disease | 7 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0002395 | Alzheimer's Disease | 6 | BeFree,GAD,LHGDN |
| umls:C0011849 | Diabetes Mellitus | 6 | BeFree,CTD_human,LHGDN |
| umls:C0018995 | Hemochromatosis | 5 | BeFree,CTD_human,LHGDN |
| umls:C0235031 | Neurologic Symptoms | 5 | BeFree |
| umls:C0268070 | Hypocupremia | 5 | BeFree |
| umls:C0002871 | Anemia | 4 | BeFree |
| umls:C0022716 | Menkes Kinky Hair Syndrome | 4 | BeFree,CTD_human |
| umls:C0023895 | Liver diseases | 4 | BeFree,LHGDN |
| umls:C0002878 | Anemia, Hemolytic | 3 | BeFree |
| umls:C0003873 | Rheumatoid Arthritis | 3 | BeFree,CTD_human,LHGDN |
| umls:C0007222 | Cardiovascular Diseases | 3 | BeFree,LHGDN |
| umls:C0011854 | Diabetes Mellitus, Insulin-Dependent | 3 | BeFree,CTD_human,GAD |
| umls:C0017638 | Glioma | 3 | BeFree,LHGDN |
| umls:C0022116 | Ischemia | 3 | BeFree,CTD_human |
| umls:C0035304 | Retinal Degeneration | 3 | BeFree,CTD_human |
| umls:C0242383 | Age related macular degeneration | 3 | BeFree |
| umls:C2931082 | Familial apoceruloplasmin deficiency | 3 | CTD_human,ORPHANET |
| umls:C0002736 | Amyotrophic Lateral Sclerosis | 2 | GAD,LHGDN |
| umls:C0004352 | Autistic Disorder | 2 | BeFree,CTD_human |
| umls:C0009375 | Colonic Neoplasms | 2 | CTD_human,LHGDN |
| umls:C0011847 | Diabetes | 2 | BeFree |
| umls:C0013362 | Dysarthria | 2 | BeFree |
| umls:C0015230 | Exanthema | 2 | BeFree |
| umls:C0017601 | Glaucoma | 2 | BeFree |
| umls:C0017661 | IGA Glomerulonephritis | 2 | BeFree,CTD_human |
| umls:C0019114 | Hemosiderosis | 2 | BeFree |
| umls:C0027765 | nervous system disorder | 2 | BeFree |
| umls:C0040822 | Tremor | 2 | BeFree,LHGDN |
| umls:C0220756 | Niemann-Pick Disease, Type C | 2 | BeFree |
| umls:C0238339 | Hereditary pancreatitis | 2 | BeFree |
| umls:C0392514 | Hereditary hemochromatosis | 2 | BeFree |
| umls:C0848332 | Spots on skin | 2 | BeFree |
| umls:C0993582 | Arthritis, Experimental | 2 | CTD_human,RGD |
| umls:C2239176 | Liver carcinoma | 2 | BeFree,CTD_human |
| umls:C0000786 | Spontaneous abortion | 1 | GAD |
| umls:C0001925 | Albuminuria | 1 | CTD_human |
| umls:C0002875 | Cooley's anemia | 1 | BeFree |
| umls:C0002965 | Angina, Unstable | 1 | LHGDN |
| umls:C0003130 | Anoxia | 1 | GAD |
| umls:C0003850 | Arteriosclerosis | 1 | BeFree |
| umls:C0004134 | Ataxia | 1 | CTD_human |
| umls:C0004153 | Atherosclerosis | 1 | BeFree |
| umls:C0005747 | Blepharospasm | 1 | BeFree |
| umls:C0006111 | Brain Diseases | 1 | CTD_human |
| umls:C0007137 | Squamous cell carcinoma | 1 | BeFree |
| umls:C0007758 | Cerebellar Ataxia | 1 | BeFree |
| umls:C0007786 | Brain Ischemia | 1 | RGD |
| umls:C0011570 | Mental Depression | 1 | GAD |
| umls:C0011853 | Diabetes Mellitus, Experimental | 1 | RGD |
| umls:C0013080 | Down Syndrome | 1 | BeFree |
| umls:C0013384 | Dyskinetic syndrome | 1 | CTD_human |
| umls:C0013421 | Dystonia | 1 | LHGDN |
| umls:C0014544 | Epilepsy | 1 | BeFree |
| umls:C0015371 | Extrapyramidal Disorders | 1 | BeFree |
| umls:C0018799 | Heart Diseases | 1 | RGD |
| umls:C0018801 | Heart failure | 1 | BeFree |
| umls:C0018802 | Congestive heart failure | 1 | BeFree |
| umls:C0019188 | Hepatitis, Animal | 1 | RGD |
| umls:C0019189 | Hepatitis, Chronic | 1 | CTD_human |
| umls:C0020179 | Huntington Disease | 1 | LHGDN |
| umls:C0020550 | Hyperthyroidism | 1 | RGD |
| umls:C0022658 | Kidney Diseases | 1 | BeFree |
| umls:C0023904 | Liver Neoplasms, Experimental | 1 | CTD_human |
| umls:C0024437 | Macular degeneration | 1 | BeFree |
| umls:C0025202 | melanoma | 1 | CTD_human |
| umls:C0026650 | Movement Disorders | 1 | BeFree |
| umls:C0027051 | Myocardial Infarction | 1 | BeFree |
| umls:C0027746 | Nerve Degeneration | 1 | CTD_human |
| umls:C0028754 | Obesity | 1 | BeFree |
| umls:C0029925 | Ovarian Carcinoma | 1 | BeFree |
| umls:C0032914 | Pre-Eclampsia | 1 | CTD_human,LHGDN |
| umls:C0033860 | Psoriasis | 1 | CTD_human |
| umls:C0034150 | Purpura | 1 | LHGDN |
| umls:C0035334 | Retinitis Pigmentosa | 1 | BeFree |
| umls:C0036341 | Schizophrenia | 1 | CTD_human |
| umls:C0036572 | Seizures | 1 | BeFree |
| umls:C0040034 | Thrombocytopenia | 1 | LHGDN |
| umls:C0040136 | Thyroid Neoplasm | 1 | RGD |
| umls:C0042384 | Vasculitis | 1 | BeFree |
| umls:C0085397 | Pasteurellaceae Infections | 1 | CTD_human |
| umls:C0151731 | Hepatic infarction | 1 | BeFree |
| umls:C0232197 | Fibrillation | 1 | BeFree |
| umls:C0235974 | Pancreatic carcinoma | 1 | BeFree |
| umls:C0240066 | Iron deficiency | 1 | BeFree |
| umls:C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 1 | BeFree |
| umls:C0341439 | Chronic liver disease | 1 | BeFree |
| umls:C0346647 | Malignant neoplasm of pancreas | 1 | BeFree |
| umls:C0374997 | Helicobacter pylori (H. pylori) infection in conditions classified elsewhere and of unspecified site | 1 | BeFree |
| umls:C0400966 | Non-alcoholic Fatty Liver Disease | 1 | BeFree |
| umls:C0403440 | Thin basement membrane disease | 1 | BeFree |
| umls:C0426980 | Motor symptoms | 1 | BeFree |
| umls:C0497327 | Dementia | 1 | CTD_human |
| umls:C0524620 | Metabolic Syndrome X | 1 | BeFree |
| umls:C0740457 | Malignant neoplasm of kidney | 1 | BeFree |
| umls:C0741923 | cardiac event | 1 | BeFree |
| umls:C0854279 | Basal ganglion degeneration | 1 | BeFree |
| umls:C0919267 | ovarian neoplasm | 1 | BeFree |
| umls:C0948089 | Acute Coronary Syndrome | 1 | BeFree |
| umls:C1140680 | Malignant neoplasm of ovary | 1 | BeFree |
| umls:C1266042 | Chromophobe Renal Cell Carcinoma | 1 | BeFree |
| umls:C1306837 | Papillary Renal Cell Carcinoma | 1 | BeFree |
| umls:C1527311 | Brain Edema | 1 | RGD |
| umls:C1833213 | Hyperferritinemia, hereditary, with congenital cataracts | 1 | BeFree |
| umls:C2930898 | Benign essential blepharospasm | 1 | BeFree |
| umls:C2931845 | Neurodegeneration with brain iron accumulation (NBIA) | 1 | BeFree |
| umls:C3714619 | Insulin resistance syndrome | 1 | BeFree |
| umls:C1858583 | HEMOSIDEROSIS, SYSTEMIC, DUE TO ACERULOPLASMINEMIA | 0 | CLINVAR |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))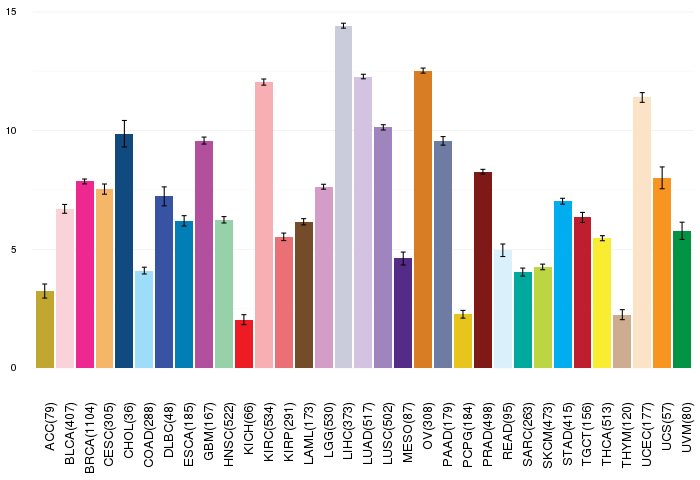
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))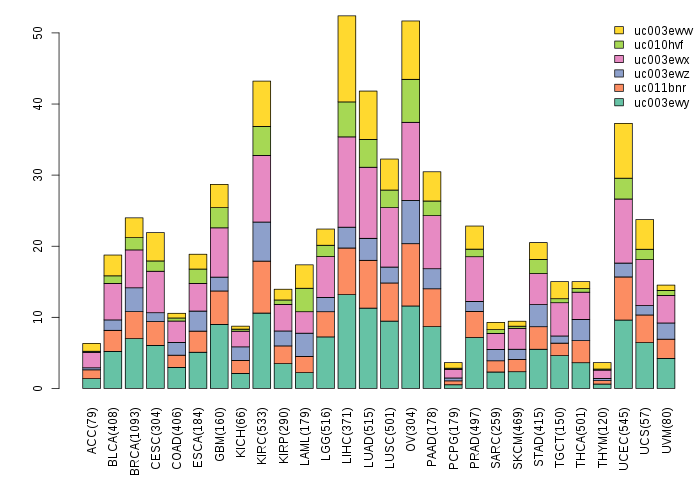
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data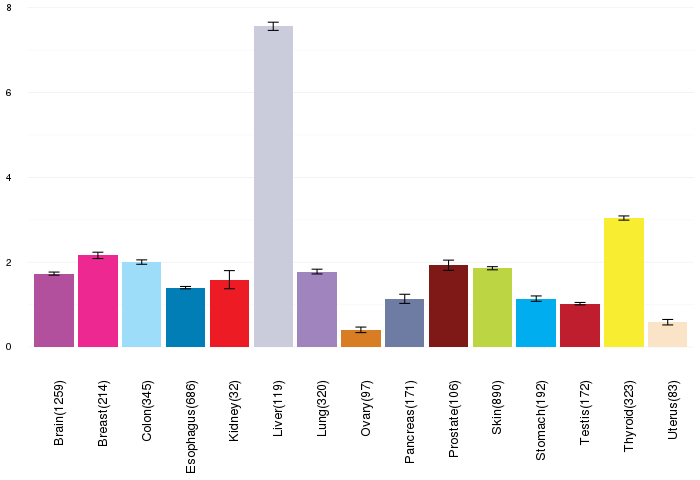
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))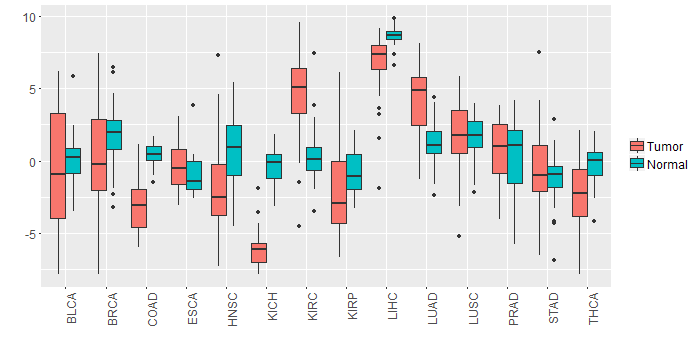
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.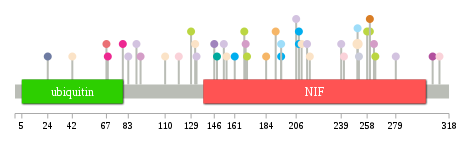

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 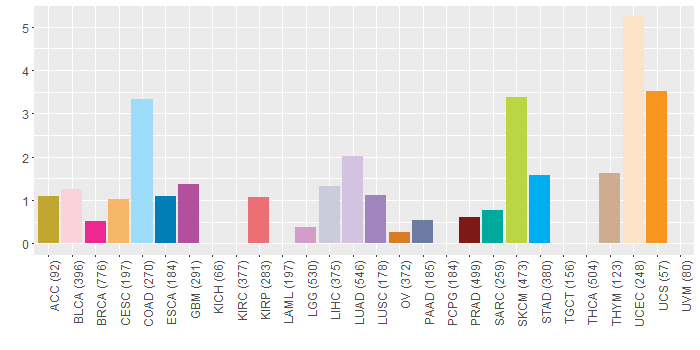
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)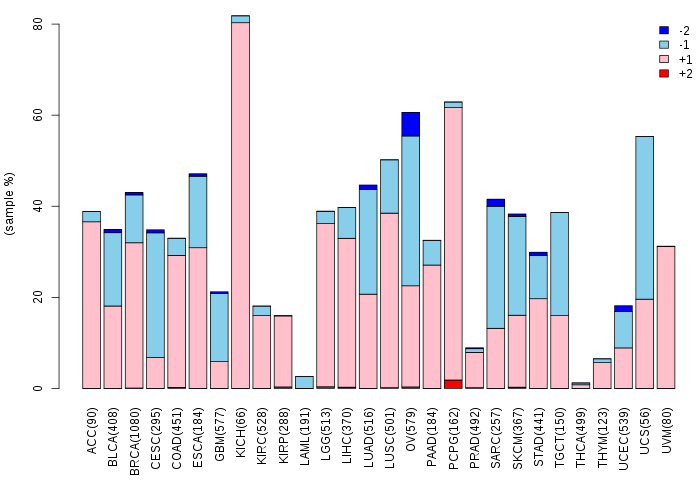
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 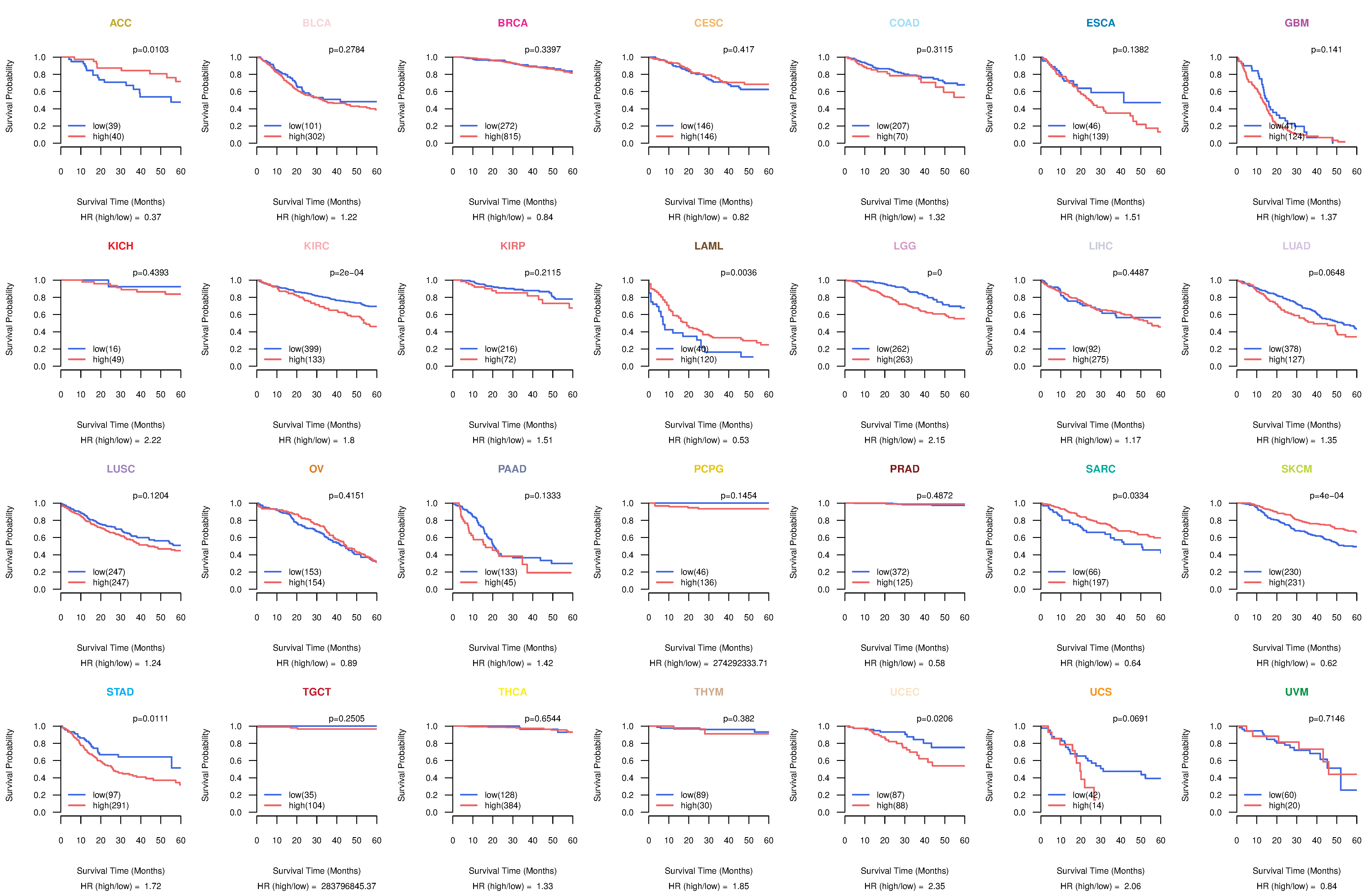
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 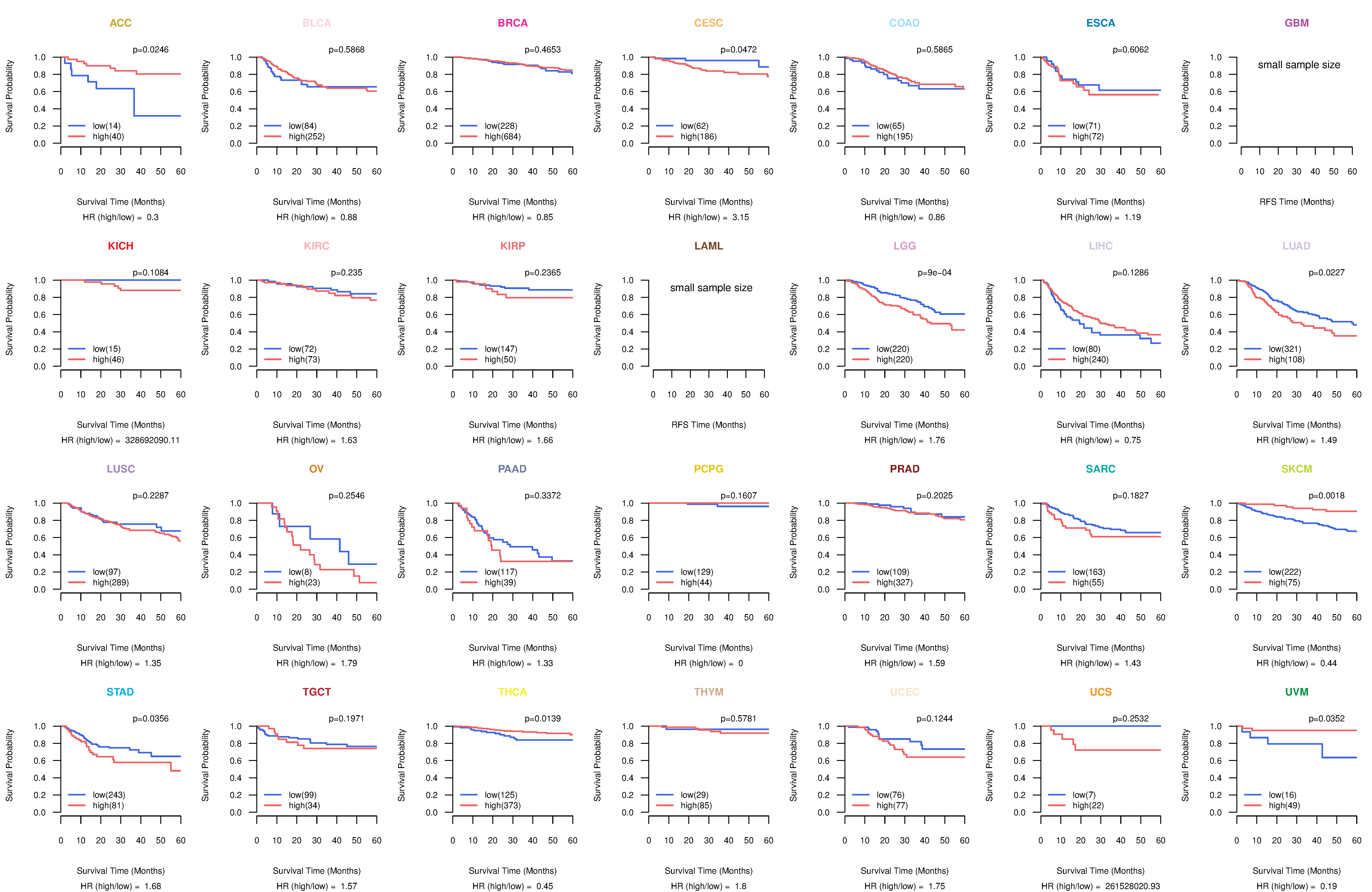
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 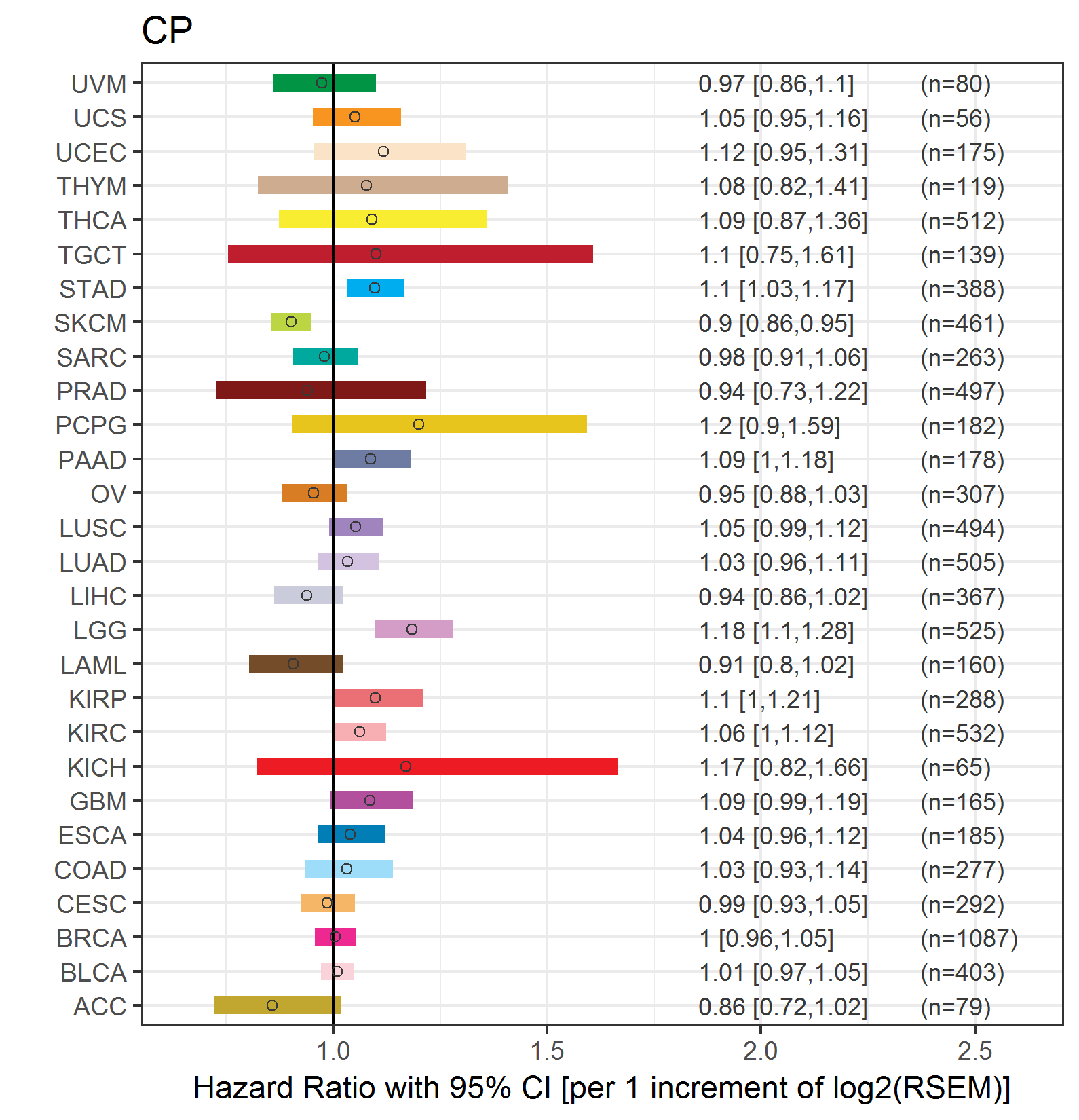
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 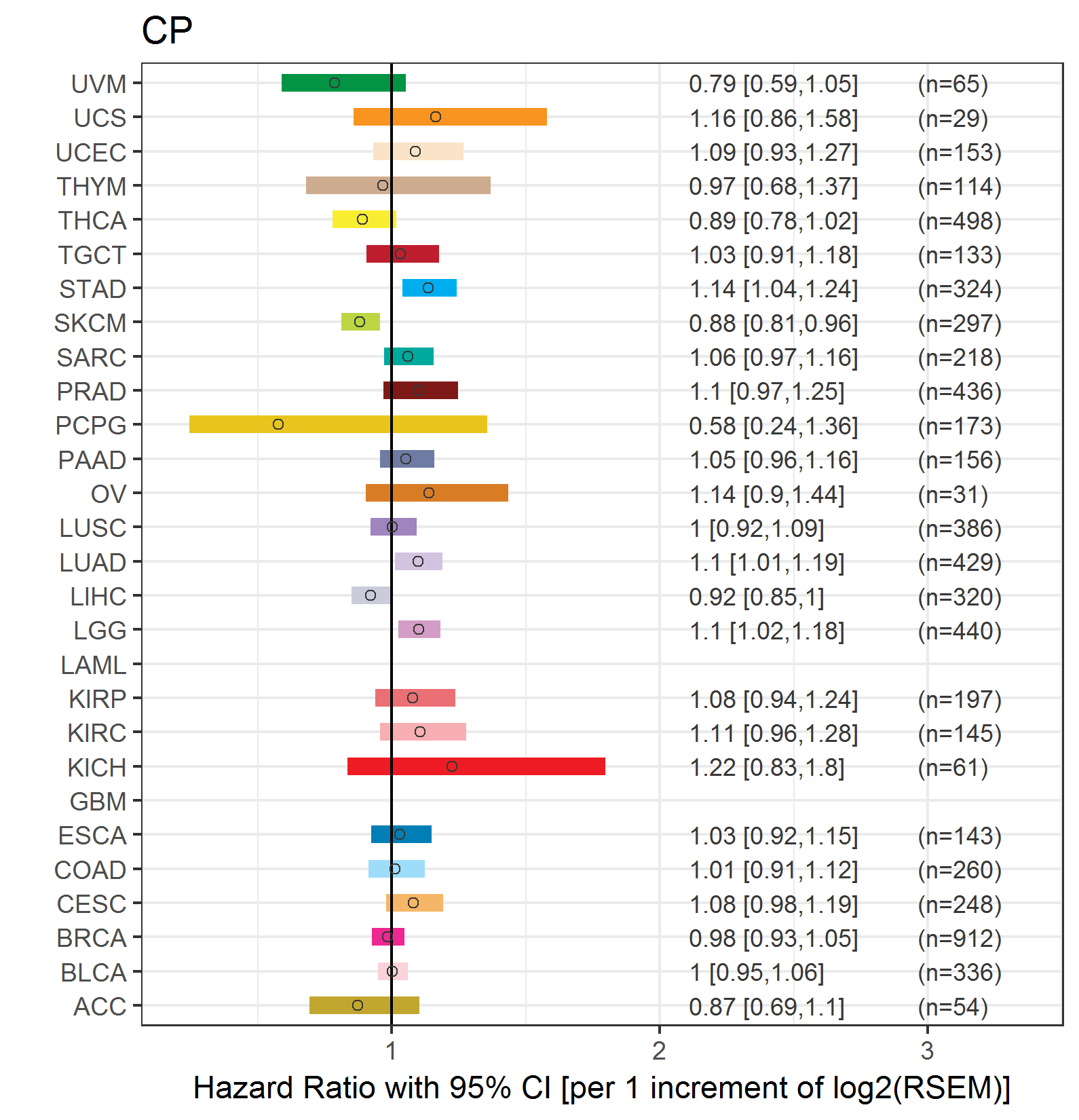
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























