| BRCA (tumor) | BRCA (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| COAD (tumor) | COAD (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| KICH (tumor) | KICH (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| KIRC (tumor) | KIRC (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| KIRP (tumor) | KIRP (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| LUAD (tumor) | LUAD (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| LUSC (tumor) | LUSC (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| PRAD (tumor) | PRAD (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| STAD (tumor) | STAD (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| THCA (tumor) | THCA (normal) |
| CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (tumor) | CPB2, HNF4A, UBC, PLG, HNF1A, A2M, F2, C5, PZP, TINAG (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0038454 | Cerebrovascular accident | 9 | BeFree,GAD,LHGDN |
| umls:C0027051 | Myocardial Infarction | 7 | BeFree,GAD |
| umls:C0010068 | Coronary heart disease | 5 | BeFree,GAD |
| umls:C0151942 | Arterial thrombosis | 5 | BeFree |
| umls:C0040053 | Thrombosis | 4 | GAD,LHGDN |
| umls:C0149871 | Deep Vein Thrombosis | 4 | BeFree,GAD |
| umls:C0007137 | Squamous cell carcinoma | 3 | BeFree,GAD |
| umls:C0010054 | Coronary Arteriosclerosis | 3 | BeFree |
| umls:C0040038 | Thromboembolism | 3 | BeFree |
| umls:C0242339 | Dyslipidemias | 3 | BeFree |
| umls:C0398623 | Thrombophilia | 3 | BeFree |
| umls:C0948008 | Ischemic stroke | 3 | BeFree |
| umls:C1956346 | Coronary Artery Disease | 3 | BeFree,GAD,LHGDN |
| umls:C3272363 | Ischemic Cerebrovascular Accident | 3 | BeFree |
| umls:C0004943 | Behcet Syndrome | 2 | BeFree,GAD,LHGDN |
| umls:C0005779 | Blood Coagulation Disorders | 2 | BeFree |
| umls:C0007222 | Cardiovascular Diseases | 2 | BeFree,GAD |
| umls:C0007785 | Cerebral Infarction | 2 | BeFree |
| umls:C0011849 | Diabetes Mellitus | 2 | BeFree,GAD |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 2 | BeFree,LHGDN |
| umls:C0014335 | Enteritis | 2 | BeFree |
| umls:C0019087 | Hemorrhagic Disorders | 2 | BeFree,GAD |
| umls:C0020443 | Hypercholesterolemia | 2 | GAD,LHGDN |
| umls:C0021368 | Inflammation | 2 | GAD,LHGDN |
| umls:C0025303 | Meningococcal Infections | 2 | BeFree |
| umls:C0025306 | Meningococcemia | 2 | BeFree |
| umls:C0026640 | Mouth Neoplasms | 2 | GAD,LHGDN |
| umls:C0034065 | Pulmonary Embolism | 2 | BeFree,LHGDN |
| umls:C0085278 | Antiphospholipid Syndrome | 2 | BeFree |
| umls:C0085409 | Polyendocrinopathies, Autoimmune | 2 | BeFree |
| umls:C0151945 | Thrombosis of cerebral veins | 2 | BeFree |
| umls:C0578159 | Antibiotic-associated diarrhea | 2 | BeFree |
| umls:C0584960 | Factor V Leiden mutation | 2 | BeFree |
| umls:C1861172 | Venous Thromboembolism | 2 | LHGDN |
| umls:C2239176 | Liver carcinoma | 2 | BeFree |
| umls:C0000809 | Abortion, Habitual | 1 | GAD |
| umls:C0001349 | Acute-Phase Reaction | 1 | LHGDN |
| umls:C0001418 | Adenocarcinoma | 1 | BeFree |
| umls:C0002453 | Amenorrhea | 1 | CTD_human |
| umls:C0002962 | Angina Pectoris | 1 | GAD |
| umls:C0002965 | Angina, Unstable | 1 | BeFree |
| umls:C0003864 | Arthritis | 1 | BeFree |
| umls:C0004153 | Atherosclerosis | 1 | LHGDN |
| umls:C0004610 | Bacteremia | 1 | RGD |
| umls:C0007786 | Brain Ischemia | 1 | GAD |
| umls:C0011991 | Diarrhea | 1 | BeFree |
| umls:C0012739 | Disseminated Intravascular Coagulation | 1 | BeFree,LHGDN |
| umls:C0013537 | Eclampsia | 1 | BeFree |
| umls:C0014072 | Experimental Autoimmune Encephalomyelitis | 1 | RGD |
| umls:C0017636 | Glioblastoma | 1 | BeFree |
| umls:C0019080 | Hemorrhage | 1 | GAD,LHGDN |
| umls:C0020538 | Hypertensive disease | 1 | GAD |
| umls:C0020550 | Hyperthyroidism | 1 | LHGDN |
| umls:C0022661 | Kidney Failure, Chronic | 1 | GAD |
| umls:C0023903 | Liver neoplasms | 1 | BeFree |
| umls:C0024121 | Lung Neoplasms | 1 | LHGDN |
| umls:C0026766 | Multiple Organ Failure | 1 | RGD |
| umls:C0028754 | Obesity | 1 | CTD_human |
| umls:C0029925 | Ovarian Carcinoma | 1 | BeFree |
| umls:C0032045 | Placenta Disorders | 1 | GAD |
| umls:C0032460 | Polycystic Ovary Syndrome | 1 | LHGDN |
| umls:C0032914 | Pre-Eclampsia | 1 | LHGDN |
| umls:C0035328 | Retinal Vein Occlusion | 1 | BeFree |
| umls:C0036983 | Septic Shock | 1 | LHGDN |
| umls:C0038525 | Subarachnoid Hemorrhage | 1 | GAD |
| umls:C0039240 | Supraventricular tachycardia | 1 | BeFree |
| umls:C0040336 | Tobacco Use Disorder | 1 | GAD |
| umls:C0042373 | Vascular Diseases | 1 | BeFree |
| umls:C0042487 | Venous Thrombosis | 1 | GAD |
| umls:C0043407 | Yersinia infections | 1 | BeFree |
| umls:C0085298 | Sudden Cardiac Death | 1 | GAD |
| umls:C0149925 | Small cell carcinoma of lung | 1 | BeFree |
| umls:C0151950 | Deep thrombophlebitis | 1 | BeFree |
| umls:C0153381 | Malignant neoplasm of mouth | 1 | BeFree |
| umls:C0162869 | Aneurysm, Ruptured | 1 | GAD |
| umls:C0220641 | Lip and Oral Cavity Carcinoma | 1 | BeFree |
| umls:C0242379 | Malignant neoplasm of lung | 1 | BeFree |
| umls:C0262584 | Carcinoma, Small Cell | 1 | BeFree |
| umls:C0524620 | Metabolic Syndrome X | 1 | GAD |
| umls:C0524702 | Pulmonary Thromboembolisms | 1 | BeFree |
| umls:C0596263 | Carcinogenesis | 1 | BeFree |
| umls:C0598608 | Hyperhomocysteinemia | 1 | BeFree |
| umls:C0600433 | Activated Protein C Resistance | 1 | BeFree,LHGDN |
| umls:C0684249 | Carcinoma of lung | 1 | BeFree |
| umls:C0687675 | Pregnancy loss | 1 | GAD |
| umls:C0836924 | Thrombocytosis | 1 | LHGDN |
| umls:C0852949 | Arteriopathic disease | 1 | BeFree |
| umls:C0860207 | Drug-Induced Liver Injury | 1 | RGD |
| umls:C0947751 | Vascular inflammations | 1 | BeFree |
| umls:C0948441 | Venoocclusive disease | 1 | BeFree |
| umls:C1140680 | Malignant neoplasm of ovary | 1 | BeFree,GAD |
| umls:C1271104 | Blood pressure finding | 1 | GAD |
| umls:C1272641 | Systemic arterial pressure | 1 | GAD |
| umls:C1273976 | First myocardial infarction | 1 | BeFree |
| umls:C1531624 | Cardioembolic stroke | 1 | BeFree |
| umls:C1867596 | Hyperprothrombinemia | 1 | BeFree |
| umls:C2584409 | Prothrombin G20210A mutation | 1 | BeFree |
| umls:C2717961 | Thrombotic Microangiopathies | 1 | BeFree |
| umls:C3495426 | Homocysteinemia | 1 | BeFree |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))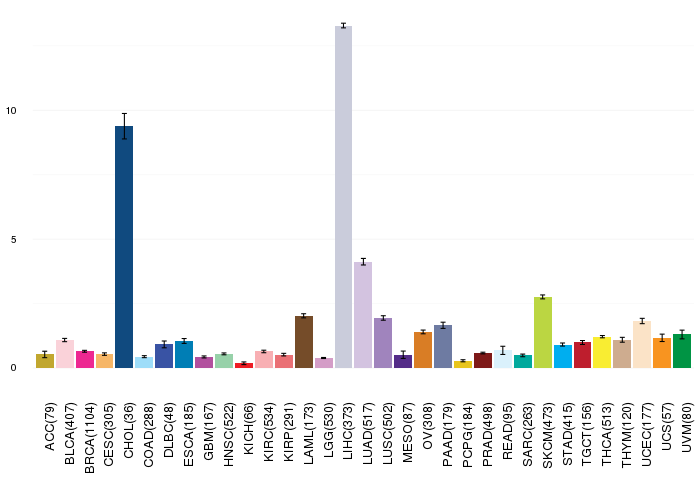
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))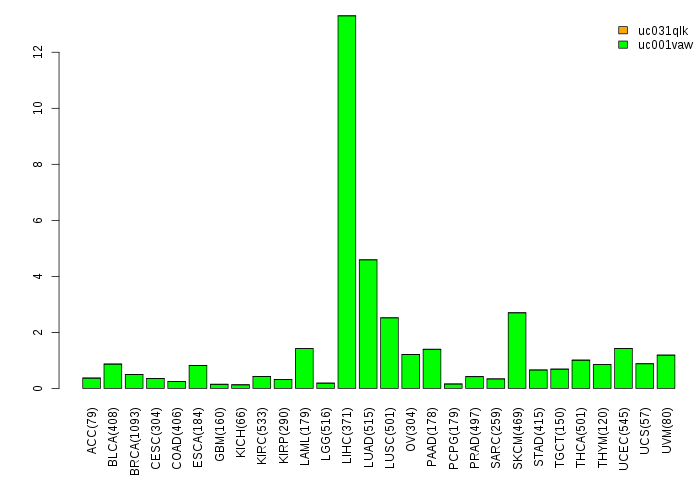
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data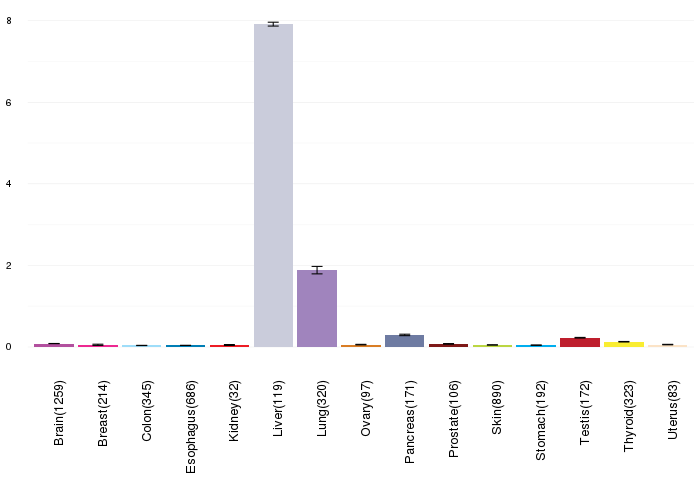
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))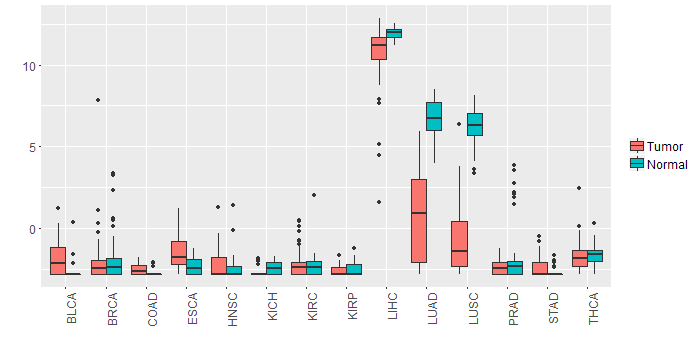
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 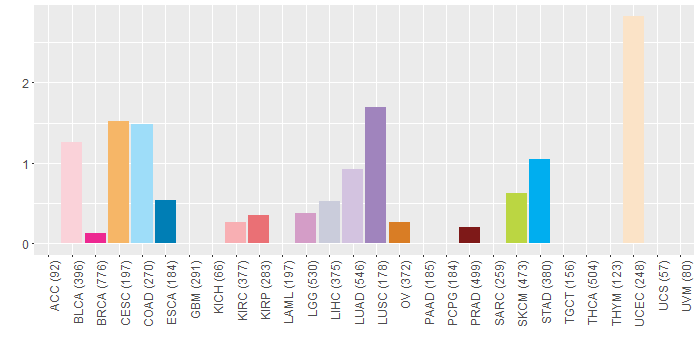
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)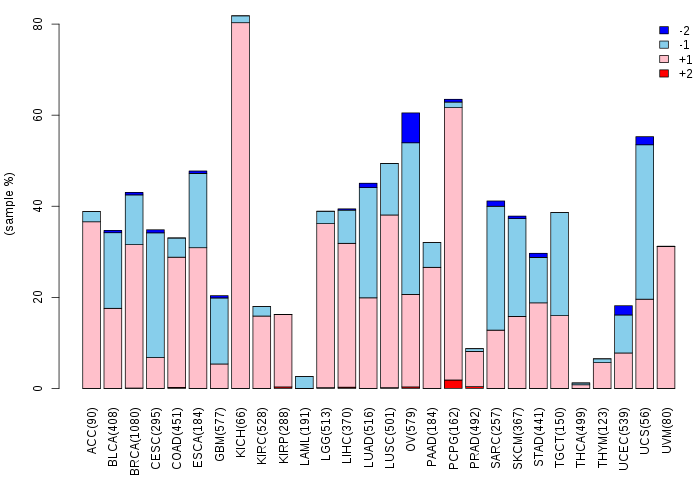
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























