| Basic gene information | Gene symbol | CYP3A5 |
| Gene name | cytochrome P450, family 3, subfamily A, polypeptide 5 |
| Synonyms | CP35|CYPIIIA5|P450PCN3|PCN3 |
| Cytomap | UCSC genome browser: 7q21.1 |
| Type of gene | protein-coding |
| RefGenes | NM_000777.4,
NM_001190484.2,NM_001291829.1,NM_001291830.1,NR_033807.2,
NR_033808.1,NR_033809.1,NR_033810.1,NR_033811.1,
NR_033812.1, |
| Description | aryl hydrocarbon hydroxylasecytochrome P450 3A5cytochrome P450 HLp2cytochrome P450, subfamily IIIA (niphedipine oxidase), polypeptide 5cytochrome P450-PCN3flavoprotein-linked monooxygenasemicrosomal monooxygenasexenobiotic monooxygenase |
| Modification date | 20141222 |
| dbXrefs | MIM : 605325 |
| HGNC : HGNC |
| Ensembl : ENSG00000106258 |
| HPRD : 05617 |
| Vega : OTTHUMG00000156724 |
| Protein | UniProt:
go to UniProt's Cross Reference DB Table |
| Expression | CleanEX: HS_CYP3A5 |
| BioGPS: 1577 |
| Pathway | NCI Pathway Interaction Database: CYP3A5 |
| KEGG: CYP3A5 |
| REACTOME: CYP3A5 |
| Pathway Commons: CYP3A5 |
| Context | iHOP: CYP3A5 |
| ligand binding site mutation search in PubMed: CYP3A5 |
| UCL Cancer Institute: CYP3A5 |
| Assigned class in TissGDB* | C |
| Included tissue-specific gene expression resources | TiGER,GTEx |
| Specific-tissues in normal samples (assigned by TissGDB using HPA, TiGER, and GTEx) | Liver |
| Cancer types related to the specific-tissues in cancer samples (assigned by TissGDB using TCGA) | LIHC |
| Reference showing the relevant tissue of CYP3A5 | |
| Description by TissGene annotations | Have significant anti-correlated miRNA
|
| BRCA (tumor) | BRCA (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
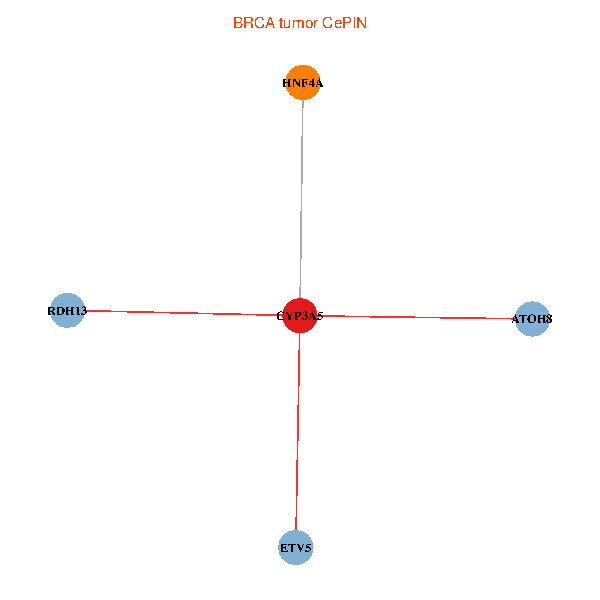 | 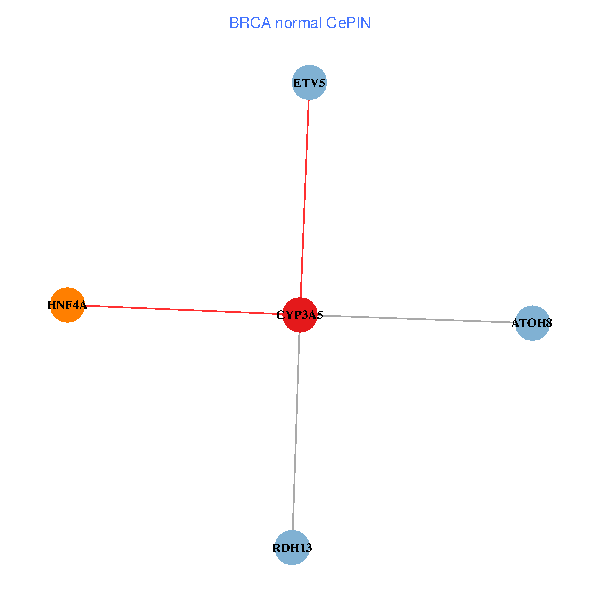 |
|
| COAD (tumor) | COAD (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
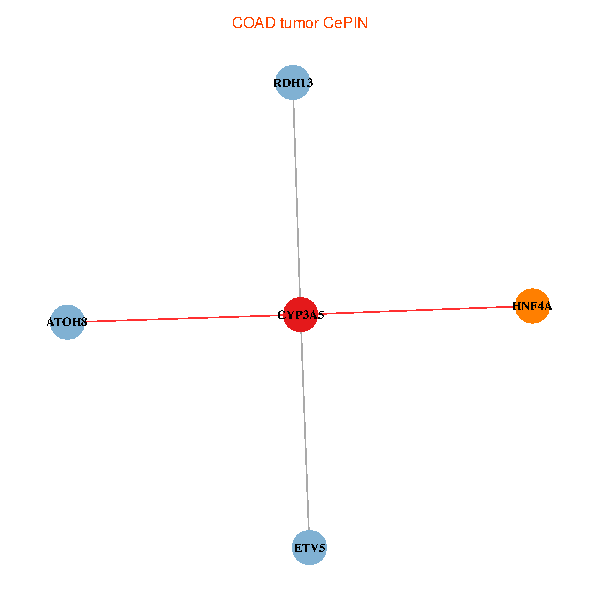 | 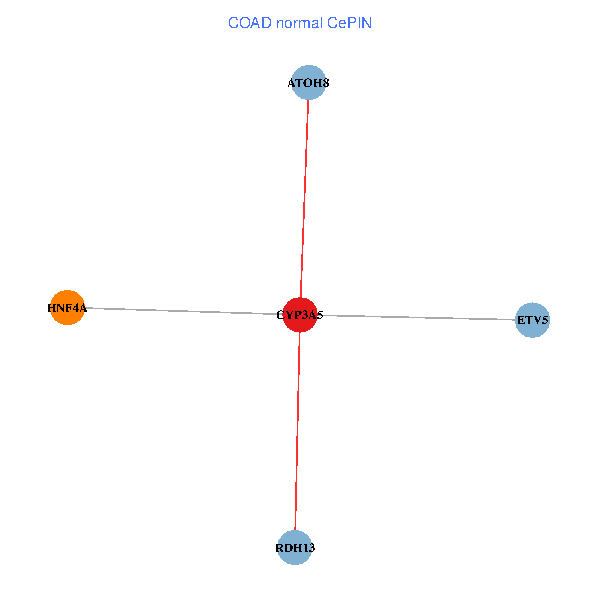 |
|
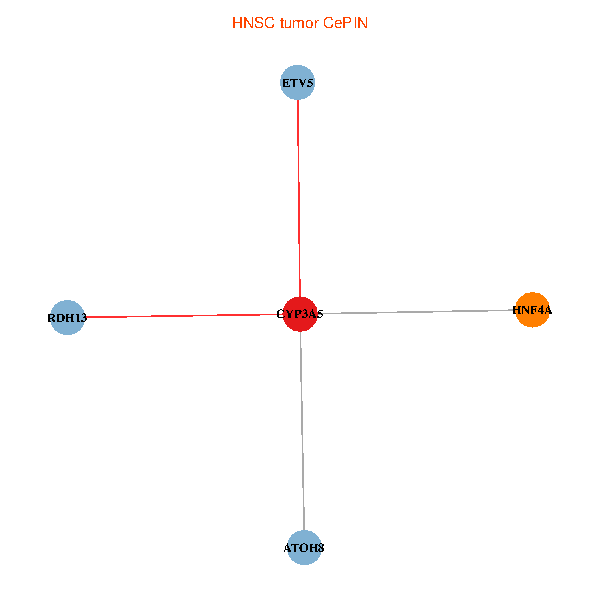
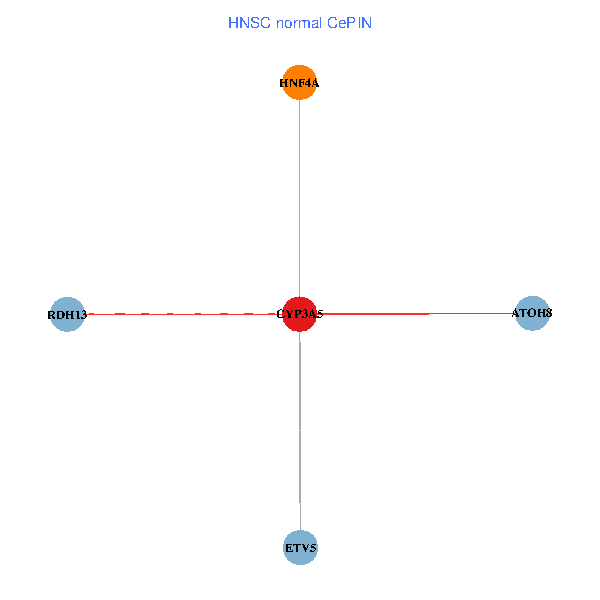
| HNSC (tumor) | HNSC (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
 |  |
|
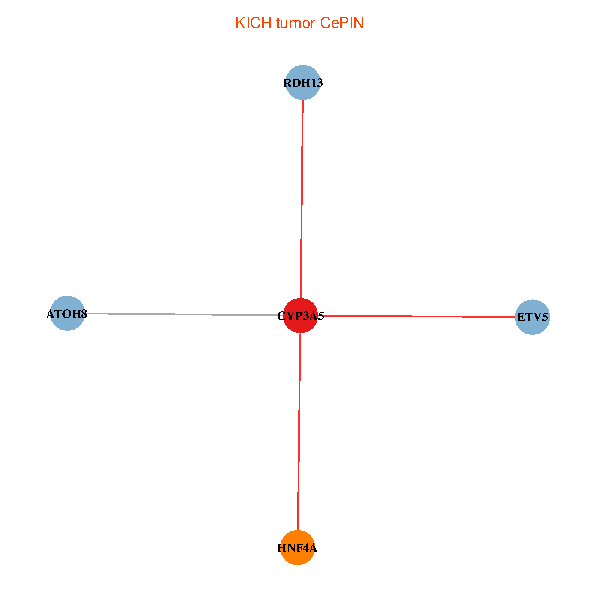
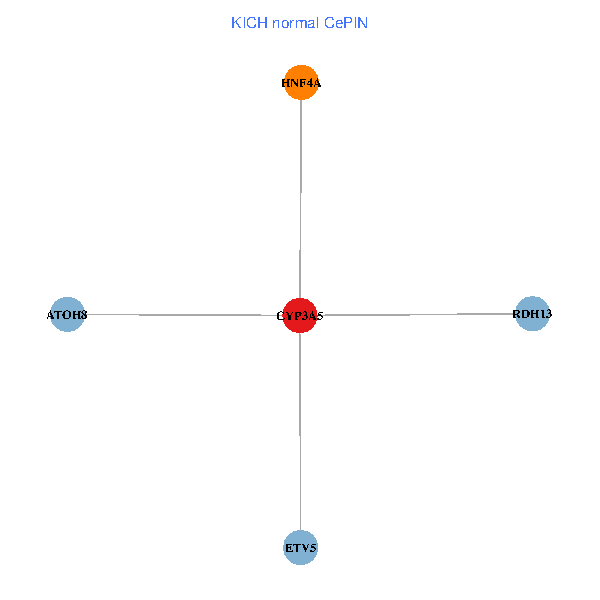
| KICH (tumor) | KICH (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
 |  |
|
| KIRC (tumor) | KIRC (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
 |  |
|
| KIRP (tumor) | KIRP (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
 |  |
|
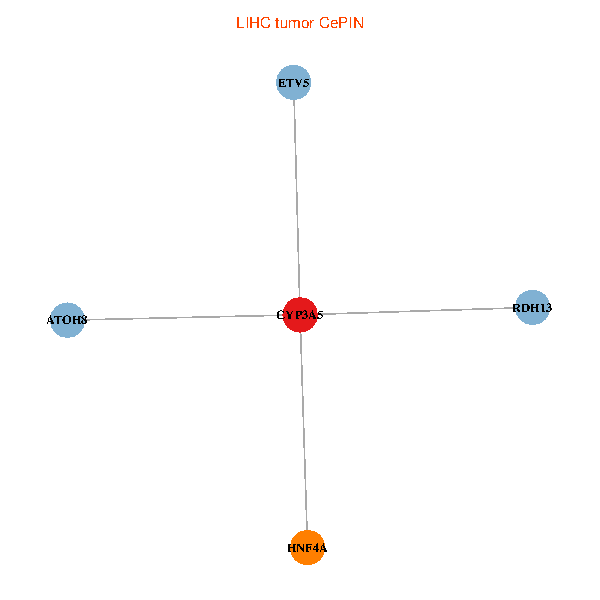
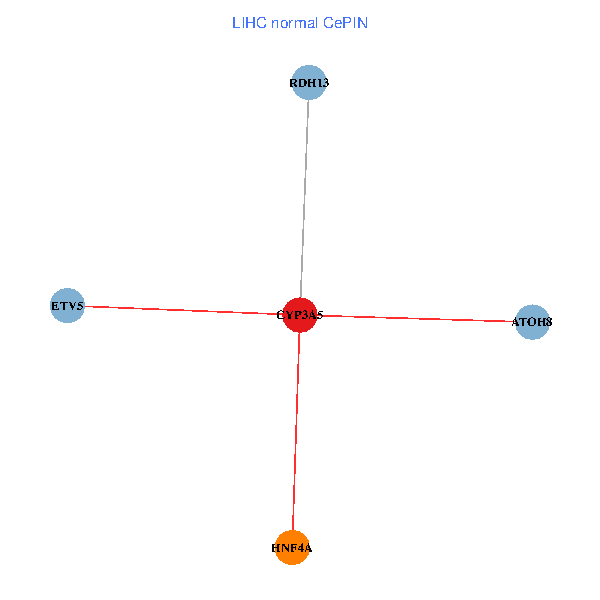
| LIHC (tumor) | LIHC (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
 |  |
|
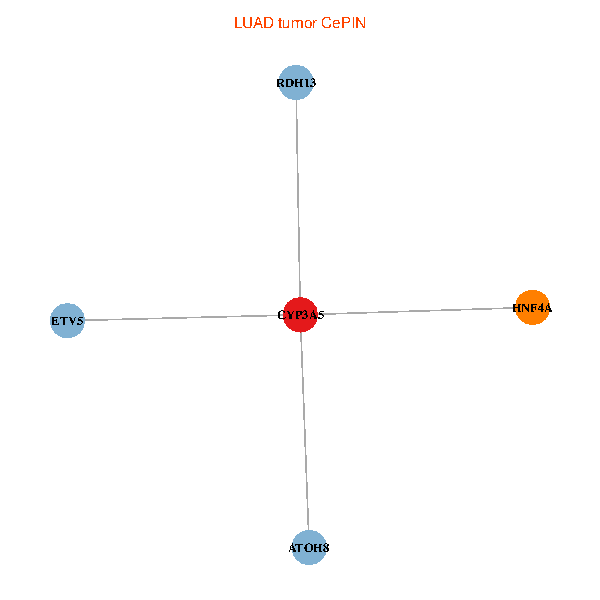
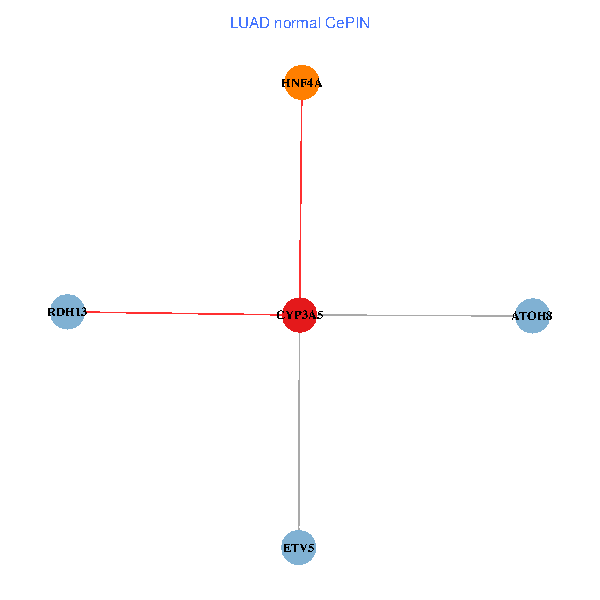
| LUAD (tumor) | LUAD (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
 |  |
|
| LUSC (tumor) | LUSC (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
 |  |
|
| PRAD (tumor) | PRAD (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
 |  |
|
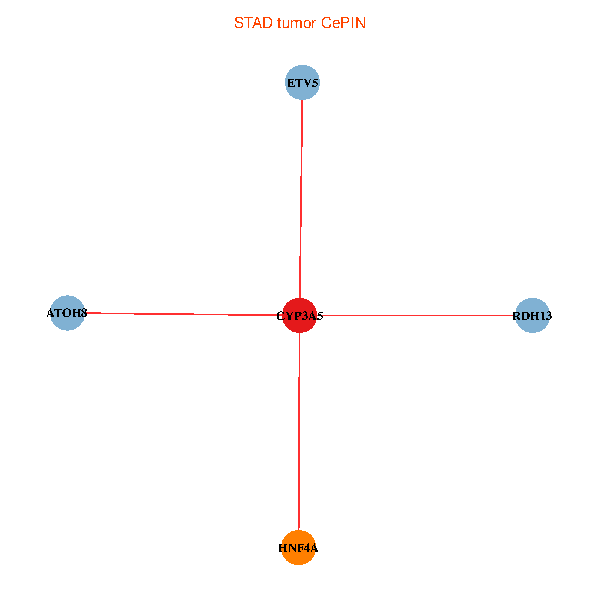
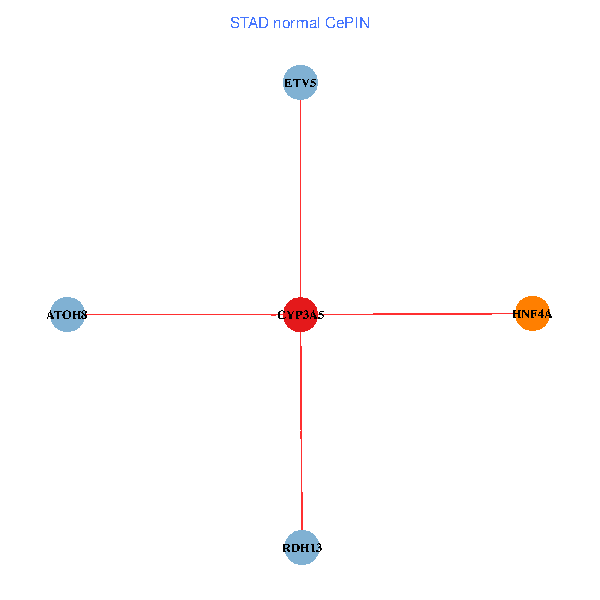
| STAD (tumor) | STAD (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
 |  |
|
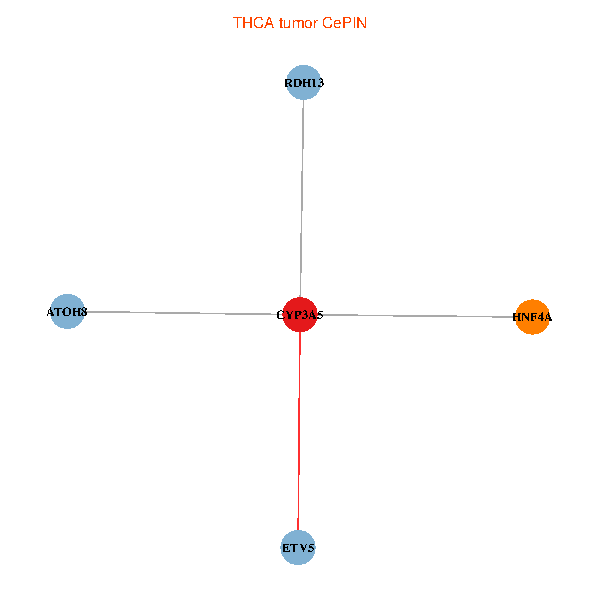
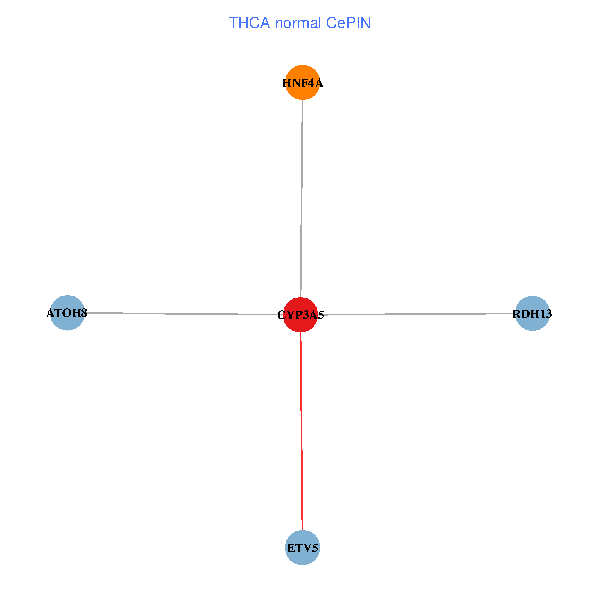
| THCA (tumor) | THCA (normal) |
| CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (tumor) | CYP3A5, HNF4A, RDH13, ETV5, ATOH8 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0006142 | Malignant neoplasm of breast | 20 | BeFree,GAD |
| umls:C0019693 | HIV Infections | 16 | BeFree,GAD |
| umls:C0020538 | Hypertensive disease | 15 | BeFree,GAD,LHGDN |
| umls:C0376358 | Malignant neoplasm of prostate | 15 | BeFree,GAD |
| umls:C0242379 | Malignant neoplasm of lung | 14 | BeFree,GAD |
| umls:C0023418 | leukemia | 7 | GAD,LHGDN |
| umls:C0023473 | Myeloid Leukemia, Chronic | 7 | BeFree,GAD |
| umls:C0600139 | Prostate carcinoma | 7 | BeFree |
| umls:C0684249 | Carcinoma of lung | 7 | BeFree |
| umls:C0020443 | Hypercholesterolemia | 6 | BeFree,GAD |
| umls:C0023449 | Acute lymphocytic leukemia | 6 | BeFree |
| umls:C0023530 | Leukopenia | 6 | BeFree,GAD |
| umls:C0027947 | Neutropenia | 6 | BeFree,GAD |
| umls:C0678222 | Breast Carcinoma | 6 | BeFree |
| umls:C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 6 | BeFree,GAD |
| umls:C0022658 | Kidney Diseases | 5 | BeFree,CTD_human,GAD |
| umls:C0948089 | Acute Coronary Syndrome | 5 | GAD |
| umls:C0033578 | Prostatic Neoplasms | 4 | CTD_human,GAD,LHGDN |
| umls:C0153594 | Malignant neoplasm of testis | 4 | BeFree,GAD |
| umls:C0546837 | Malignant neoplasm of esophagus | 4 | BeFree,GAD |
| umls:C1527249 | Colorectal Cancer | 4 | BeFree,GAD |
| umls:C2239176 | Liver carcinoma | 4 | BeFree |
| umls:C0007137 | Squamous cell carcinoma | 3 | BeFree,GAD |
| umls:C0009324 | Ulcerative Colitis | 3 | BeFree,GAD |
| umls:C0014544 | Epilepsy | 3 | BeFree,GAD |
| umls:C0023452 | Leukemia, Lymphocytic, Acute, L1 | 3 | BeFree |
| umls:C0027051 | Myocardial Infarction | 3 | GAD |
| umls:C0036341 | Schizophrenia | 3 | BeFree,GAD,LHGDN |
| umls:C0153381 | Malignant neoplasm of mouth | 3 | GAD |
| umls:C0302592 | Cervix carcinoma | 3 | GAD |
| umls:C0345904 | Malignant neoplasm of liver | 3 | GAD |
| umls:C0549473 | Thyroid carcinoma | 3 | GAD |
| umls:C0919267 | ovarian neoplasm | 3 | GAD |
| umls:C1272641 | Systemic arterial pressure | 3 | GAD |
| umls:C1956346 | Coronary Artery Disease | 3 | BeFree,GAD |
| umls:C0007131 | Non-Small Cell Lung Carcinoma | 2 | BeFree,GAD |
| umls:C0007134 | Renal Cell Carcinoma | 2 | BeFree,GAD |
| umls:C0007222 | Cardiovascular Diseases | 2 | GAD |
| umls:C0009402 | Colorectal Carcinoma | 2 | BeFree |
| umls:C0010054 | Coronary Arteriosclerosis | 2 | BeFree |
| umls:C0010068 | Coronary heart disease | 2 | BeFree |
| umls:C0011849 | Diabetes Mellitus | 2 | BeFree,GAD |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 2 | BeFree,GAD |
| umls:C0011991 | Diarrhea | 2 | BeFree,GAD |
| umls:C0018671 | Head and Neck Neoplasms | 2 | GAD |
| umls:C0023470 | Myeloid Leukemia | 2 | BeFree,GAD,LHGDN |
| umls:C0023903 | Liver neoplasms | 2 | BeFree |
| umls:C0024117 | Chronic Obstructive Airway Disease | 2 | BeFree,GAD |
| umls:C0024143 | Lupus Nephritis | 2 | GAD |
| umls:C0026848 | Myopathy | 2 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 2 | BeFree |
| umls:C0027658 | Neoplasms, Germ Cell and Embryonal | 2 | GAD |
| umls:C0031117 | Peripheral Neuropathy | 2 | BeFree |
| umls:C0038454 | Cerebrovascular accident | 2 | GAD |
| umls:C0040053 | Thrombosis | 2 | CTD_human,GAD |
| umls:C0085215 | Ovarian Failure, Premature | 2 | GAD |
| umls:C0279626 | Squamous cell carcinoma of esophagus | 2 | BeFree |
| umls:C0403447 | Chronic Kidney Insufficiency | 2 | BeFree |
| umls:C0596263 | Carcinogenesis | 2 | BeFree |
| umls:C1458155 | Mammary Neoplasms | 2 | GAD |
| umls:C1561643 | Chronic Kidney Diseases | 2 | BeFree |
| umls:C1704272 | Benign Prostatic Hyperplasia | 2 | BeFree |
| umls:C0003873 | Rheumatoid Arthritis | 1 | BeFree |
| umls:C0004153 | Atherosclerosis | 1 | GAD |
| umls:C0005684 | Malignant neoplasm of urinary bladder | 1 | GAD |
| umls:C0008370 | Cholestasis | 1 | CTD_mouse |
| umls:C0009404 | Colorectal Neoplasms | 1 | GAD |
| umls:C0011206 | Delirium | 1 | BeFree |
| umls:C0011853 | Diabetes Mellitus, Experimental | 1 | CTD_mouse |
| umls:C0012860 | DNA Damage | 1 | GAD |
| umls:C0013182 | Drug Allergy | 1 | GAD |
| umls:C0015230 | Exanthema | 1 | GAD |
| umls:C0018133 | Graft-vs-Host Disease | 1 | GAD |
| umls:C0019193 | Hepatitis, Toxic | 1 | GAD |
| umls:C0020473 | Hyperlipidemia | 1 | GAD |
| umls:C0022660 | Kidney Failure, Acute | 1 | BeFree |
| umls:C0022661 | Kidney Failure, Chronic | 1 | GAD |
| umls:C0022665 | Kidney Neoplasm | 1 | GAD |
| umls:C0023467 | Leukemia, Myelocytic, Acute | 1 | GAD |
| umls:C0023474 | Leukemia, Myeloid, Chronic-Phase | 1 | BeFree,GAD |
| umls:C0023485 | Precursor B-Cell Lymphoblastic Leukemia-Lymphoma | 1 | GAD |
| umls:C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_mouse |
| umls:C0023895 | Liver diseases | 1 | GAD |
| umls:C0023904 | Liver Neoplasms, Experimental | 1 | CTD_mouse |
| umls:C0024121 | Lung Neoplasms | 1 | GAD |
| umls:C0024530 | Malaria | 1 | GAD |
| umls:C0024620 | Primary malignant neoplasm of liver | 1 | BeFree |
| umls:C0027686 | Pathologic Neovascularization | 1 | GAD |
| umls:C0027720 | Nephrosis | 1 | CTD_rat |
| umls:C0027819 | Neuroblastoma | 1 | BeFree |
| umls:C0029172 | Oral Submucous Fibrosis | 1 | BeFree |
| umls:C0032787 | Postoperative Complications | 1 | GAD |
| umls:C0035126 | Reperfusion Injury | 1 | CTD_rat |
| umls:C0035220 | Respiratory Distress Syndrome, Newborn | 1 | BeFree |
| umls:C0035222 | Respiratory Distress Syndrome, Adult | 1 | BeFree |
| umls:C0037286 | Skin Neoplasms | 1 | GAD |
| umls:C0040038 | Thromboembolism | 1 | BeFree |
| umls:C0042065 | Genitourinary Neoplasms | 1 | GAD |
| umls:C0042076 | Urologic Neoplasms | 1 | GAD |
| umls:C0042749 | Viremia | 1 | BeFree |
| umls:C0079773 | Lymphoma, T-Cell, Cutaneous | 1 | GAD |
| umls:C0079774 | Peripheral T-Cell Lymphoma | 1 | GAD |
| umls:C0149925 | Small cell carcinoma of lung | 1 | BeFree |
| umls:C0151650 | Renal fibrosis | 1 | BeFree |
| umls:C0152115 | Lingual-Facial-Buccal Dyskinesia | 1 | GAD |
| umls:C0206720 | Squamous Cell Neoplasms | 1 | GAD |
| umls:C0235032 | Neurotoxicity Syndromes | 1 | CTD_human,GAD |
| umls:C0238198 | Gastrointestinal Stromal Tumors | 1 | GAD |
| umls:C0240035 | Interstitial fibrosis | 1 | BeFree |
| umls:C0242383 | Age related macular degeneration | 1 | GAD |
| umls:C0242992 | Multiple Chemical Sensitivity | 1 | GAD |
| umls:C0280100 | Solid tumour | 1 | BeFree |
| umls:C0280280 | stage, prostate cancer | 1 | BeFree |
| umls:C0344307 | Absence of pain sensation | 1 | BeFree |
| umls:C0392525 | Nephrolithiasis | 1 | BeFree |
| umls:C0442833 | Arteriolar hyalinosis | 1 | BeFree |
| umls:C0442874 | Neuropathy | 1 | BeFree |
| umls:C0524909 | Hepatitis B, Chronic | 1 | GAD |
| umls:C0677886 | Epithelial ovarian cancer | 1 | GAD |
| umls:C0685938 | Malignant neoplasm of gastrointestinal tract | 1 | BeFree |
| umls:C0686347 | Tardive Dyskinesia | 1 | GAD |
| umls:C0700095 | Central neuroblastoma | 1 | BeFree |
| umls:C0746883 | Febrile Neutropenia | 1 | BeFree |
| umls:C0809983 | Schizophrenia and related disorders | 1 | BeFree |
| umls:C0855197 | Testicular malignant germ cell tumor | 1 | BeFree |
| umls:C0860207 | Drug-Induced Liver Injury | 1 | GAD |
| umls:C0948380 | Colorectal cancer metastatic | 1 | BeFree |
| umls:C1140680 | Malignant neoplasm of ovary | 1 | GAD |
| umls:C1279296 | Chronic leukemia (category) | 1 | BeFree |
| umls:C1565489 | Renal Insufficiency | 1 | GAD |
| umls:C1566590 | Delayed Graft Function | 1 | GAD |
| umls:C1841972 | Glucocorticoid Receptor Deficiency | 1 | BeFree |
| umls:C1861172 | Venous Thromboembolism | 1 | GAD |
| umls:C2937421 | Prostatic Hyperplasia | 1 | GAD |
| umls:C0085580 | Essential Hypertension | 0 | CTD_human |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))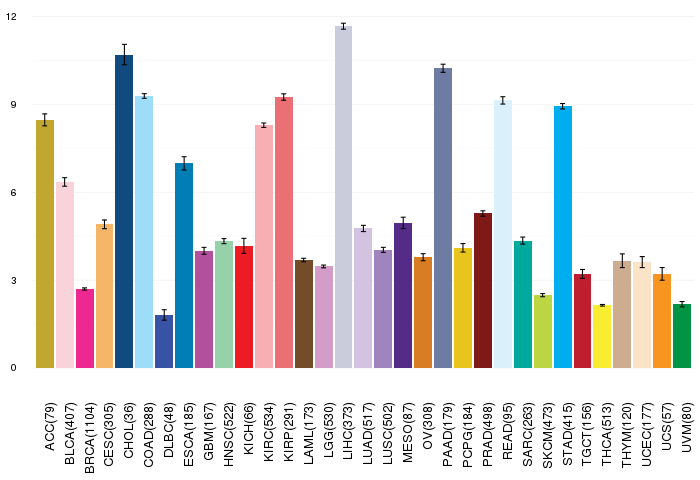
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))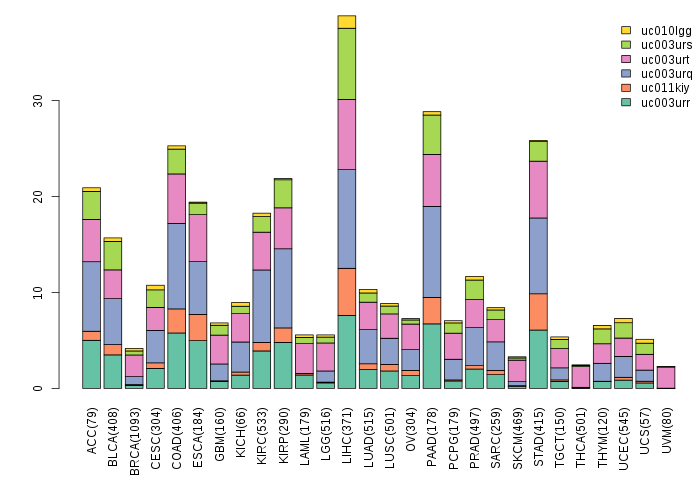
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data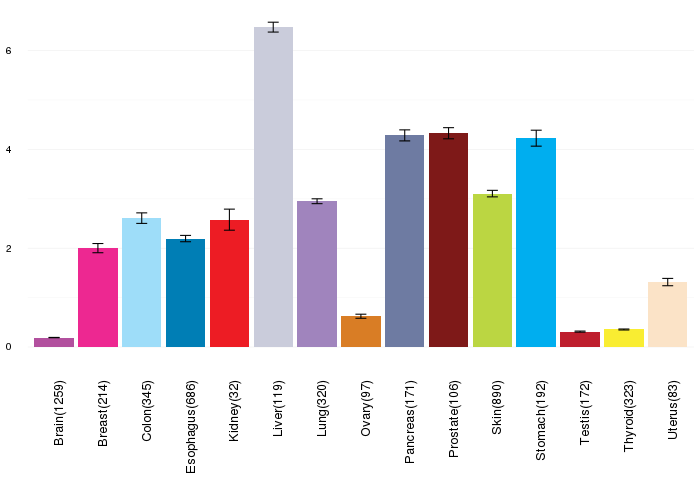
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))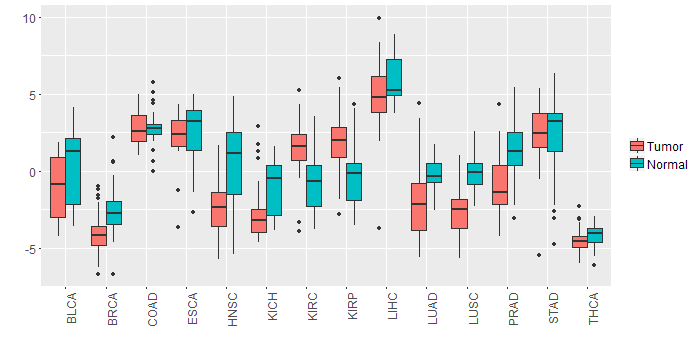
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.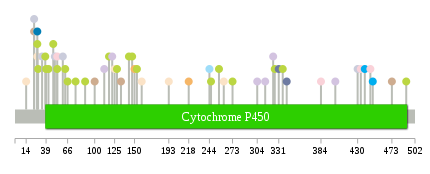

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 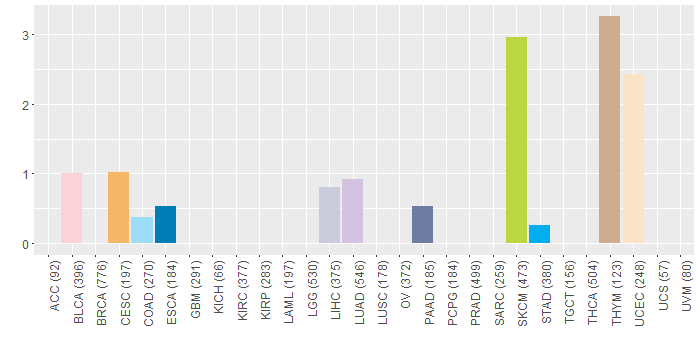
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)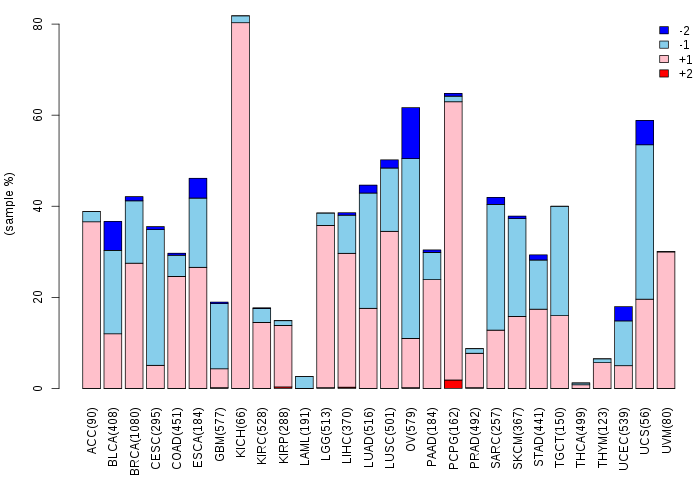
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 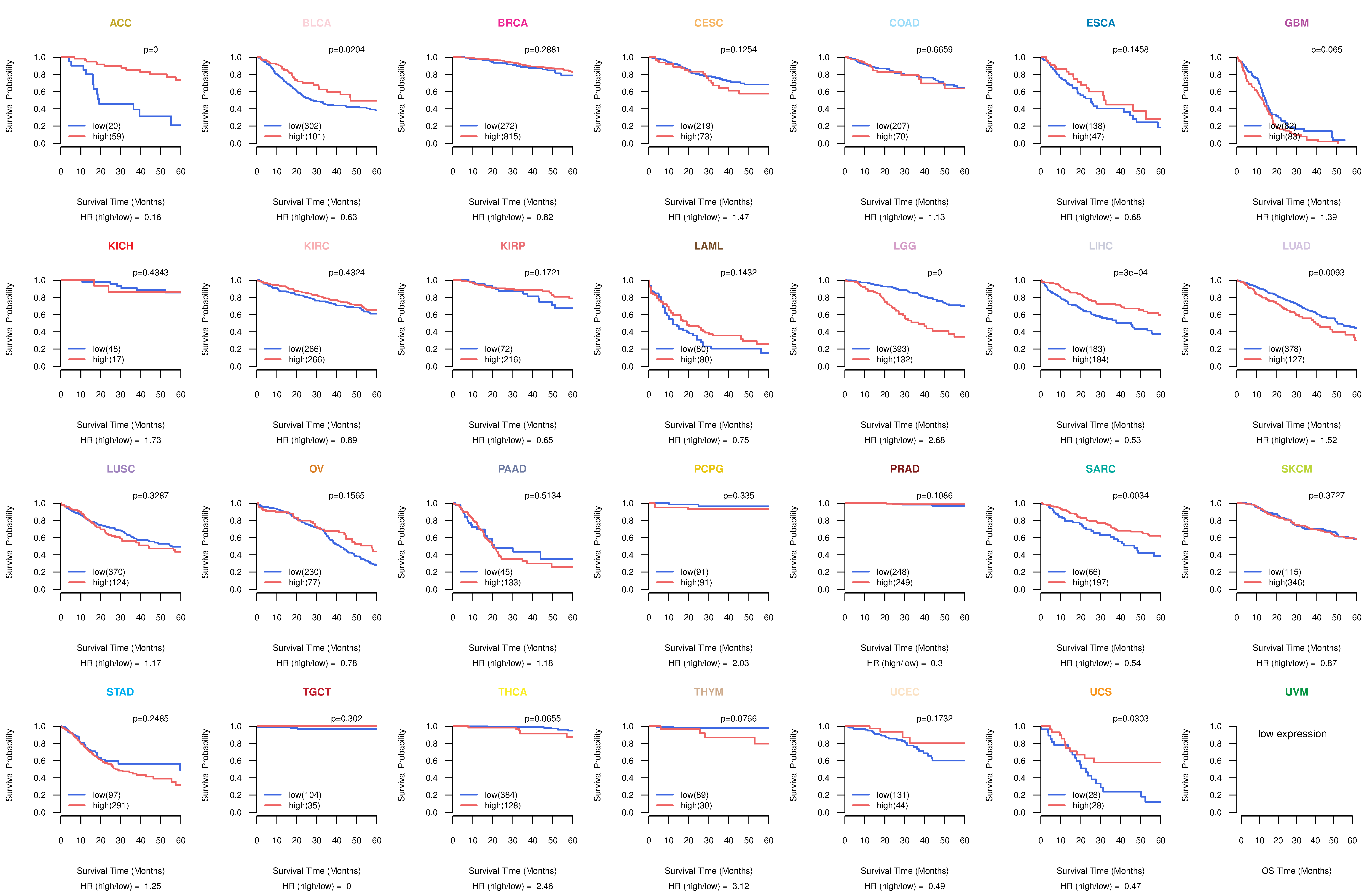
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 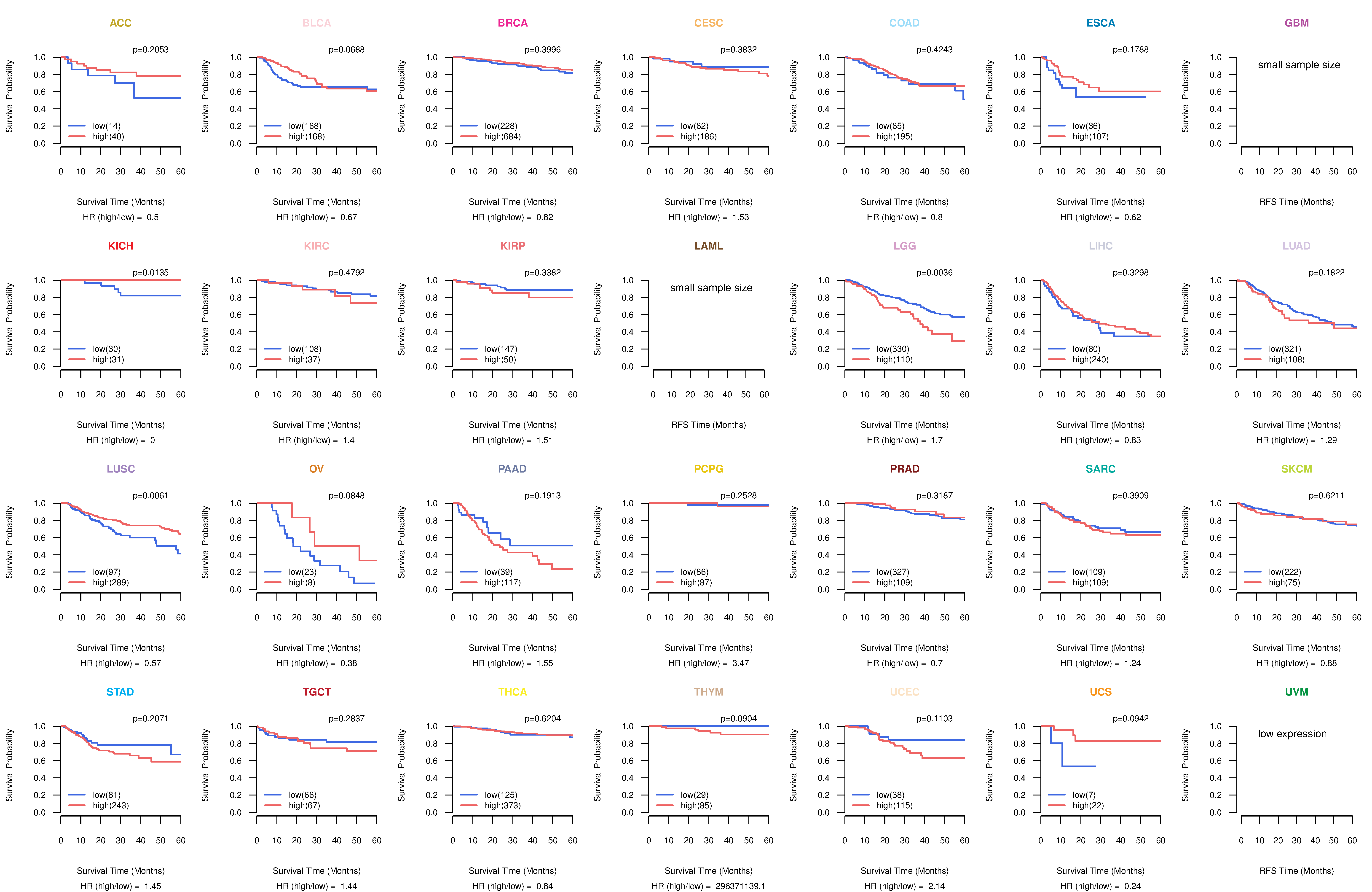
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 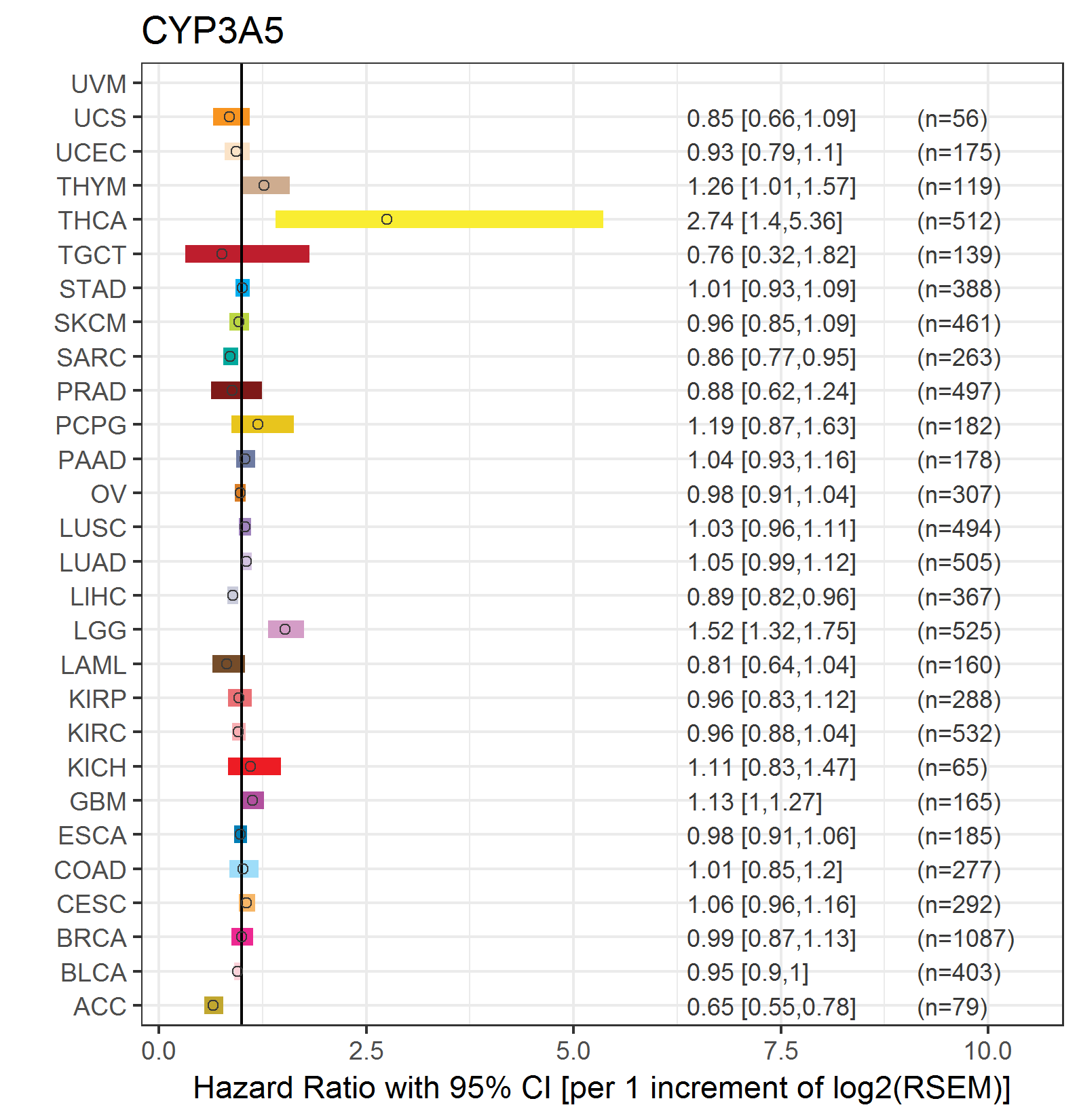
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 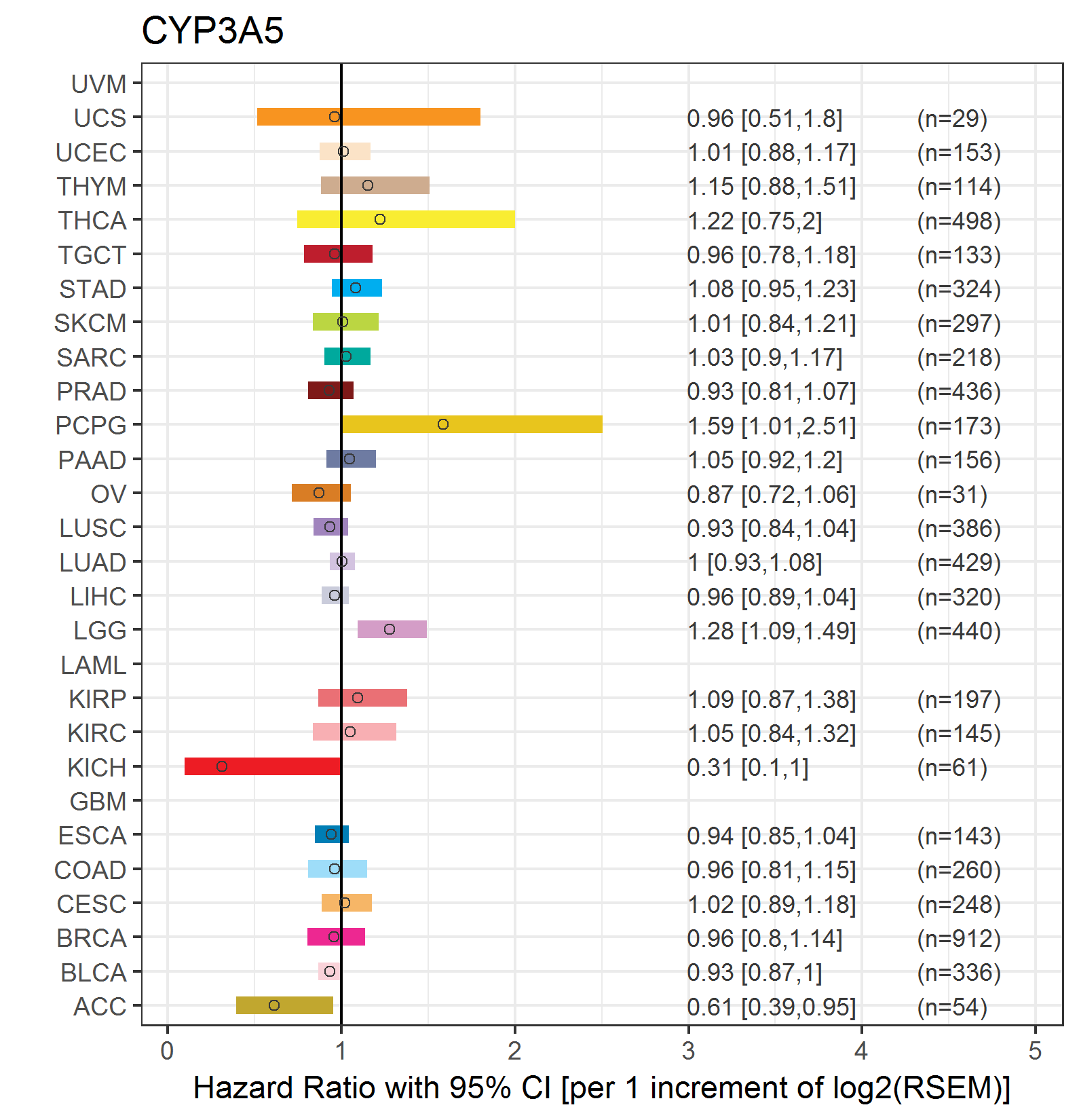
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























