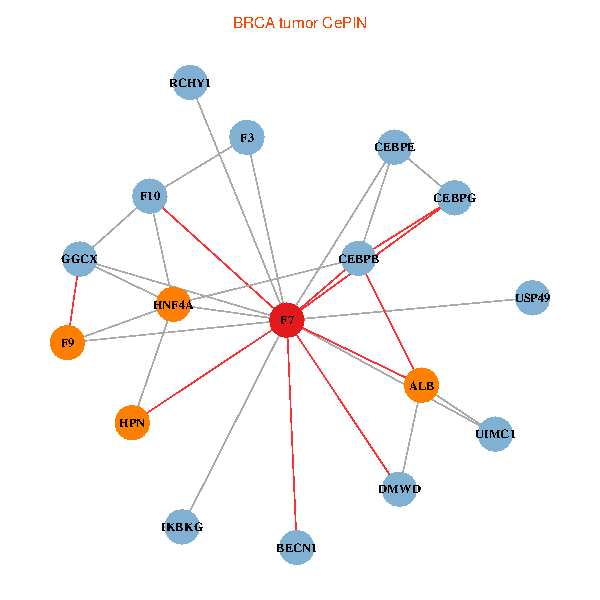
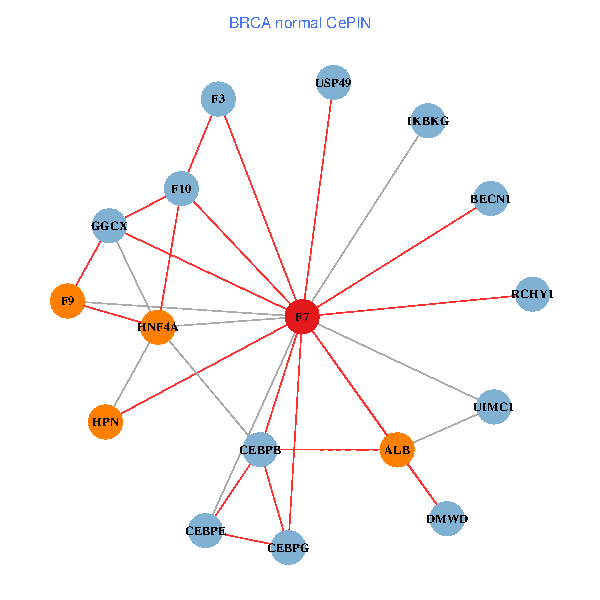
| BRCA (tumor) | BRCA (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
 |  |
|
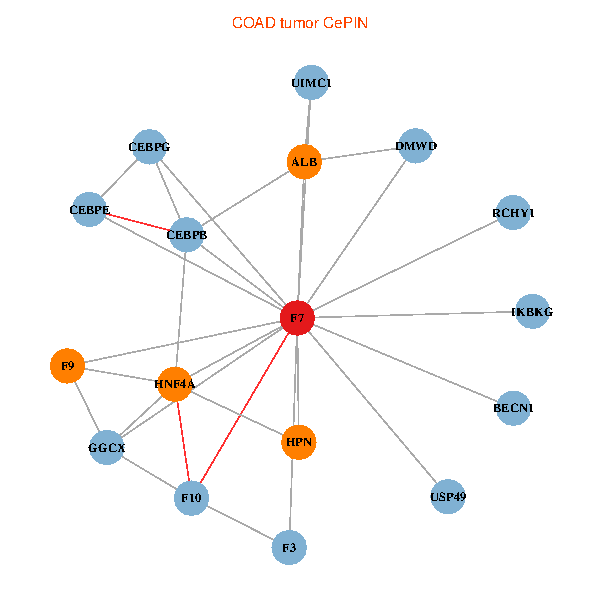
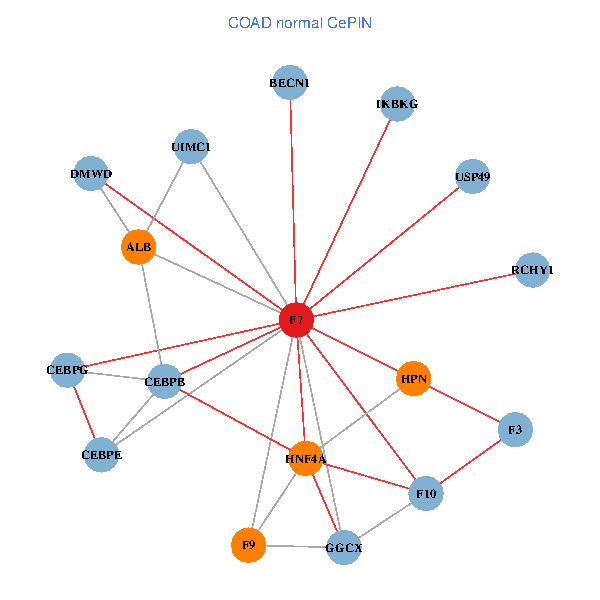
| COAD (tumor) | COAD (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
 |  |
|
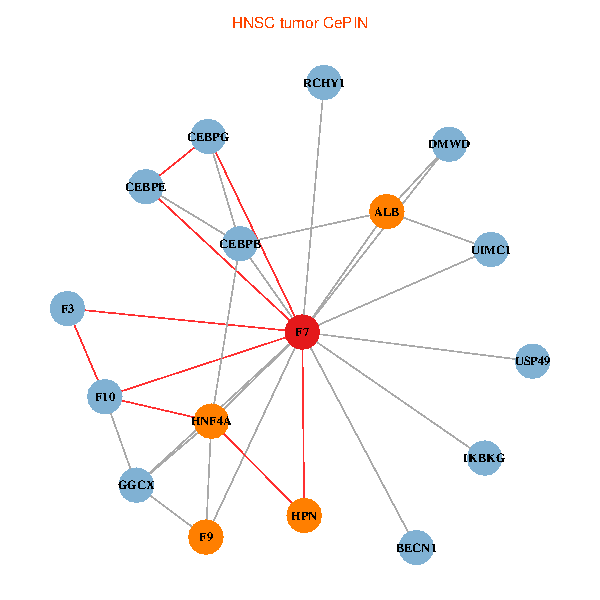
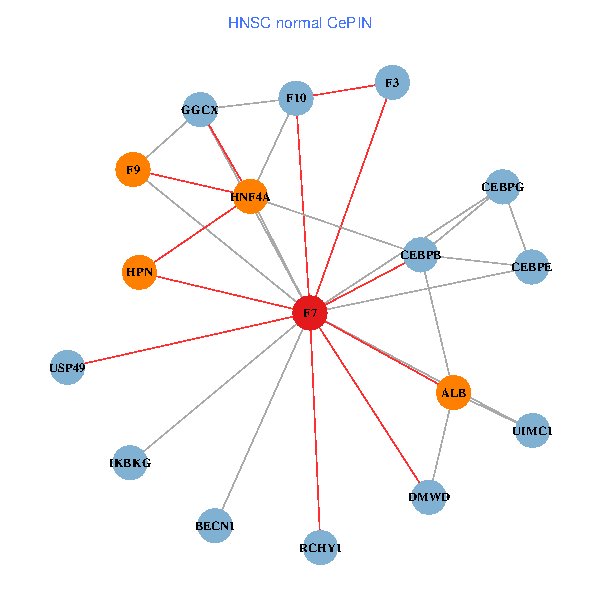
| HNSC (tumor) | HNSC (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
 |  |
|
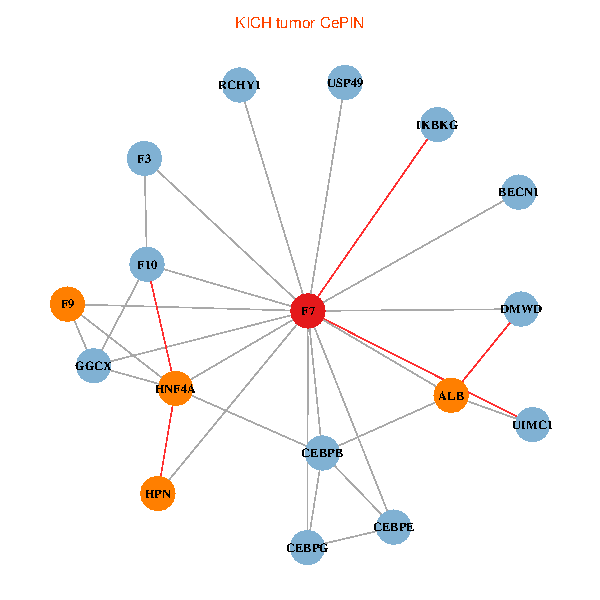
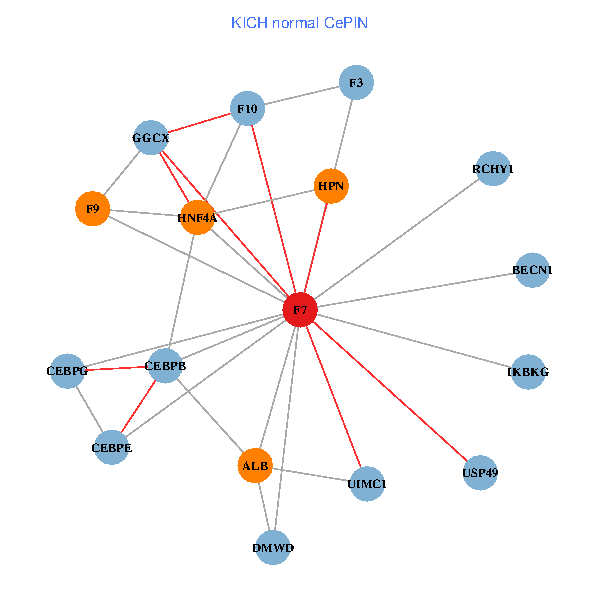
| KICH (tumor) | KICH (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
 |  |
|
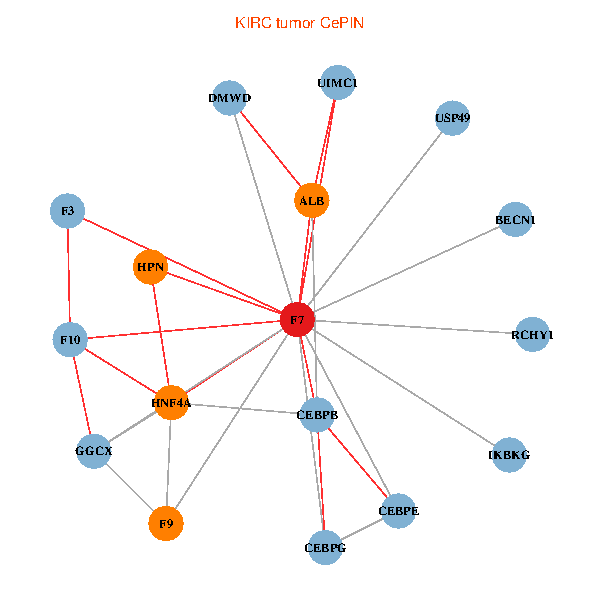
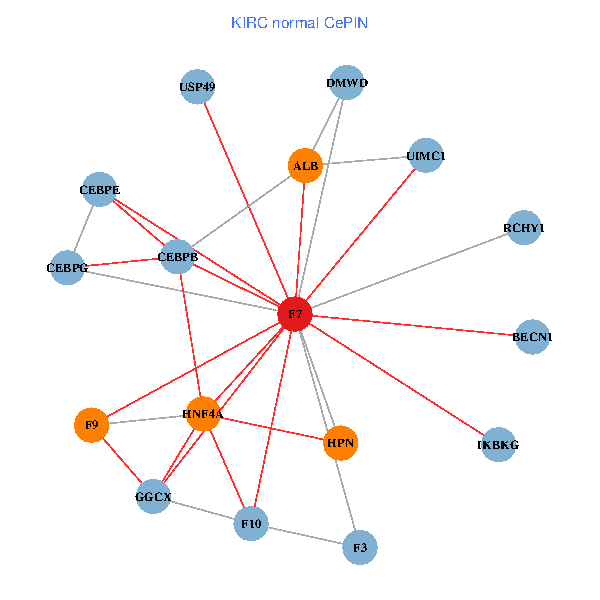
| KIRC (tumor) | KIRC (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
 |  |
|
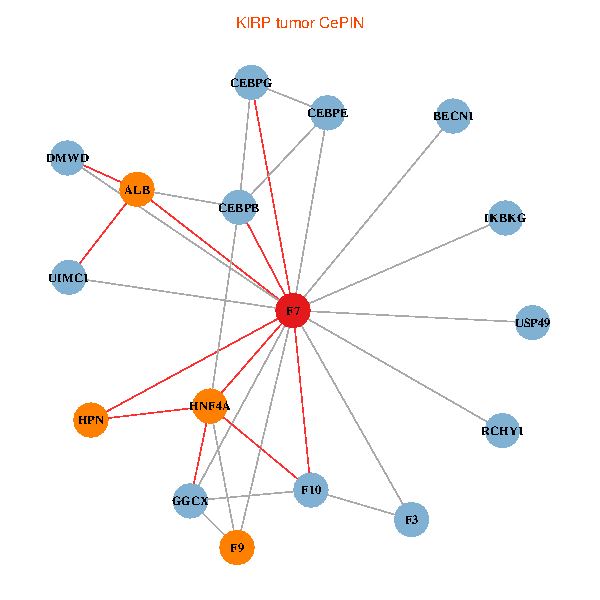
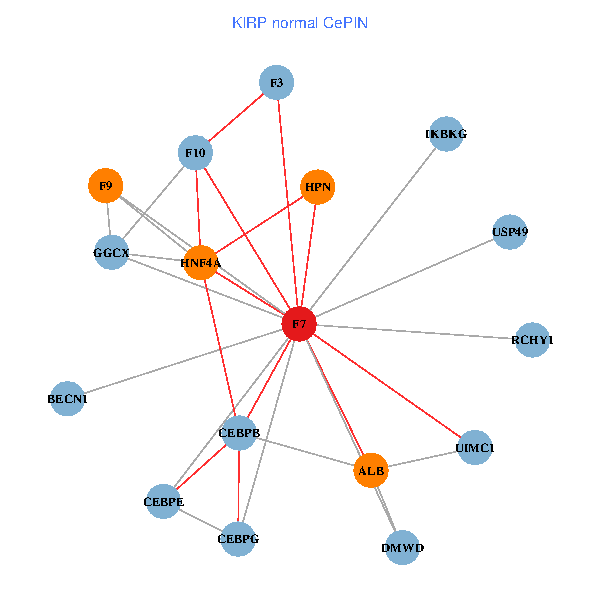
| KIRP (tumor) | KIRP (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
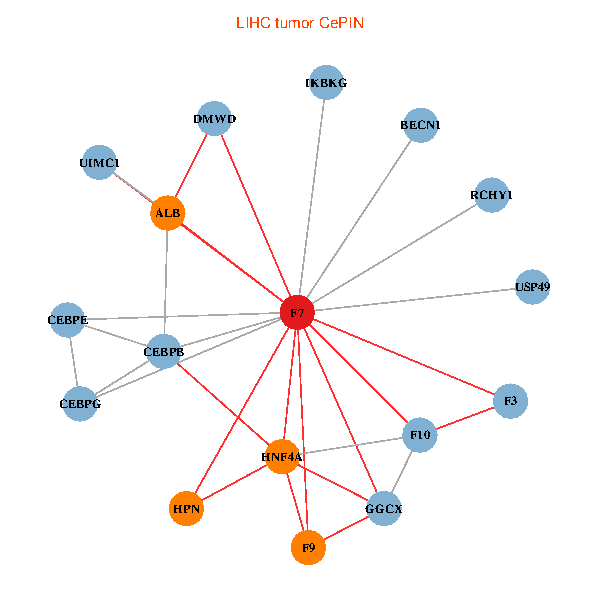 | 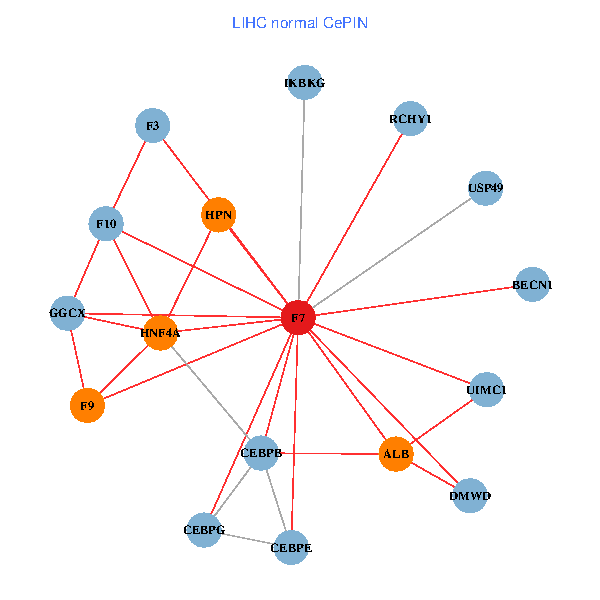 |
|
| LUAD (tumor) | LUAD (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
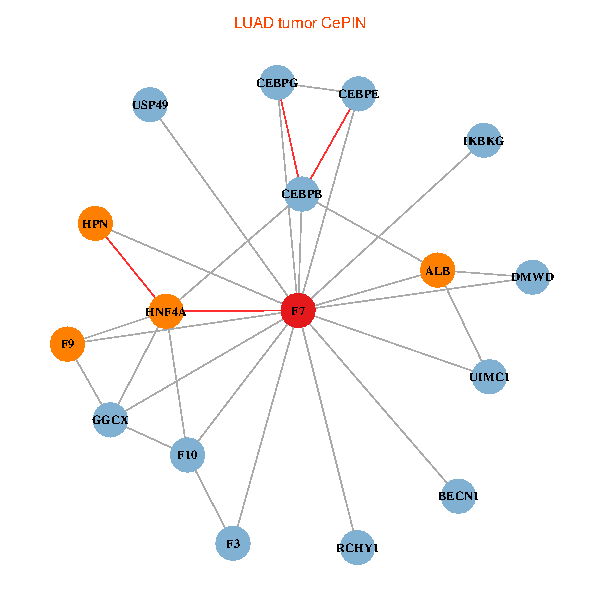 | 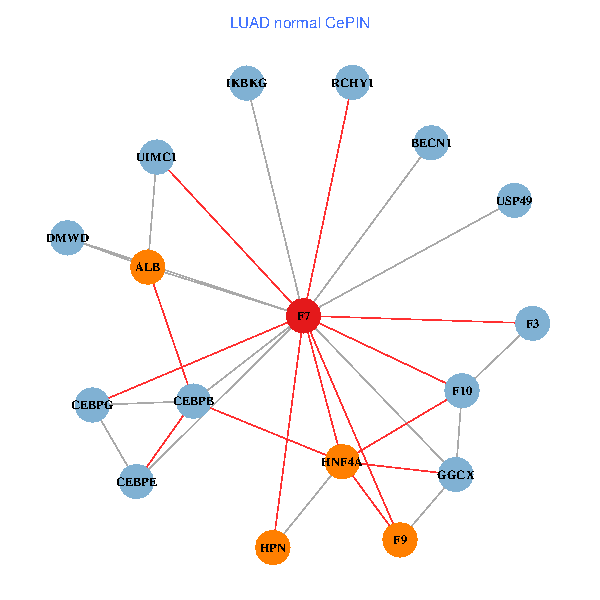 |
|
| LUSC (tumor) | LUSC (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
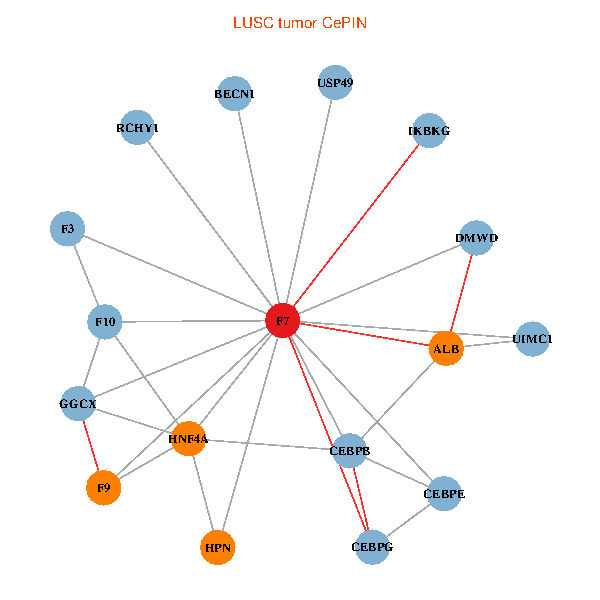 | 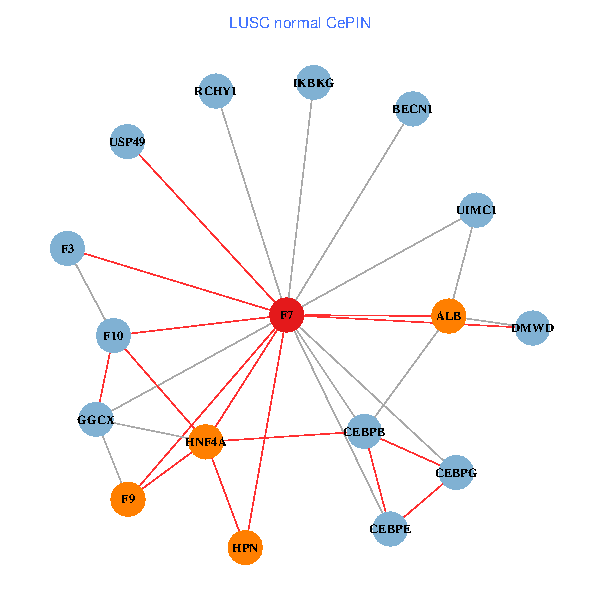 |
|
| PRAD (tumor) | PRAD (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
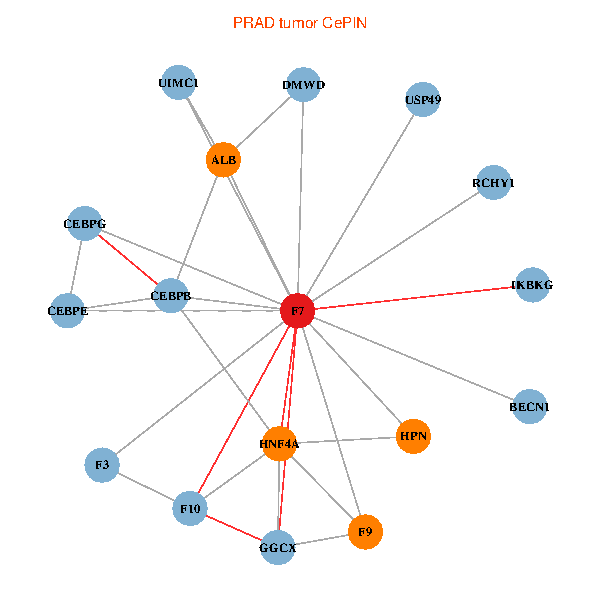 | 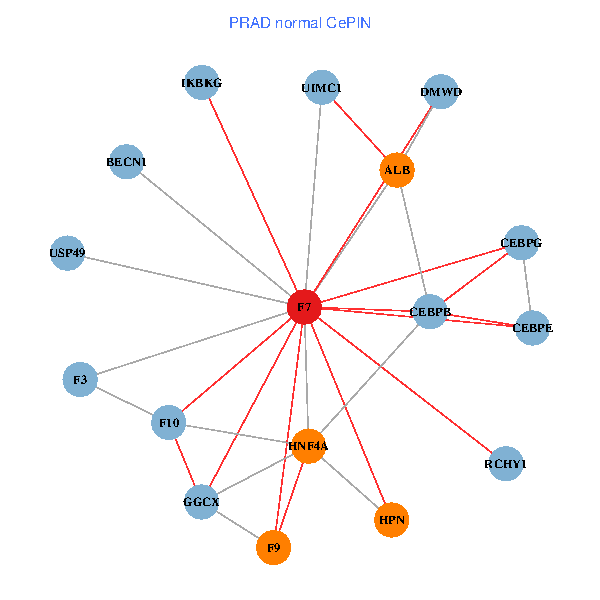 |
|
| STAD (tumor) | STAD (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
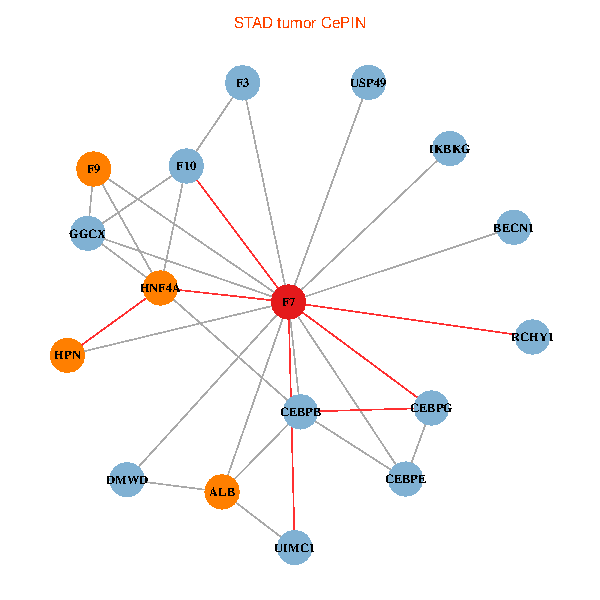 | 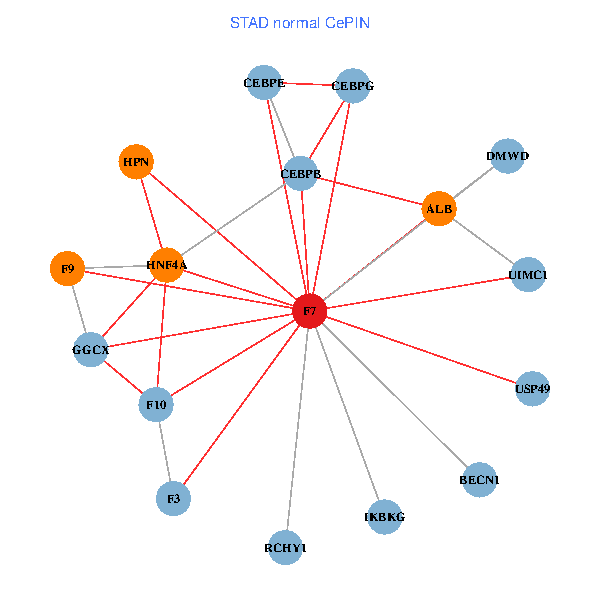 |
|
| THCA (tumor) | THCA (normal) |
| F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (tumor) | F7, HNF4A, IKBKG, CEBPB, CEBPE, UIMC1, ALB, F3, BECN1, CEBPG, USP49, DMWD, RCHY1, F10, GGCX, F9, HPN (normal) |
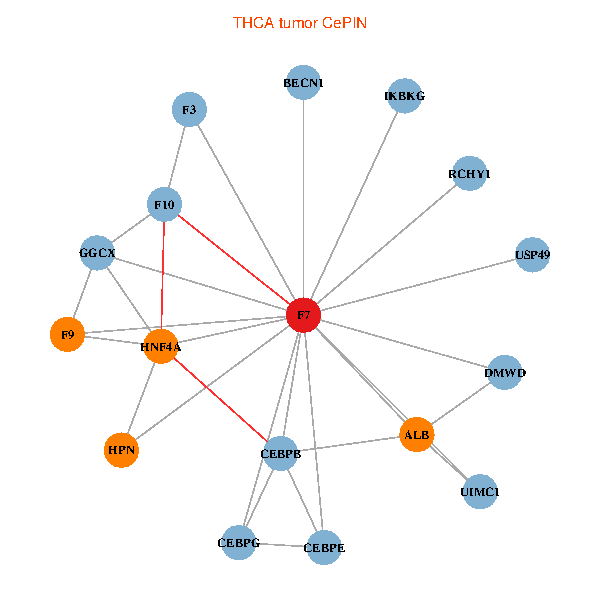 | 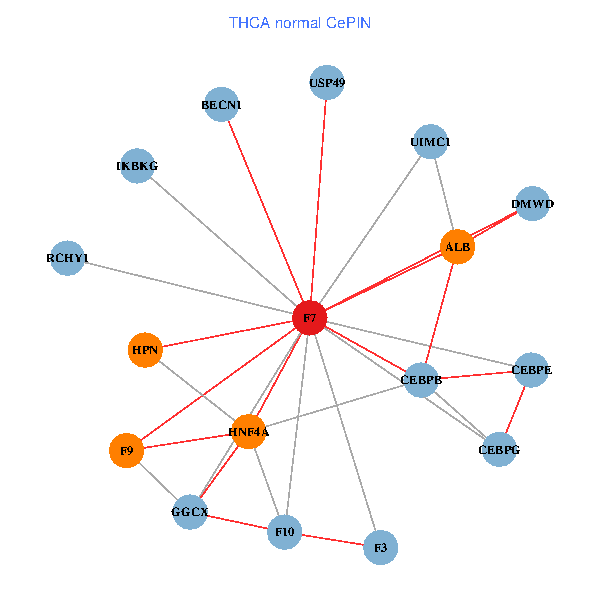 |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0015503 | Factor VII Deficiency | 44 | BeFree,CLINVAR,CTD_human,GAD,LHGDN,MGD,ORPHANET,UNIPROT |
| umls:C0010054 | Coronary Arteriosclerosis | 36 | BeFree,GAD |
| umls:C0010068 | Coronary heart disease | 36 | BeFree,GAD,LHGDN |
| umls:C0005779 | Blood Coagulation Disorders | 34 | BeFree,GAD,LHGDN |
| umls:C0027051 | Myocardial Infarction | 34 | BeFree,GAD,LHGDN |
| umls:C1956346 | Coronary Artery Disease | 22 | BeFree,GAD |
| umls:C0151744 | Myocardial Ischemia | 21 | BeFree,GAD |
| umls:C0007222 | Cardiovascular Diseases | 19 | BeFree,GAD |
| umls:C0019069 | Hemophilia A | 16 | BeFree,GAD |
| umls:C0038454 | Cerebrovascular accident | 13 | BeFree,GAD,LHGDN |
| umls:C0684275 | Hemophilia, NOS | 12 | BeFree |
| umls:C0004153 | Atherosclerosis | 8 | BeFree,GAD,LHGDN |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 6 | BeFree,GAD,RGD |
| umls:C0019080 | Hemorrhage | 6 | CTD_human,GAD,LHGDN |
| umls:C0003850 | Arteriosclerosis | 5 | BeFree |
| umls:C0008533 | Hemophilia B | 5 | BeFree,GAD |
| umls:C0020538 | Hypertensive disease | 5 | BeFree,GAD,LHGDN,RGD |
| umls:C0028754 | Obesity | 5 | BeFree,GAD |
| umls:C0151699 | Intracranial Hemorrhages | 5 | CTD_human |
| umls:C0155626 | Acute myocardial infarction | 5 | BeFree |
| umls:C0948008 | Ischemic stroke | 5 | BeFree,GAD |
| umls:C2239176 | Liver carcinoma | 5 | BeFree |
| umls:C3272363 | Ischemic Cerebrovascular Accident | 5 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 4 | BeFree |
| umls:C0007789 | Cerebral Palsy | 4 | GAD |
| umls:C0019087 | Hemorrhagic Disorders | 4 | BeFree,CTD_human,GAD |
| umls:C0027627 | Neoplasm Metastasis | 4 | BeFree,LHGDN |
| umls:C0040038 | Thromboembolism | 4 | BeFree,CTD_human,GAD |
| umls:C0042487 | Venous Thrombosis | 4 | CTD_human,GAD |
| umls:C0151526 | Premature Birth | 4 | GAD |
| umls:C0584960 | Factor V Leiden mutation | 4 | BeFree |
| umls:C0678222 | Breast Carcinoma | 4 | BeFree |
| umls:C0948089 | Acute Coronary Syndrome | 4 | BeFree,GAD |
| umls:C1458155 | Mammary Neoplasms | 4 | BeFree,LHGDN |
| umls:C2608079 | WARFARIN SENSITIVITY (disorder) | 4 | BeFree,GAD |
| umls:C0007785 | Cerebral Infarction | 3 | BeFree,GAD,LHGDN |
| umls:C0011849 | Diabetes Mellitus | 3 | BeFree,RGD |
| umls:C0015519 | Factor X Deficiency | 3 | BeFree |
| umls:C0021368 | Inflammation | 3 | GAD,LHGDN |
| umls:C0022116 | Ischemia | 3 | BeFree,RGD |
| umls:C0023903 | Liver neoplasms | 3 | BeFree |
| umls:C0042974 | von Willebrand Disease | 3 | BeFree |
| umls:C0151942 | Arterial thrombosis | 3 | BeFree |
| umls:C0243026 | Sepsis | 3 | BeFree,LHGDN,RGD |
| umls:C0272320 | Hereditary factor VII deficiency disease | 3 | BeFree |
| umls:C1861172 | Venous Thromboembolism | 3 | GAD,LHGDN |
| umls:C2937358 | Cerebral Hemorrhage | 3 | CTD_human,GAD,LHGDN |
| umls:C0002895 | Anemia, Sickle Cell | 2 | BeFree,GAD |
| umls:C0007786 | Brain Ischemia | 2 | GAD |
| umls:C0009402 | Colorectal Carcinoma | 2 | BeFree |
| umls:C0011853 | Diabetes Mellitus, Experimental | 2 | RGD |
| umls:C0015523 | Factor XI Deficiency | 2 | BeFree |
| umls:C0020473 | Hyperlipidemia | 2 | BeFree,RGD |
| umls:C0023890 | Liver Cirrhosis | 2 | BeFree |
| umls:C0023895 | Liver diseases | 2 | BeFree |
| umls:C0040053 | Thrombosis | 2 | GAD |
| umls:C0042880 | Vitamin K Deficiency | 2 | BeFree,LHGDN |
| umls:C0149871 | Deep Vein Thrombosis | 2 | BeFree,GAD |
| umls:C0242339 | Dyslipidemias | 2 | BeFree |
| umls:C0398623 | Thrombophilia | 2 | BeFree,GAD |
| umls:C0856761 | Budd-Chiari Syndrome | 2 | GAD,RGD |
| umls:C1096116 | Acquired haemophilia | 2 | BeFree |
| umls:C1527249 | Colorectal Cancer | 2 | BeFree |
| umls:C1623038 | Cirrhosis | 2 | BeFree |
| umls:C0000744 | Abetalipoproteinemia | 1 | BeFree |
| umls:C0001815 | Primary Myelofibrosis | 1 | BeFree |
| umls:C0002395 | Alzheimer's Disease | 1 | GAD |
| umls:C0002736 | Amyotrophic Lateral Sclerosis | 1 | GAD |
| umls:C0003130 | Anoxia | 1 | LHGDN |
| umls:C0004238 | Atrial Fibrillation | 1 | GAD,LHGDN |
| umls:C0006287 | Bronchopulmonary Dysplasia | 1 | BeFree |
| umls:C0007102 | Malignant tumor of colon | 1 | BeFree |
| umls:C0007282 | Carotid Stenosis | 1 | GAD |
| umls:C0007820 | Cerebrovascular Disorders | 1 | GAD |
| umls:C0008495 | Chorioamnionitis | 1 | GAD |
| umls:C0009782 | Connective Tissue Diseases | 1 | GAD |
| umls:C0010324 | Crigler Najjar syndrome, type 1 | 1 | BeFree |
| umls:C0011875 | Diabetic Angiopathies | 1 | GAD |
| umls:C0012739 | Disseminated Intravascular Coagulation | 1 | CTD_human |
| umls:C0013080 | Down Syndrome | 1 | BeFree |
| umls:C0015929 | Fetal Diseases | 1 | GAD |
| umls:C0015944 | Fetal Membranes, Premature Rupture | 1 | GAD |
| umls:C0017327 | Generalized atherosclerosis | 1 | GAD |
| umls:C0017638 | Glioma | 1 | BeFree |
| umls:C0018965 | Hematuria | 1 | CTD_human |
| umls:C0019158 | Hepatitis | 1 | GAD |
| umls:C0020433 | Hyperbilirubinemia | 1 | RGD |
| umls:C0020640 | Inherited Factor II deficiency | 1 | RGD |
| umls:C0020676 | Hypothyroidism | 1 | RGD |
| umls:C0021655 | Insulin Resistance | 1 | GAD |
| umls:C0022658 | Kidney Diseases | 1 | BeFree |
| umls:C0022661 | Kidney Failure, Chronic | 1 | GAD |
| umls:C0022876 | Premature Obstetric Labor | 1 | GAD |
| umls:C0023418 | leukemia | 1 | BeFree |
| umls:C0025517 | Metabolic Diseases | 1 | BeFree |
| umls:C0026827 | Muscle hypotonia | 1 | BeFree |
| umls:C0026857 | Musculoskeletal Diseases | 1 | GAD |
| umls:C0027660 | Neoplasms, Glandular and Epithelial | 1 | RGD |
| umls:C0029925 | Ovarian Carcinoma | 1 | BeFree |
| umls:C0031090 | Periodontal Diseases | 1 | GAD |
| umls:C0032787 | Postoperative Complications | 1 | CTD_human |
| umls:C0032914 | Pre-Eclampsia | 1 | GAD |
| umls:C0032964 | Pregnancy Complications, Hematologic | 1 | GAD |
| umls:C0036690 | Septicemia | 1 | BeFree |
| umls:C0037274 | Dermatologic disorders | 1 | GAD |
| umls:C0040028 | Thrombocythemia, Essential | 1 | BeFree |
| umls:C0042373 | Vascular Diseases | 1 | BeFree |
| umls:C0085096 | Peripheral Vascular Diseases | 1 | BeFree,GAD,LHGDN |
| umls:C0085110 | Severe Combined Immunodeficiency | 1 | BeFree |
| umls:C0149931 | Migraine Disorders | 1 | BeFree,GAD |
| umls:C0162316 | Iron deficiency anemia | 1 | BeFree |
| umls:C0232197 | Fibrillation | 1 | BeFree |
| umls:C0242231 | Coronary Stenosis | 1 | GAD |
| umls:C0263675 | Chronic arthropathy | 1 | BeFree |
| umls:C0268542 | Ornithine carbamoyltransferase deficiency | 1 | BeFree |
| umls:C0273058 | Traumatic intracranial hemorrhage | 1 | CTD_human |
| umls:C0280100 | Solid tumour | 1 | BeFree |
| umls:C0282666 | Very Low Birth Weight | 1 | BeFree |
| umls:C0333186 | Restenosis | 1 | GAD |
| umls:C0333203 | Occlusive thrombus | 1 | BeFree |
| umls:C0340288 | Stable angina | 1 | BeFree |
| umls:C0340708 | Deep vein thrombosis of lower limb | 1 | BeFree |
| umls:C0341439 | Chronic liver disease | 1 | BeFree,GAD |
| umls:C0342257 | Complications of Diabetes Mellitus | 1 | GAD |
| umls:C0376358 | Malignant neoplasm of prostate | 1 | BeFree |
| umls:C0518010 | body mass | 1 | GAD |
| umls:C0553692 | Brain hemorrhage | 1 | GAD |
| umls:C0577631 | Carotid Atherosclerosis | 1 | BeFree |
| umls:C0598608 | Hyperhomocysteinemia | 1 | BeFree |
| umls:C0600518 | Choroidal Neovascularization | 1 | RGD |
| umls:C0687675 | Pregnancy loss | 1 | GAD |
| umls:C0699790 | Colon Carcinoma | 1 | BeFree |
| umls:C0699885 | Carcinoma of bladder | 1 | BeFree |
| umls:C0749098 | Hematoma, Subdural, Acute | 1 | CTD_human |
| umls:C0751956 | Acute Cerebrovascular Accidents | 1 | BeFree |
| umls:C0752140 | Intracranial Embolism | 1 | GAD |
| umls:C0810006 | Acute cerebrovascular disease | 1 | BeFree |
| umls:C0948480 | Coronary Restenosis | 1 | GAD |
| umls:C1140680 | Malignant neoplasm of ovary | 1 | BeFree |
| umls:C1272641 | Systemic arterial pressure | 1 | GAD |
| umls:C1519666 | Tumor-Associated Vasculature | 1 | BeFree |
| umls:C1704436 | Peripheral Arterial Diseases | 1 | BeFree |
| umls:C2919032 | Infection of amniotic sac and membranes, unspecified, unspecified trimester, not applicable or unspecified | 1 | GAD |
| umls:C2936179 | Obesity, Visceral | 1 | BeFree |
| umls:C3495426 | Homocysteinemia | 1 | BeFree |
| umls:C3714514 | Infection | 1 | GAD |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))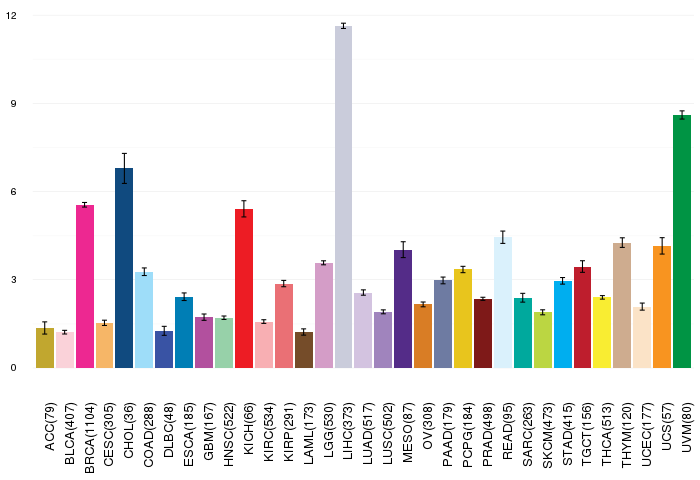
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))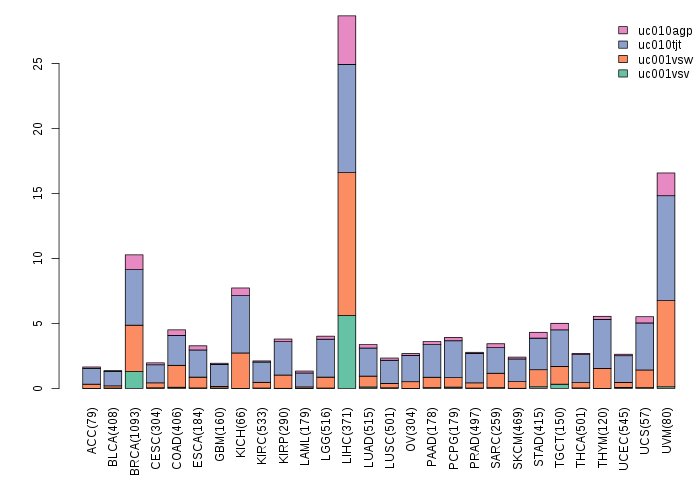
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data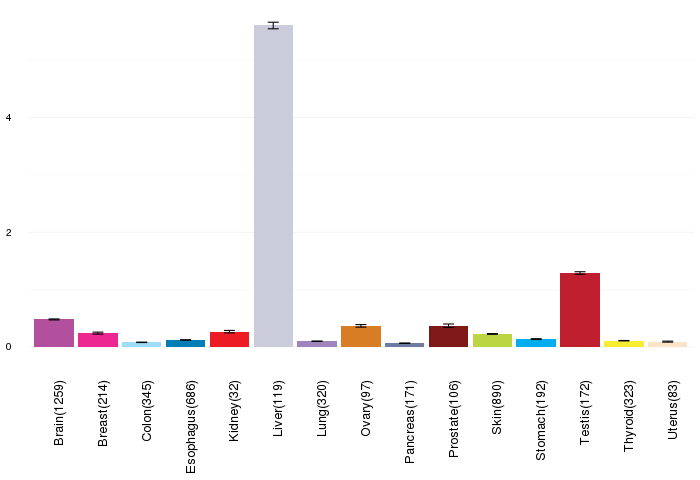
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))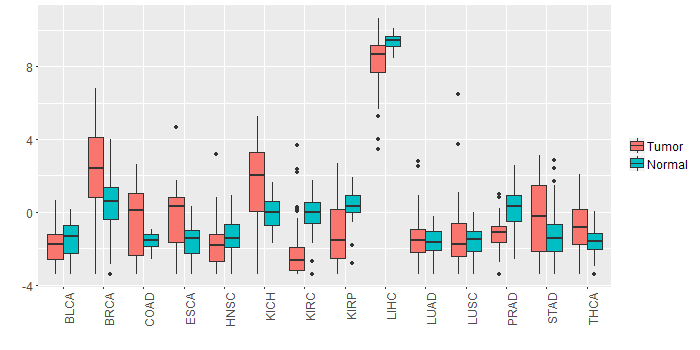
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.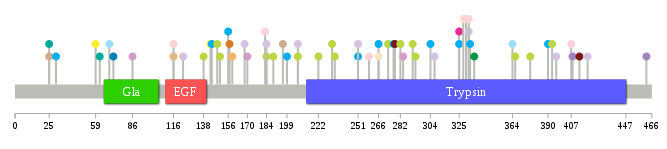

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 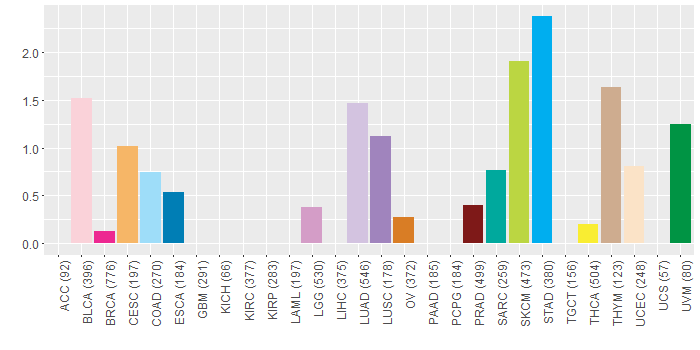
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)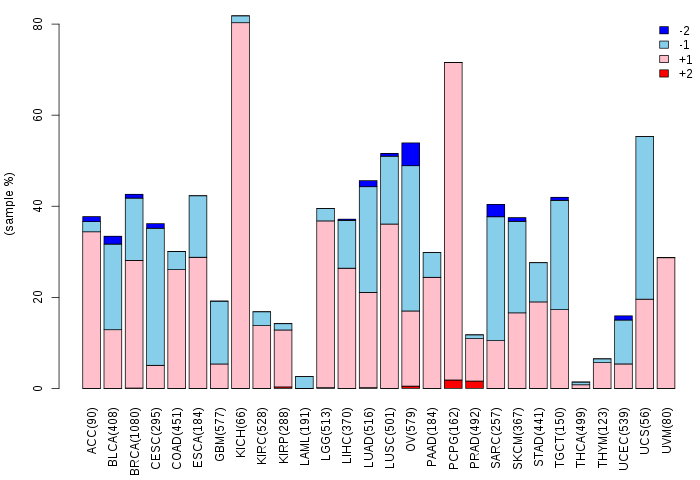
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 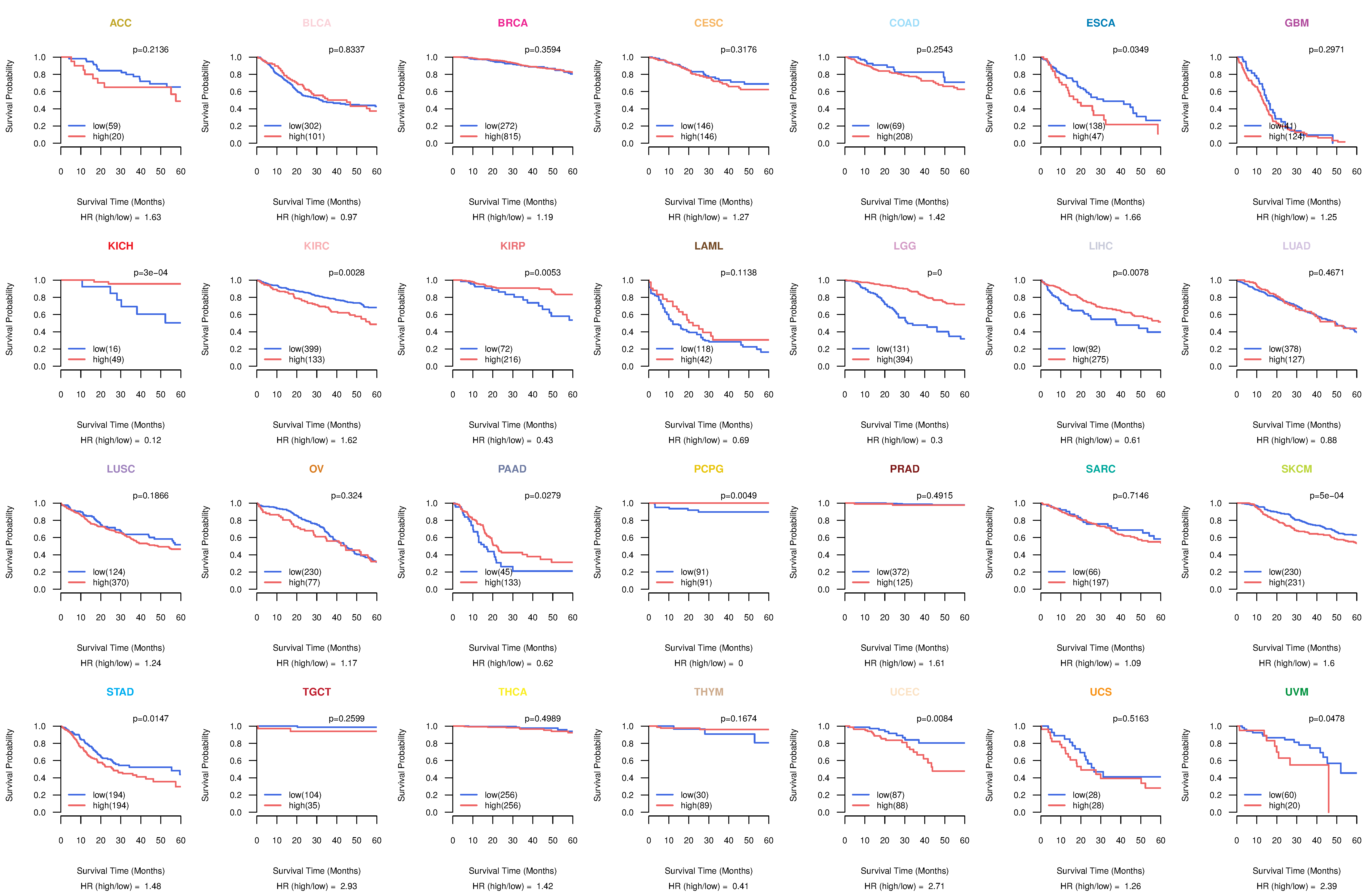
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 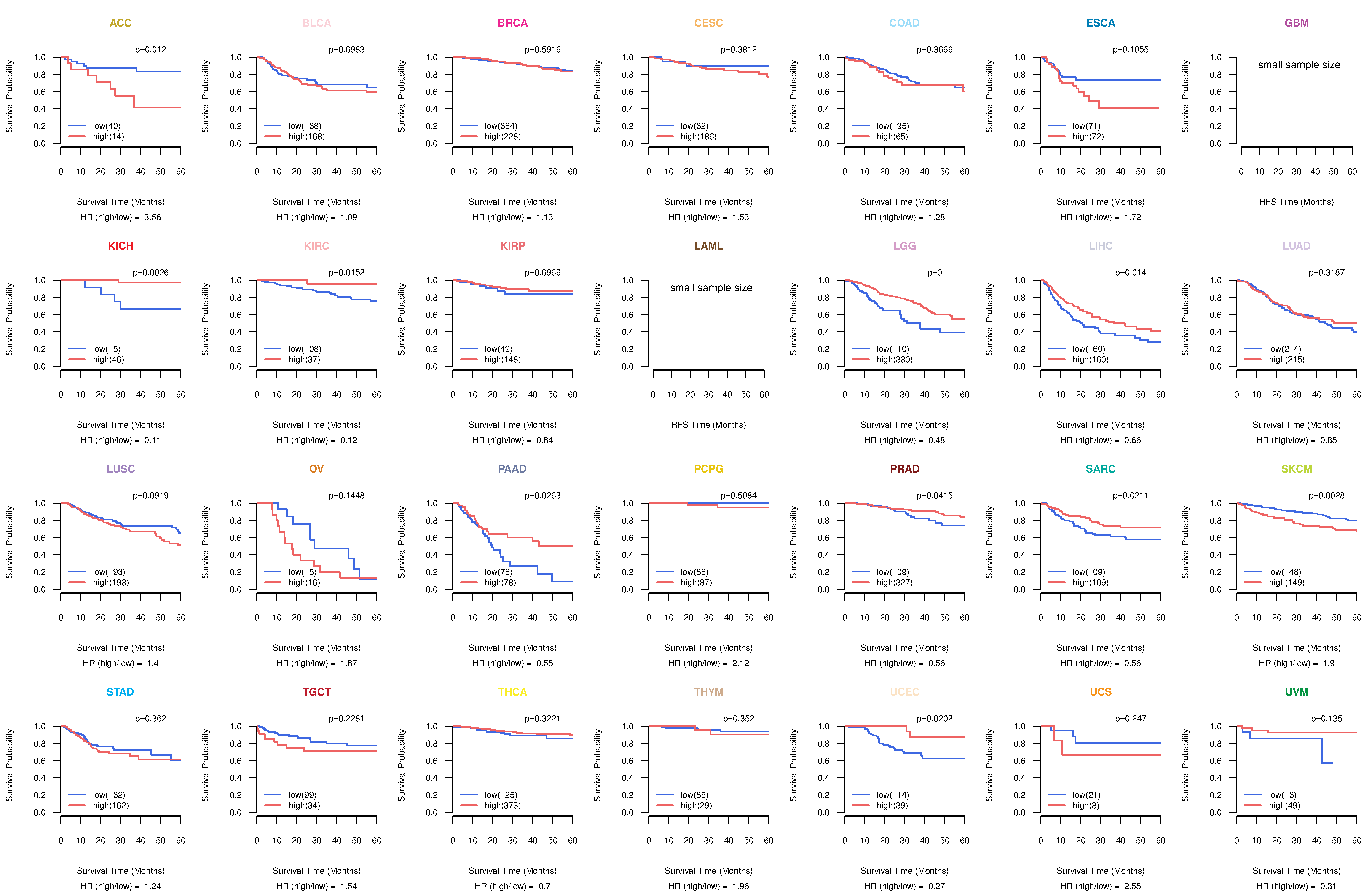
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 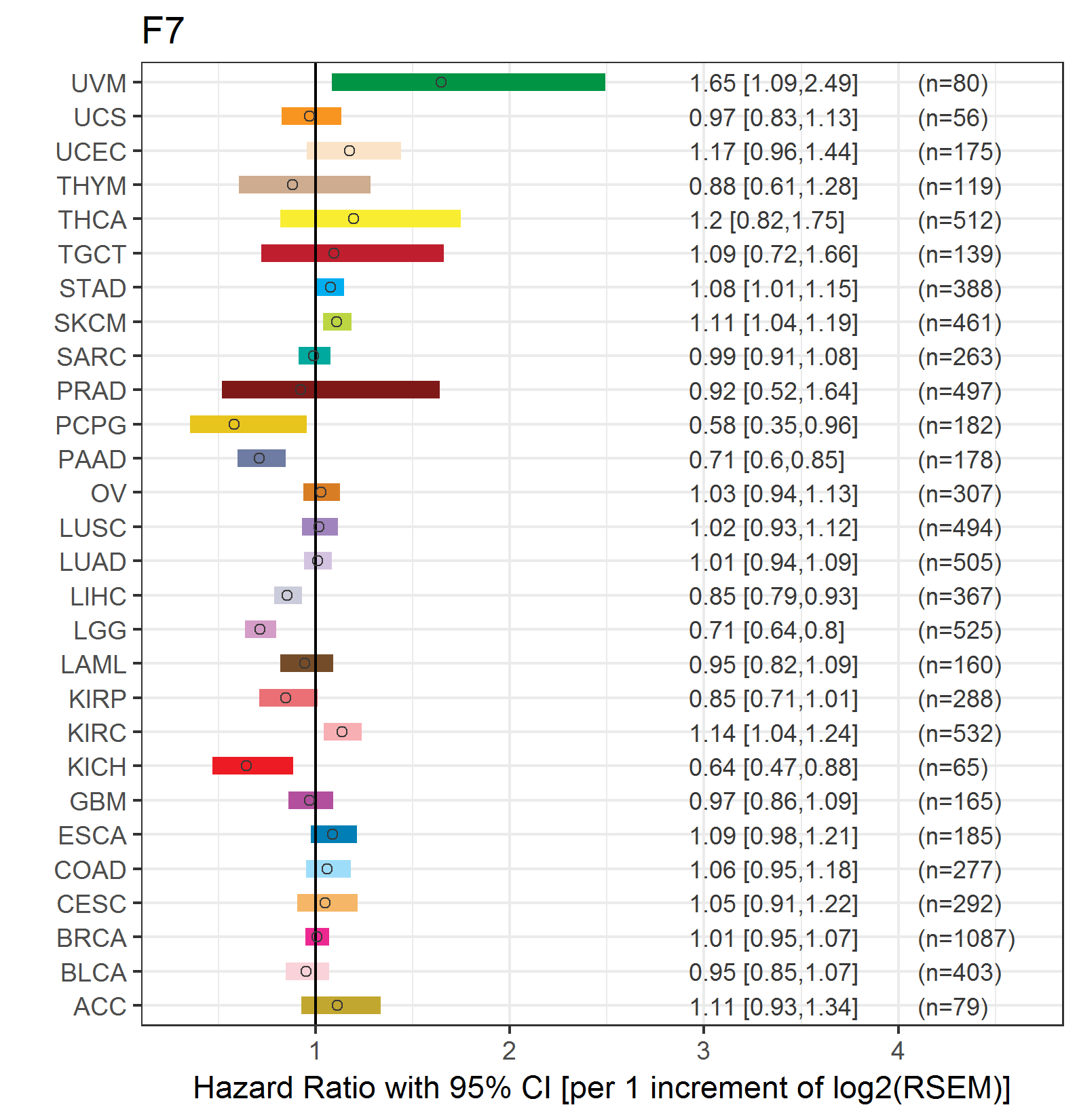
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 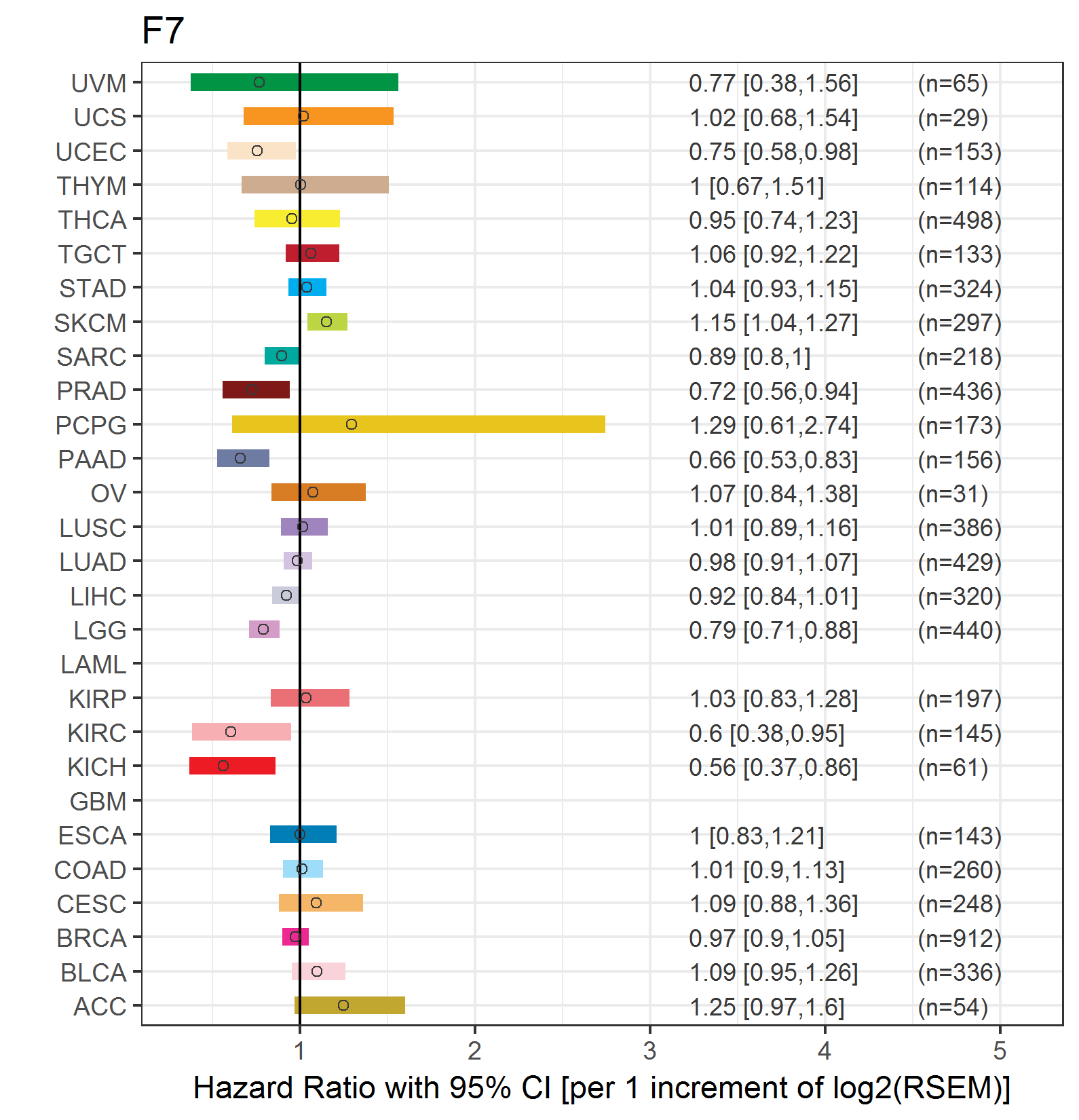
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























