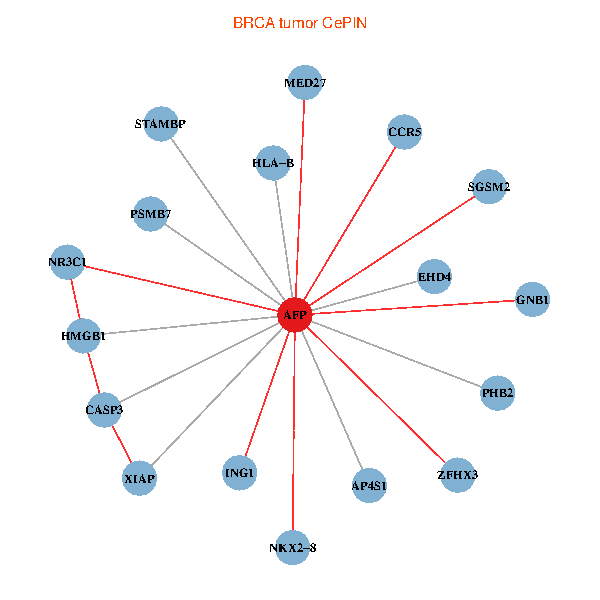
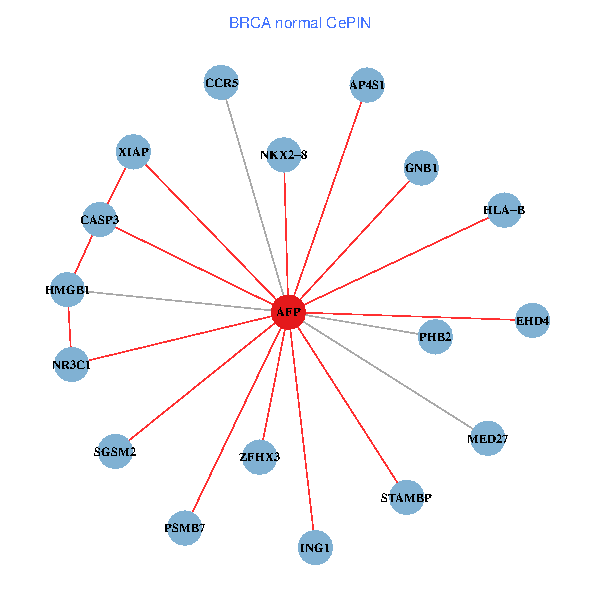
| BRCA (tumor) | BRCA (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
 |  |
|
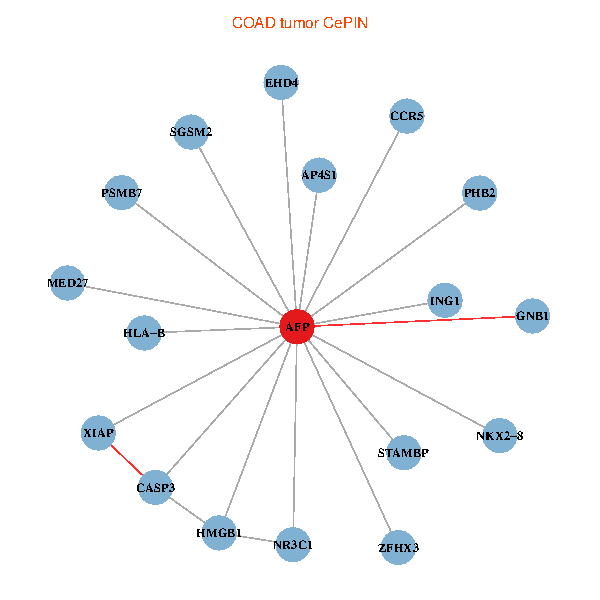
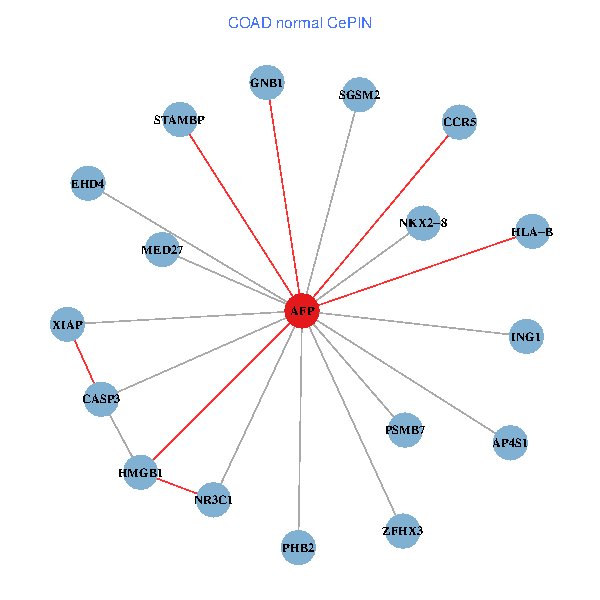
| COAD (tumor) | COAD (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
 |  |
|
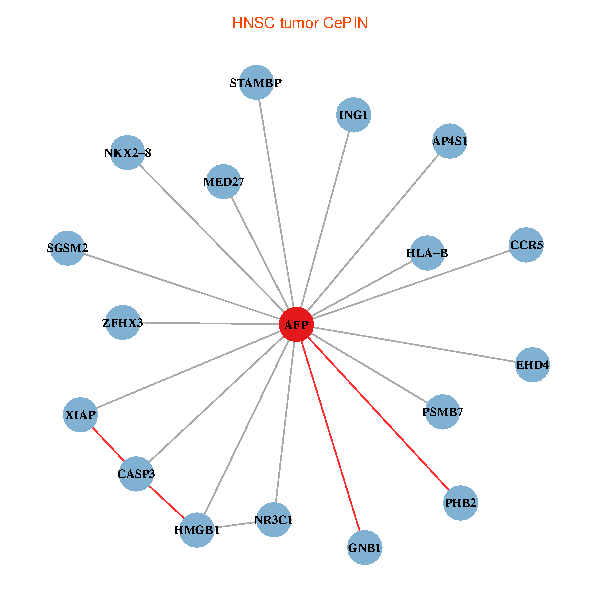
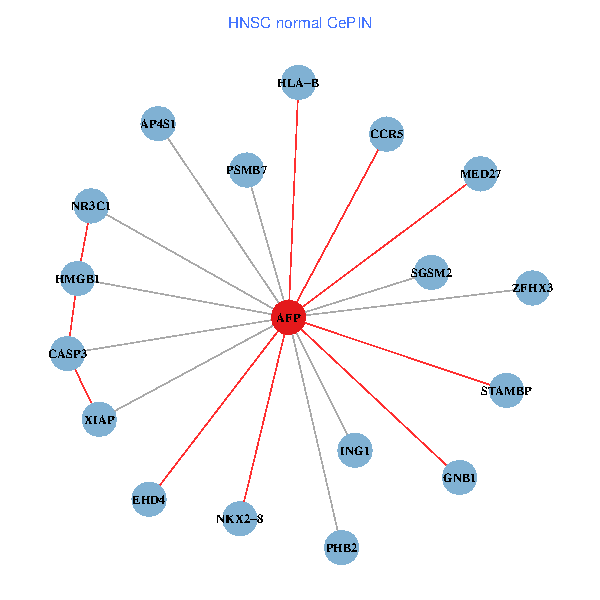
| HNSC (tumor) | HNSC (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
 |  |
|
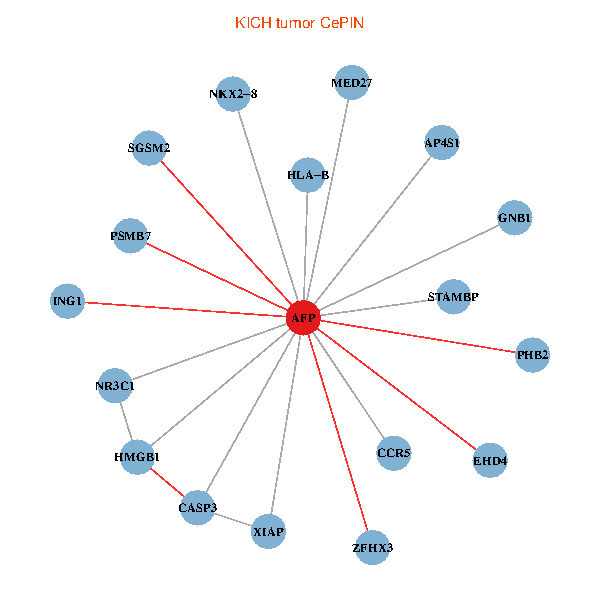
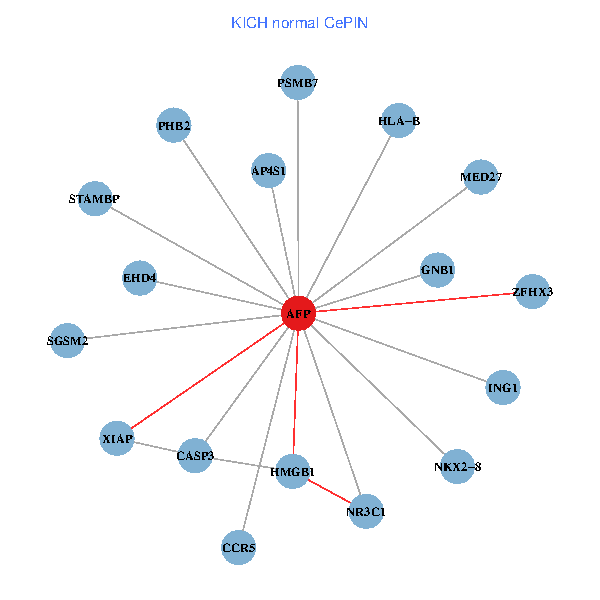
| KICH (tumor) | KICH (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
 |  |
|
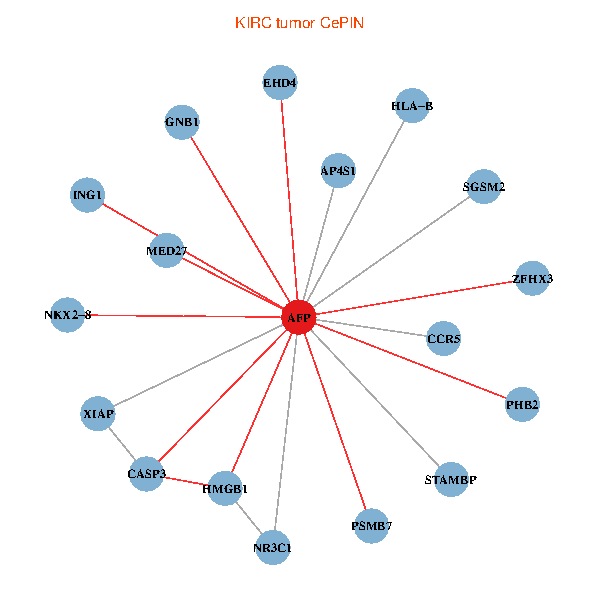
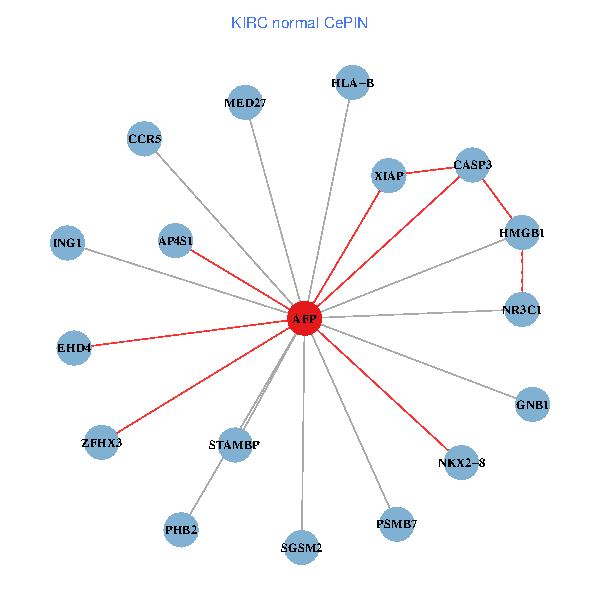
| KIRC (tumor) | KIRC (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
 |  |
|
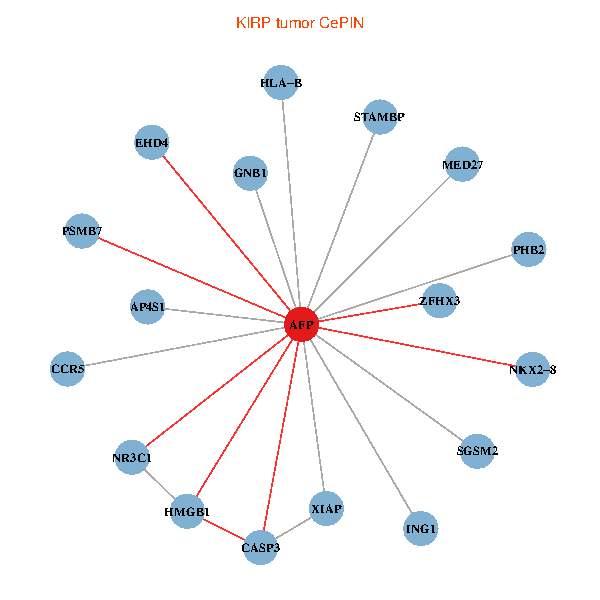
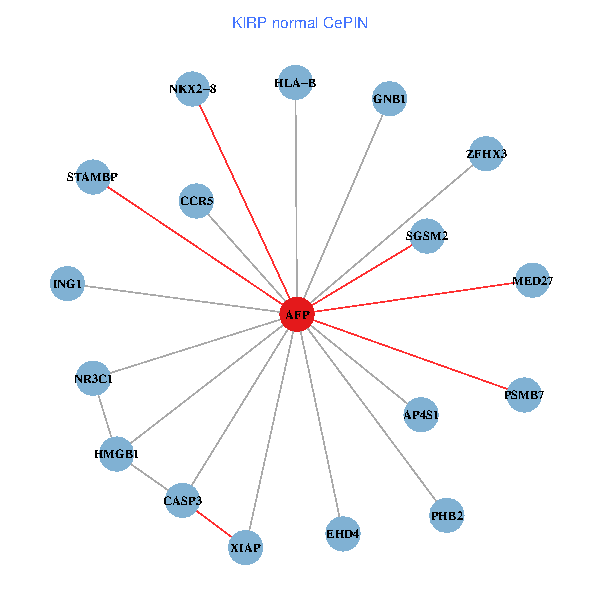
| KIRP (tumor) | KIRP (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
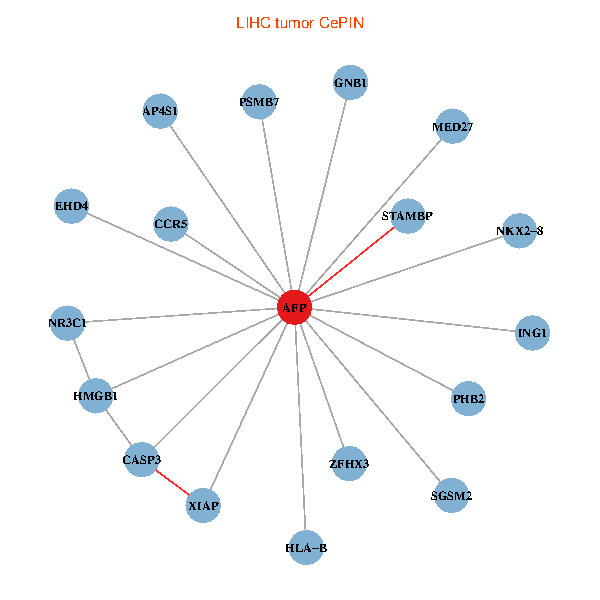 | 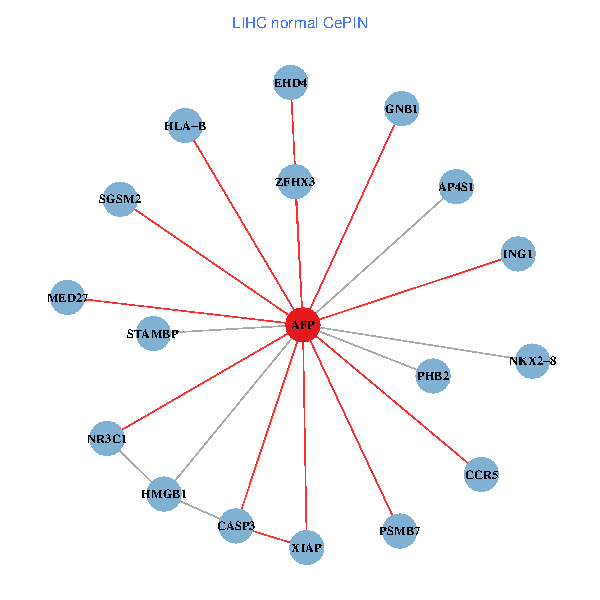 |
|
| LUAD (tumor) | LUAD (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
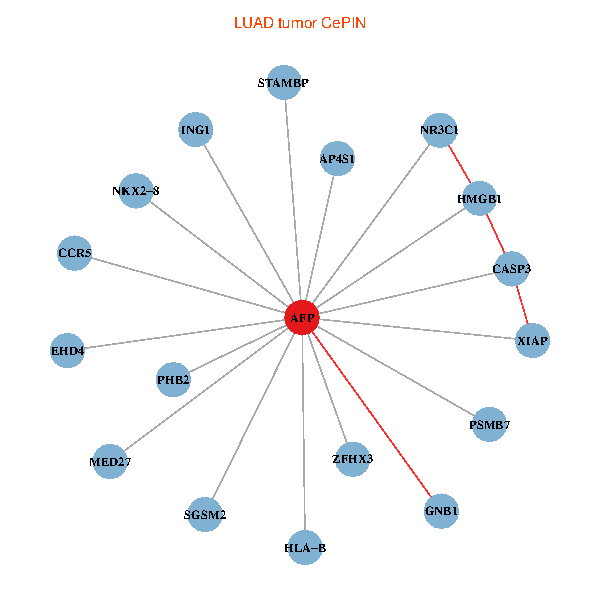 | 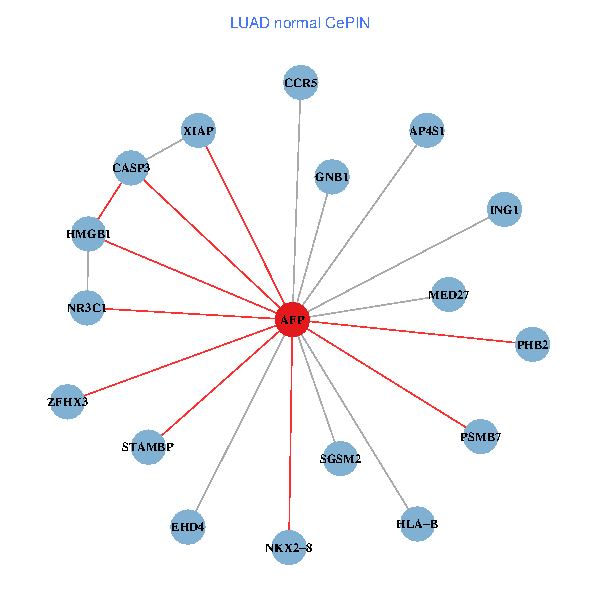 |
|
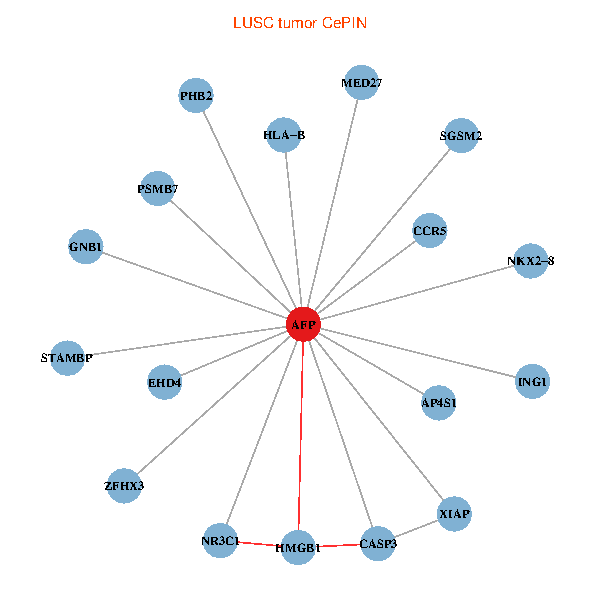
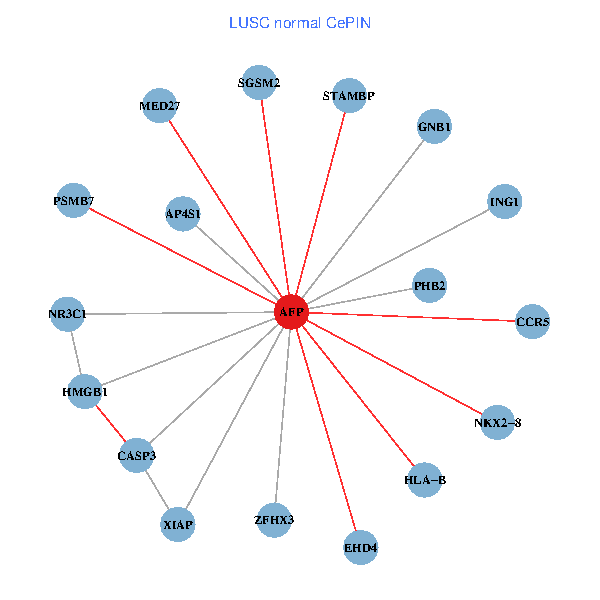
| LUSC (tumor) | LUSC (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
 |  |
|
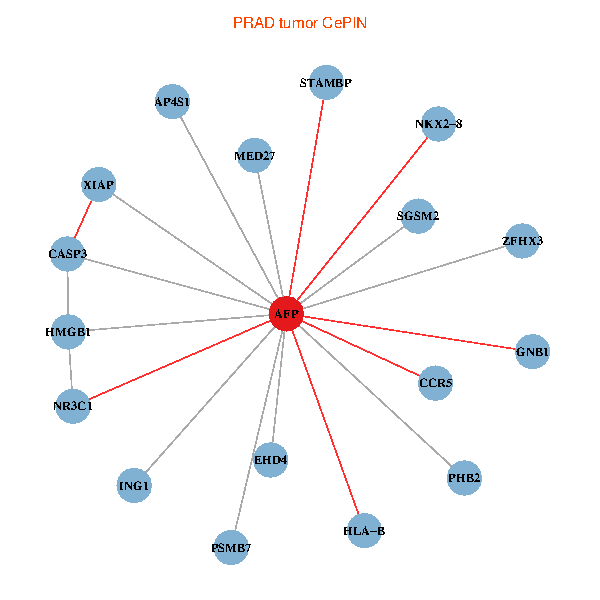
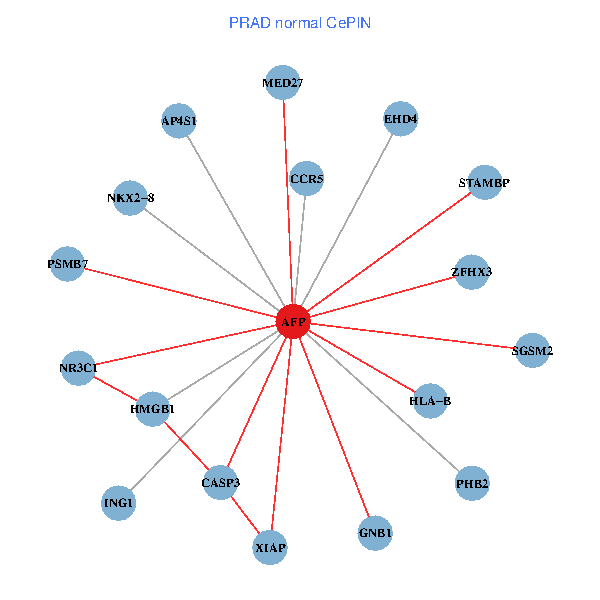
| PRAD (tumor) | PRAD (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
 |  |
|
| STAD (tumor) | STAD (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
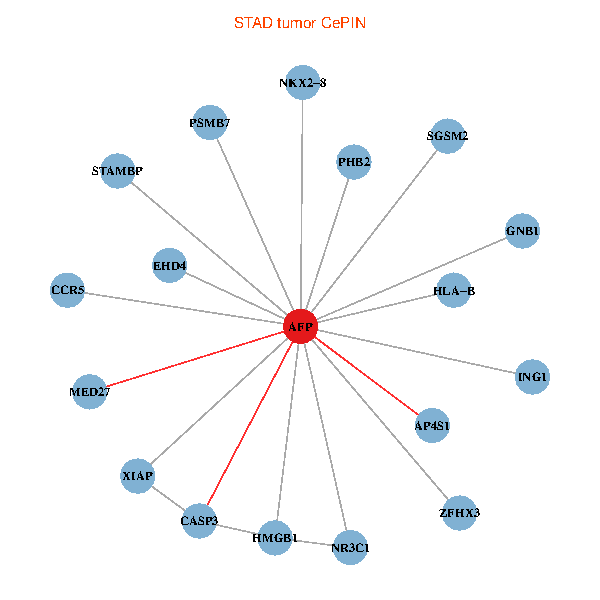 | 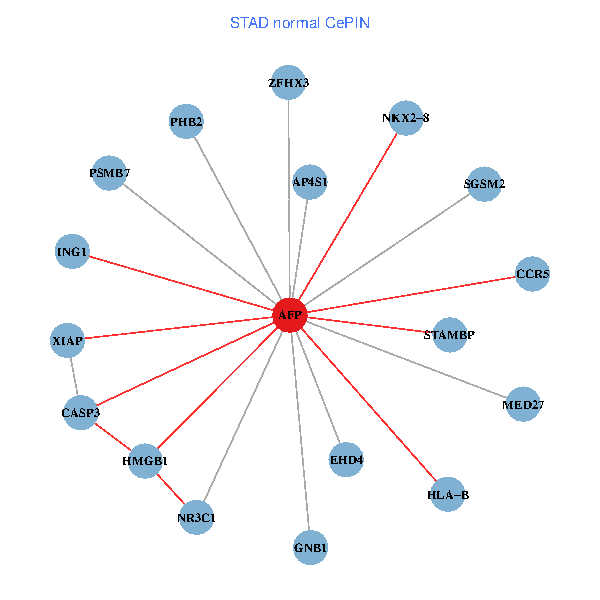 |
|
| THCA (tumor) | THCA (normal) |
| AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (tumor) | AFP, HMGB1, NR3C1, CASP3, HLA-B, PSMB7, ING1, XIAP, SGSM2, GNB1, PHB2, STAMBP, MED27, CCR5, ZFHX3, EHD4, AP4S1, NKX2-8 (normal) |
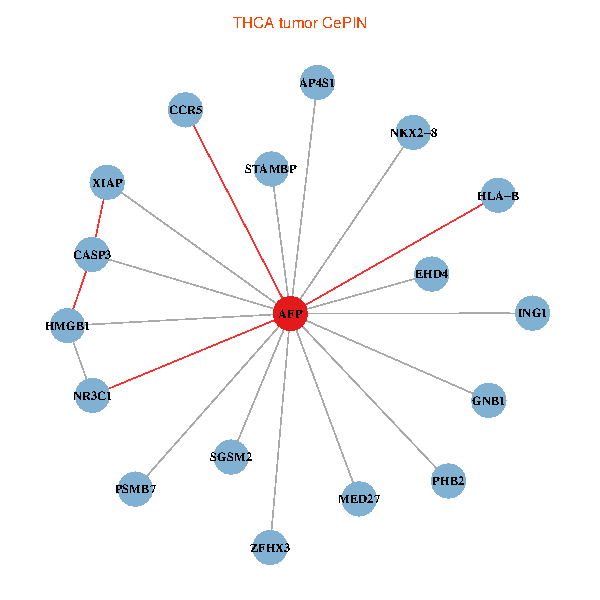 | 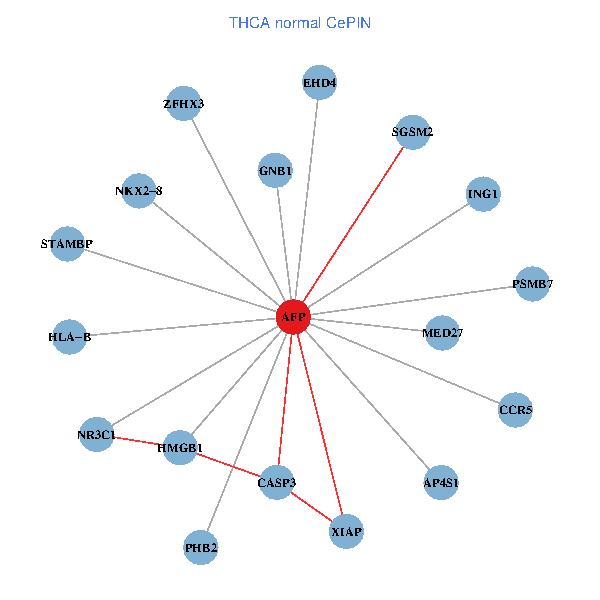 |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C2239176 | Liver carcinoma | 425 | BeFree,CTD_human,LHGDN |
| umls:C0023903 | Liver neoplasms | 83 | BeFree,LHGDN |
| umls:C0019163 | Hepatitis B | 30 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 30 | BeFree |
| umls:C0345904 | Malignant neoplasm of liver | 30 | BeFree |
| umls:C0023890 | Liver Cirrhosis | 26 | BeFree |
| umls:C0013080 | Down Syndrome | 21 | BeFree |
| umls:C0279000 | Liver and Intrahepatic Biliary Tract Carcinoma | 19 | BeFree |
| umls:C0596263 | Carcinogenesis | 15 | BeFree |
| umls:C1623038 | Cirrhosis | 15 | BeFree |
| umls:C0521158 | Recurrent tumor | 12 | BeFree |
| umls:C0699791 | Stomach Carcinoma | 12 | BeFree |
| umls:C0004135 | Ataxia Telangiectasia | 11 | BeFree |
| umls:C0023895 | Liver diseases | 10 | BeFree,CTD_human |
| umls:C0024623 | Malignant neoplasm of stomach | 10 | BeFree |
| umls:C1512409 | Hepatocarcinogenesis | 10 | BeFree |
| umls:C0019348 | Herpes Simplex Infections | 9 | BeFree |
| umls:C0205851 | Germ cell tumor | 9 | BeFree |
| umls:C0004134 | Ataxia | 8 | BeFree |
| umls:C0041107 | Trisomy | 8 | BeFree |
| umls:C0152096 | Complete trisomy 18 syndrome | 8 | BeFree |
| umls:C0206624 | Hepatoblastoma | 8 | BeFree |
| umls:C0524910 | Hepatitis C, Chronic | 8 | BeFree |
| umls:C0019158 | Hepatitis | 7 | BeFree |
| umls:C0019159 | Hepatitis A | 7 | BeFree |
| umls:C0008626 | Congenital chromosomal disease | 6 | BeFree |
| umls:C0019189 | Hepatitis, Chronic | 6 | BeFree |
| umls:C0206698 | Cholangiocarcinoma | 5 | BeFree,LHGDN |
| umls:C0494165 | Secondary malignant neoplasm of liver | 5 | BeFree |
| umls:C0001418 | Adenocarcinoma | 4 | BeFree |
| umls:C0019196 | Hepatitis C | 4 | BeFree |
| umls:C0178874 | Tumor Progression | 4 | BeFree |
| umls:C0206659 | Embryonal Carcinoma | 4 | BeFree |
| umls:C0278701 | Gastric Adenocarcinoma | 4 | BeFree |
| umls:C0000768 | Congenital Abnormality | 3 | BeFree |
| umls:C0014145 | Yolk Sac Tumor | 3 | BeFree |
| umls:C0024299 | Lymphoma | 3 | BeFree |
| umls:C0024620 | Primary malignant neoplasm of liver | 3 | BeFree |
| umls:C0036631 | Seminoma | 3 | BeFree |
| umls:C0039446 | Telangiectasis | 3 | BeFree |
| umls:C0042769 | Virus Diseases | 3 | BeFree |
| umls:C0260037 | Multiple tumors | 3 | BeFree |
| umls:C0271270 | Oculovestibuloauditory syndrome | 3 | BeFree |
| umls:C0346647 | Malignant neoplasm of pancreas | 3 | BeFree |
| umls:C1318485 | Liver regeneration disorder | 3 | BeFree |
| umls:C3163918 | Tumor thrombus | 3 | BeFree |
| umls:C3489733 | Oculomotor apraxia | 3 | BeFree |
| umls:C3501848 | Nephrosis, congenital | 3 | BeFree |
| umls:C0004903 | Beckwith-Wiedemann Syndrome | 2 | BeFree,LHGDN |
| umls:C0006142 | Malignant neoplasm of breast | 2 | BeFree |
| umls:C0007758 | Cerebellar Ataxia | 2 | BeFree |
| umls:C0012546 | Diphtheria | 2 | BeFree |
| umls:C0029925 | Ovarian Carcinoma | 2 | BeFree |
| umls:C0031117 | Peripheral Neuropathy | 2 | BeFree |
| umls:C0036868 | Sex Chromosome Aberrations | 2 | BeFree |
| umls:C0039538 | Teratoma | 2 | BeFree |
| umls:C0149925 | Small cell carcinoma of lung | 2 | BeFree |
| umls:C0153594 | Malignant neoplasm of testis | 2 | BeFree |
| umls:C0235974 | Pancreatic carcinoma | 2 | BeFree |
| umls:C0239946 | Fibrosis, Liver | 2 | BeFree |
| umls:C0266159 | Pyloric Atresia | 2 | BeFree |
| umls:C0275524 | Coinfection | 2 | BeFree |
| umls:C0280100 | Solid tumour | 2 | BeFree |
| umls:C0333693 | Triploidy syndrome | 2 | BeFree |
| umls:C0334520 | Teratoma, Malignant | 2 | BeFree |
| umls:C0442874 | Neuropathy | 2 | BeFree |
| umls:C0677886 | Epithelial ovarian cancer | 2 | BeFree |
| umls:C0678222 | Breast Carcinoma | 2 | BeFree |
| umls:C0740279 | Cerebellar atrophy | 2 | BeFree |
| umls:C0740380 | Varicella zoster | 2 | BeFree |
| umls:C0795801 | trisomy 2 | 2 | BeFree |
| umls:C0855197 | Testicular malignant germ cell tumor | 2 | BeFree |
| umls:C1266090 | Hepatoid adenocarcinoma | 2 | BeFree |
| umls:C1336708 | Testicular Germ Cell Tumor | 2 | BeFree |
| umls:C1519680 | Tumor Immunity | 2 | BeFree |
| umls:C1853761 | SPINOCEREBELLAR ATAXIA, AUTOSOMAL RECESSIVE 1 | 2 | BeFree |
| umls:C0001430 | Adenoma | 1 | BeFree |
| umls:C0002793 | Anaplasia | 1 | BeFree |
| umls:C0003130 | Anoxia | 1 | LHGDN |
| umls:C0003635 | Apraxias | 1 | BeFree |
| umls:C0005411 | Biliary Atresia | 1 | BeFree |
| umls:C0007102 | Malignant tumor of colon | 1 | BeFree |
| umls:C0007137 | Squamous cell carcinoma | 1 | BeFree |
| umls:C0009207 | Cockayne Syndrome | 1 | BeFree |
| umls:C0010674 | Cystic Fibrosis | 1 | BeFree |
| umls:C0011849 | Diabetes Mellitus | 1 | BeFree |
| umls:C0013377 | Dysgerminoma | 1 | BeFree |
| umls:C0013930 | Embolism, Tumor | 1 | BeFree |
| umls:C0018050 | Gonadal Disorders | 1 | BeFree |
| umls:C0019322 | Umbilical hernia | 1 | BeFree |
| umls:C0020443 | Hypercholesterolemia | 1 | BeFree |
| umls:C0020615 | Hypoglycemia | 1 | BeFree |
| umls:C0022658 | Kidney Diseases | 1 | BeFree |
| umls:C0022660 | Kidney Failure, Acute | 1 | BeFree |
| umls:C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| umls:C0023904 | Liver Neoplasms, Experimental | 1 | CTD_human |
| umls:C0024668 | Mammary Neoplasms, Experimental | 1 | CTD_human |
| umls:C0026650 | Movement Disorders | 1 | BeFree |
| umls:C0027613 | Neonatal hepatitis | 1 | BeFree |
| umls:C0027658 | Neoplasms, Germ Cell and Embryonal | 1 | LHGDN |
| umls:C0027773 | Nesidioblastosis | 1 | BeFree |
| umls:C0028860 | Oculocerebrorenal Syndrome | 1 | BeFree |
| umls:C0032580 | Adenomatous Polyposis Coli | 1 | BeFree |
| umls:C0035369 | Retroviridae Infections | 1 | BeFree |
| umls:C0038356 | Stomach Neoplasms | 1 | CTD_human |
| umls:C0038454 | Cerebrovascular accident | 1 | BeFree |
| umls:C0042721 | Viral hepatitis | 1 | BeFree |
| umls:C0042963 | Vomiting | 1 | BeFree |
| umls:C0080178 | Spina Bifida | 1 | BeFree |
| umls:C0085548 | Autosomal Recessive Polycystic Kidney Disease | 1 | BeFree |
| umls:C0085605 | Liver Failure | 1 | BeFree |
| umls:C0151747 | Renal tubular disorder | 1 | BeFree |
| umls:C0162530 | Porphyria, Erythropoietic | 1 | BeFree |
| umls:C0162872 | Aortic Aneurysm, Thoracic | 1 | BeFree |
| umls:C0205824 | Liposarcoma, Dedifferentiated | 1 | BeFree |
| umls:C0206664 | Teratocarcinoma | 1 | BeFree |
| umls:C0206666 | Trophoblastic Tumor, Placental Site | 1 | BeFree |
| umls:C0206716 | Ganglioglioma | 1 | BeFree |
| umls:C0206754 | Neuroendocrine Tumors | 1 | LHGDN |
| umls:C0235325 | Gastric hemorrhage | 1 | BeFree |
| umls:C0235833 | Congenital diaphragmatic hernia | 1 | BeFree |
| umls:C0265706 | Gastroschisis | 1 | BeFree |
| umls:C0270246 | Meconium plug syndrome | 1 | BeFree |
| umls:C0279661 | Acinar cell carcinoma of pancreas | 1 | BeFree |
| umls:C0332890 | Congenital hemihypertrophy | 1 | BeFree |
| umls:C0334529 | Hydatidiform Mole, Partial | 1 | BeFree |
| umls:C0341439 | Chronic liver disease | 1 | BeFree |
| umls:C0346627 | Intestinal Cancer | 1 | BeFree |
| umls:C0400936 | Autoimmune liver disease | 1 | BeFree |
| umls:C0443306 | Spastic | 1 | BeFree |
| umls:C0496870 | Benign neoplasm of liver | 1 | BeFree |
| umls:C0524909 | Hepatitis B, Chronic | 1 | BeFree |
| umls:C0546982 | Cystic fibrosis with meconium ileus | 1 | BeFree |
| umls:C0677607 | Hashimoto Disease | 1 | BeFree |
| umls:C0699790 | Colon Carcinoma | 1 | BeFree |
| umls:C0796154 | SIMPSON-GOLABI-BEHMEL SYNDROME, TYPE 1 | 1 | BeFree |
| umls:C0877373 | Advanced cancer | 1 | BeFree |
| umls:C0920350 | Autoimmune thyroiditis | 1 | BeFree |
| umls:C0950121 | Denys-Drash Syndrome | 1 | BeFree |
| umls:C1266158 | Nonseminomatous germ cell tumor | 1 | BeFree |
| umls:C1285291 | Fetal ascites | 1 | BeFree |
| umls:C1306503 | Congenital exomphalos | 1 | BeFree |
| umls:C1368683 | Epithelioma | 1 | BeFree |
| umls:C1384670 | Single umbilical artery | 1 | BeFree |
| umls:C1389280 | Basal ganglia calcification | 1 | BeFree |
| umls:C1458155 | Mammary Neoplasms | 1 | CTD_human |
| umls:C1519670 | Tumor Angiogenesis | 1 | BeFree |
| umls:C1833561 | UV-Sensitive Syndrome | 1 | BeFree |
| umls:C1840311 | Laryngeal cleft | 1 | BeFree |
| umls:C1856184 | HEMIHYPERPLASIA, ISOLATED | 1 | BeFree |
| umls:C2939175 | Meconium ileus | 1 | BeFree |
| umls:C2986703 | Overgrowth Syndrome | 1 | BeFree |
| umls:C1863080 | ALPHA-FETOPROTEIN, HEREDITARY PERSISTENCE OF | 0 | CLINVAR,ORPHANET |
| umls:C1863081 | alpha-Fetoprotein Deficiency | 0 | CLINVAR,ORPHANET |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))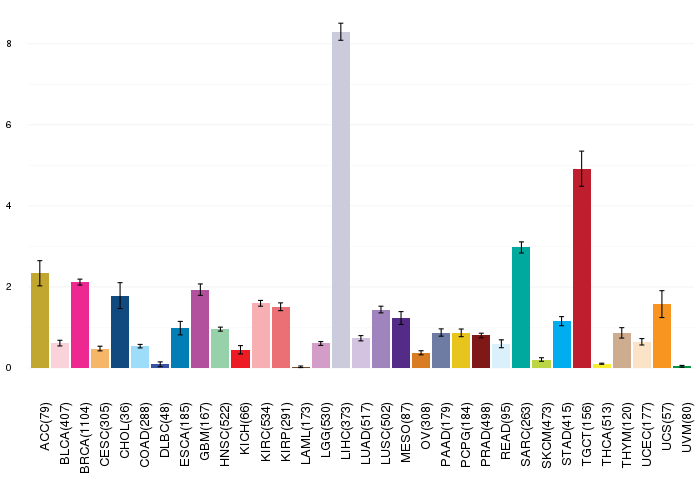
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))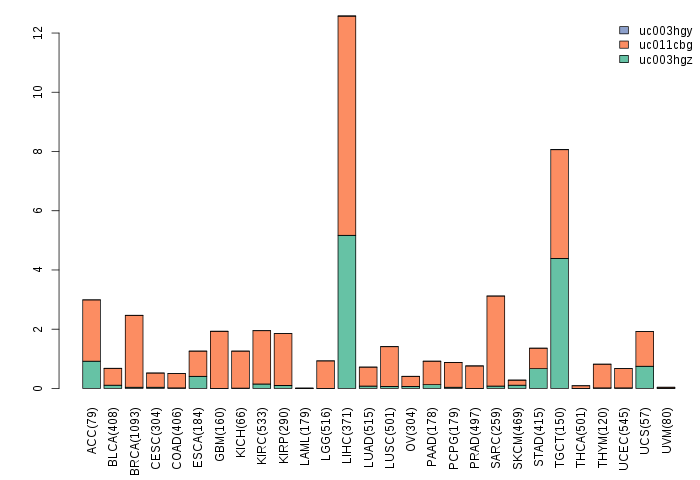
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data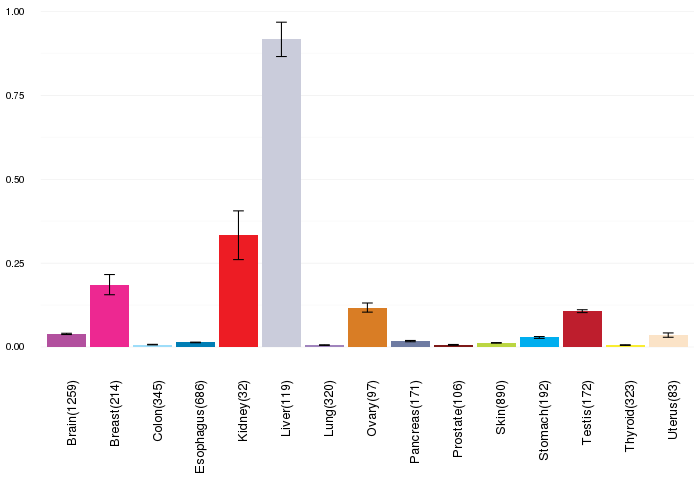
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))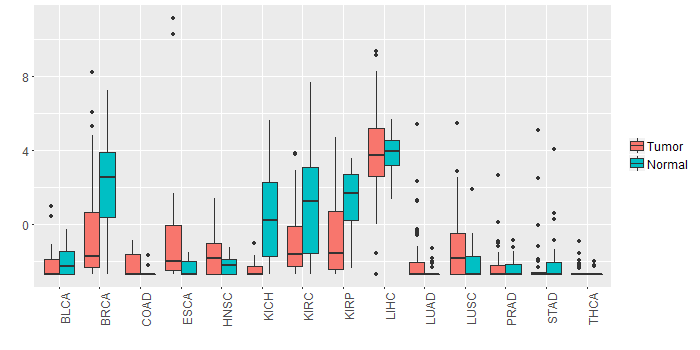
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.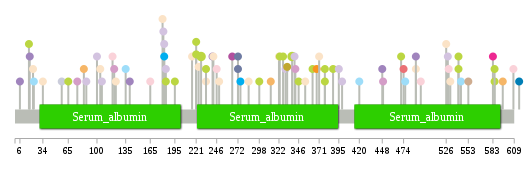

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 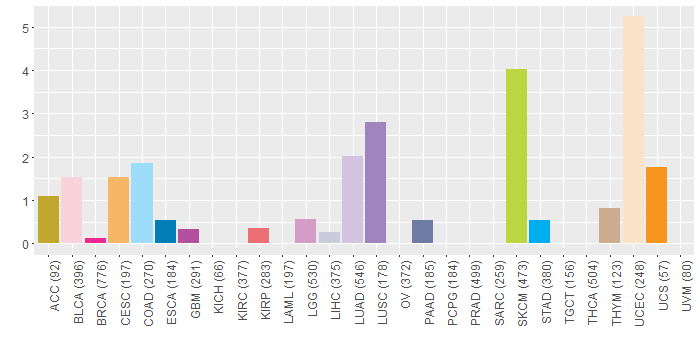
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)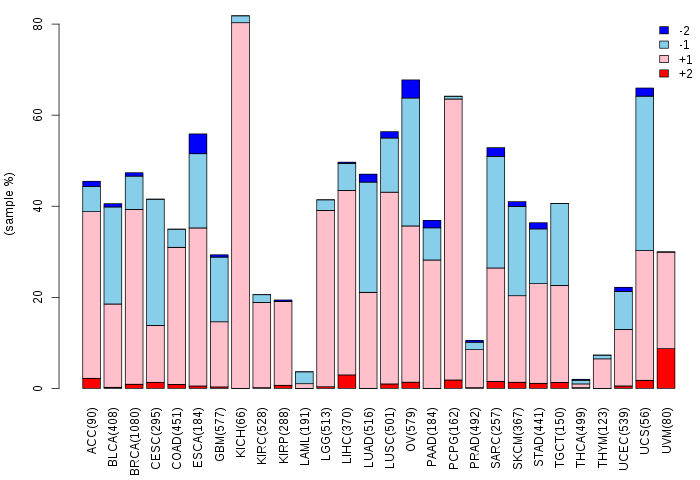
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 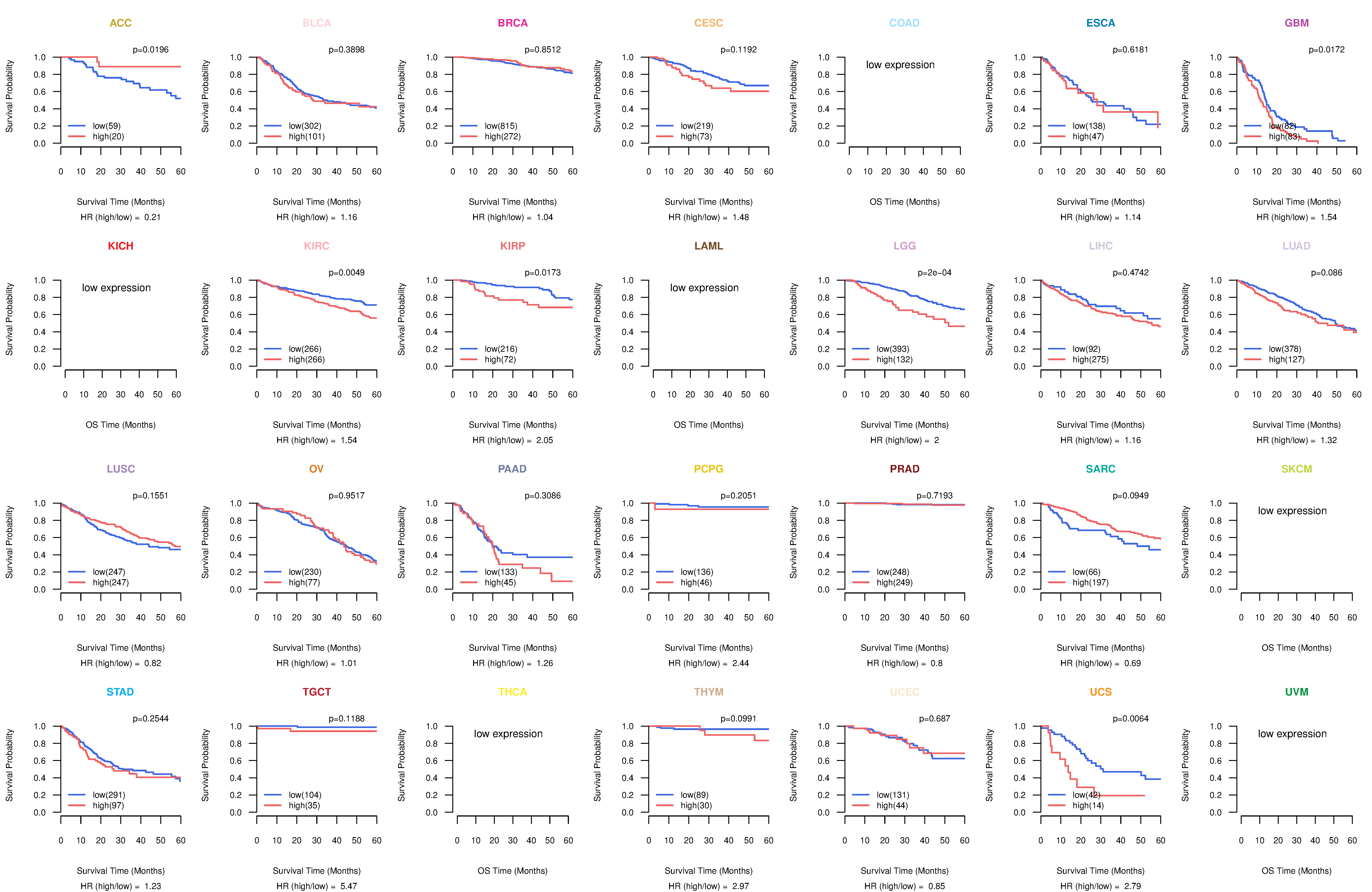
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 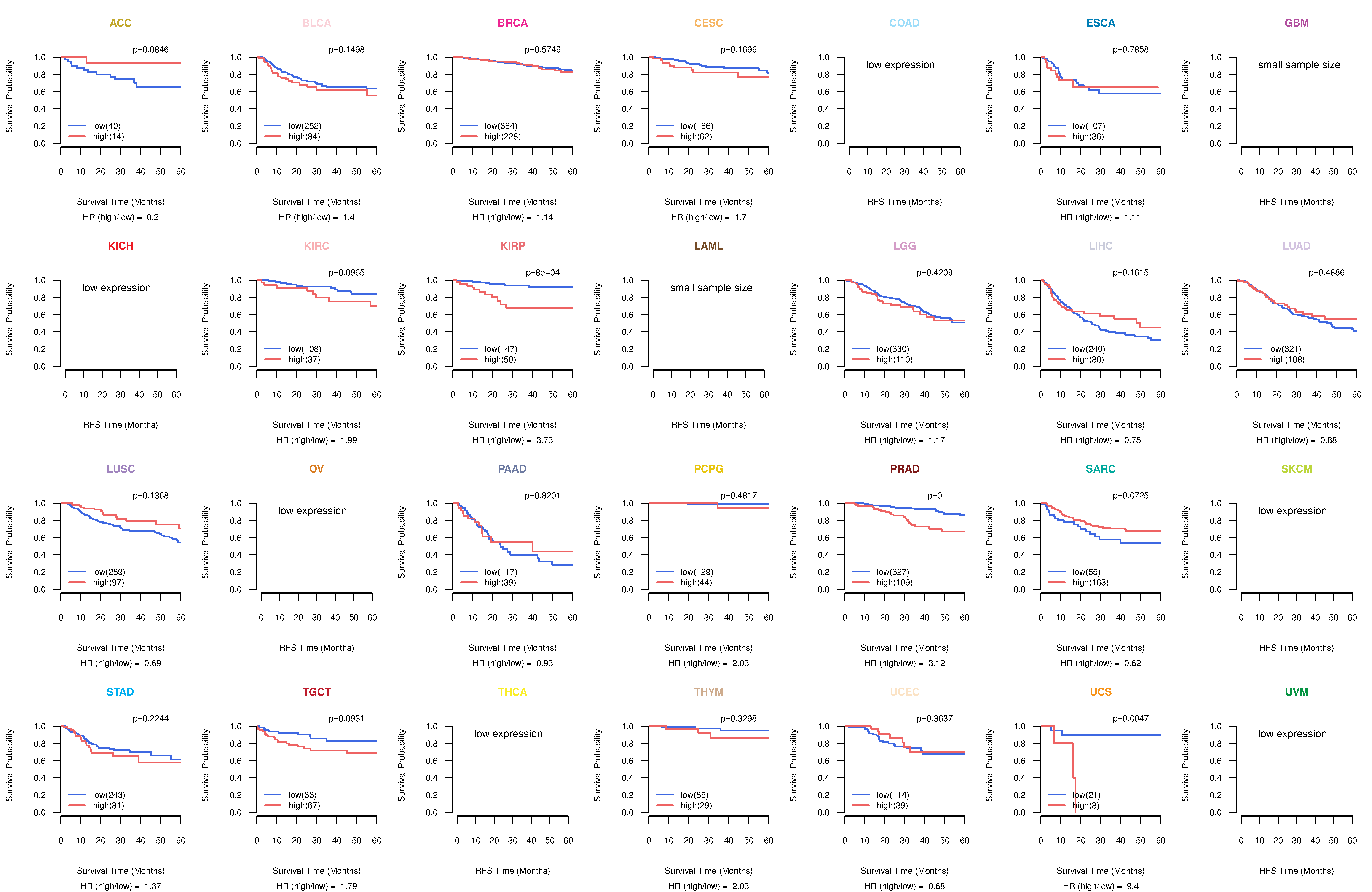
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 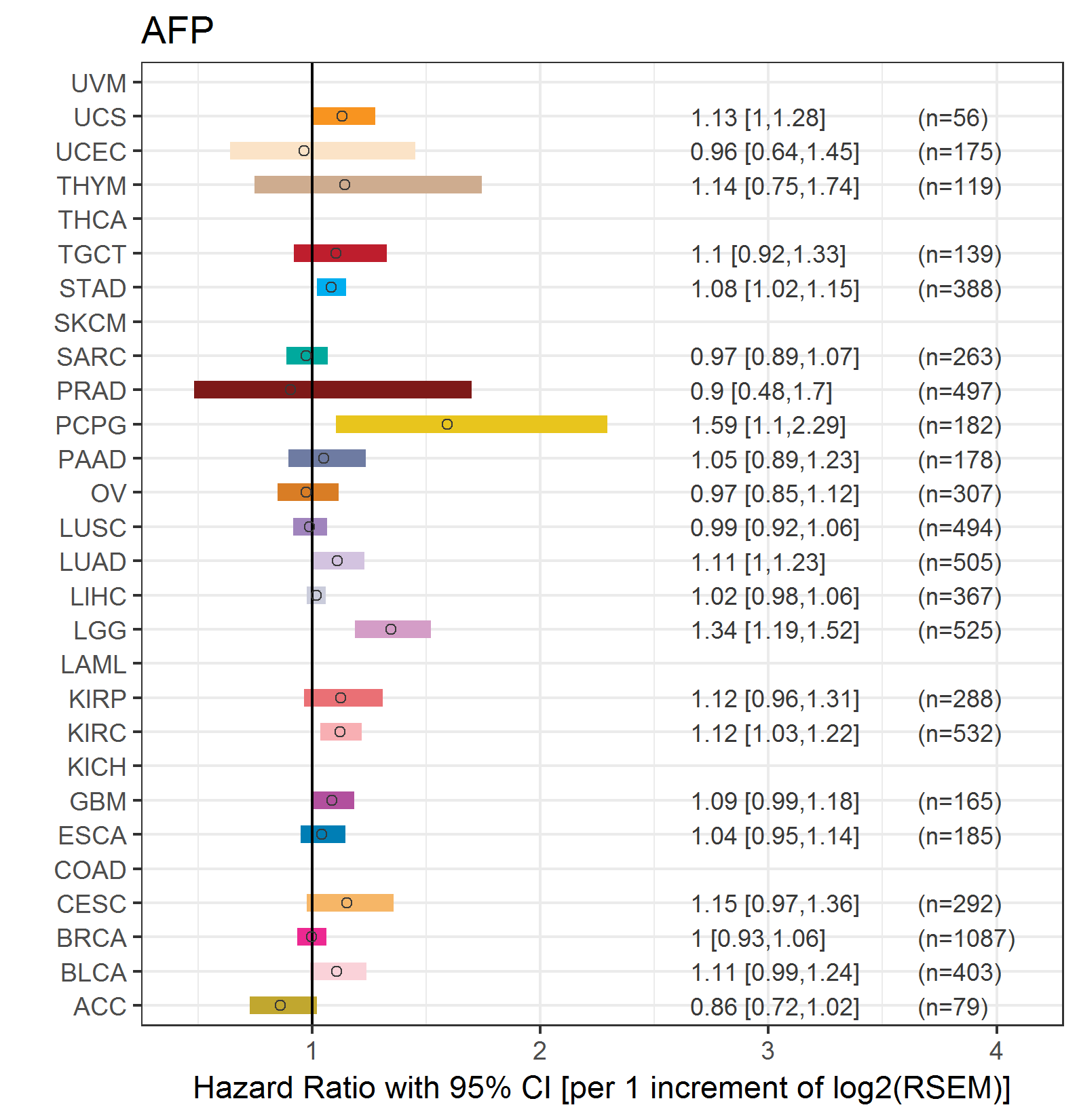
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 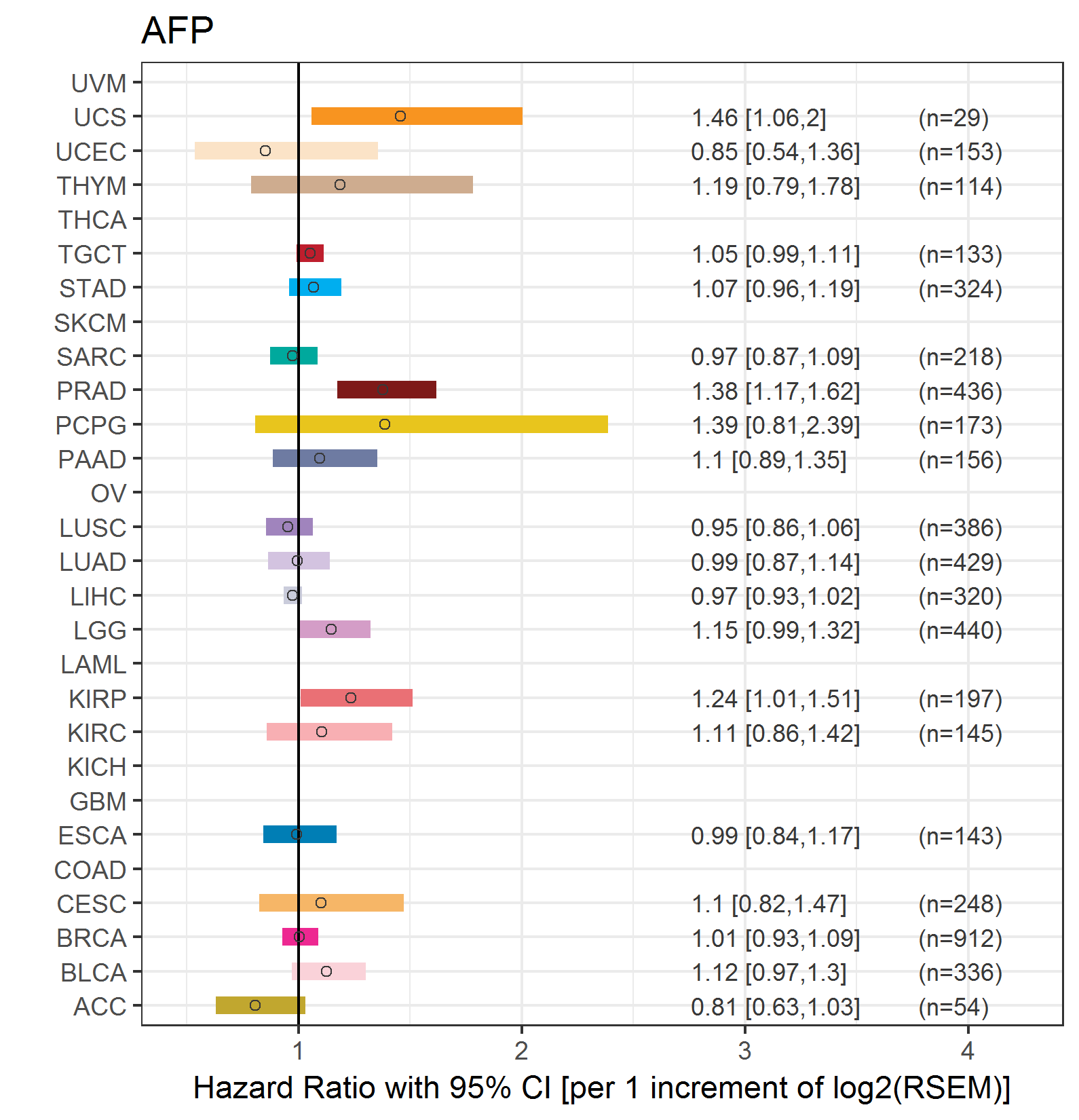
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























