| BRCA (tumor) | BRCA (normal) |
| FBN1, FBN2, MFAP2, SERPINB2, MYOC, FBLN5, HSPG2, ATXN7, CXCL11, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, RHOB, CXCL9, LOX, MFAP5 (tumor) | FBN1, FBN2, MFAP2, SERPINB2, MYOC, CALR, FBLN5, HSPG2, ATXN7, CXCL11, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, FBLN2, RHOB, CXCL9, LOX, MFAP5 (normal) |
 |  |
|
| COAD (tumor) | COAD (normal) |
| FBN1, FBN2, MFAP2, MYOC, FBLN5, HSPG2, ATXN7, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, RHOB, CXCL9, CXCL10, LOX, MFAP5 (tumor) | FBN1, FBN2, MFAP2, SERPINB2, MYOC, FBLN5, HSPG2, ATXN7, CXCL11, VCAN, ELN, EFEMP2, LTBP1, DCN, FBLN2, CXCL13, RHOB, CXCL9, CXCL10, LOX, MFAP5 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| FBN1, MFAP2, SERPINB2, MYOC, CALR, FBLN5, HSPG2, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, RHOB, CXCL9, CXCL10, LOX, MFAP5 (tumor) | FBN1, FBN2, MFAP2, SERPINB2, MYOC, FBLN5, HSPG2, ATXN7, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, RHOB, CXCL9, LOX, MFAP5 (normal) |
 |  |
|
| KICH (tumor) | KICH (normal) |
| FBN1, FBN2, MFAP2, SERPINB2, MYOC, FBLN5, HSPG2, ATXN7, CXCL11, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, FBLN2, RHOB, CXCL9, CXCL10, LOX, MFAP5 (tumor) | FBN1, FBN2, MFAP2, SERPINB2, MYOC, HSPG2, ATXN7, CXCL11, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, CXCL9, CXCL10, LOX, MFAP5 (normal) |
 |  |
|
| KIRC (tumor) | KIRC (normal) |
| FBN1, FBN2, MFAP2, SERPINB2, MYOC, FBLN5, HSPG2, CXCL11, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, RHOB, CXCL9, LOX, MFAP5 (tumor) | FBN1, FBN2, MFAP2, SERPINB2, FBLN5, HSPG2, ATXN7, CXCL11, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, RHOB, CXCL9, LOX, MFAP5 (normal) |
 |  |
|
| KIRP (tumor) | KIRP (normal) |
| FBN1, FBN2, MFAP2, SERPINB2, FBLN5, HSPG2, ATXN7, CXCL11, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, RHOB, CXCL9, CXCL10, MFAP5 (tumor) | FBN1, FBN2, MFAP2, SERPINB2, MYOC, CALR, FBLN5, HSPG2, ATXN7, CXCL11, VCAN, ELN, LTBP1, DCN, FBLN2, CXCL13, RHOB, CXCL9, CXCL10, LOX, MFAP5 (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| FBN1, FBN2, MFAP2, SERPINB2, CALR, FBLN5, HSPG2, ATXN7, CXCL11, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, CXCL9, CXCL10, LOX, MFAP5 (tumor) | FBN1, FBN2, MFAP2, SERPINB2, FBLN5, HSPG2, ATXN7, CXCL11, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, CXCL9, CXCL10, LOX, MFAP5 (normal) |
 |  |
|
| LUAD (tumor) | LUAD (normal) |
| FBN1, FBN2, MFAP2, SERPINB2, MYOC, FBLN5, HSPG2, CXCL11, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, RHOB, CXCL9, CXCL10, LOX, MFAP5 (tumor) | FBN1, FBN2, MFAP2, SERPINB2, MYOC, FBLN5, HSPG2, ATXN7, CXCL11, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, CXCL9, CXCL10, LOX, MFAP5 (normal) |
 |  |
|
| LUSC (tumor) | LUSC (normal) |
| FBN1, FBN2, MFAP2, SERPINB2, MYOC, CALR, FBLN5, HSPG2, ATXN7, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, RHOB, CXCL9, LOX, MFAP5 (tumor) | FBN1, FBN2, MFAP2, MYOC, FBLN5, HSPG2, ATXN7, CXCL11, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, FBLN2, CXCL13, RHOB, CXCL9, CXCL10, LOX, MFAP5 (normal) |
 |  |
|
| PRAD (tumor) | PRAD (normal) |
| FBN1, FBN2, MFAP2, SERPINB2, MYOC, CALR, FBLN5, HSPG2, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, RHOB, CXCL9, LOX, MFAP5 (tumor) | FBN1, FBN2, MFAP2, MYOC, CALR, FBLN5, HSPG2, ATXN7, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, FBLN2, CXCL13, RHOB, CXCL9, CXCL10, LOX, MFAP5 (normal) |
 |  |
|
| STAD (tumor) | STAD (normal) |
| FBN1, MFAP2, MYOC, CALR, FBLN5, HSPG2, ATXN7, CXCL11, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, PF4, FBLN2, CXCL13, RHOB, CXCL9, CXCL10, MFAP5 (tumor) | FBN1, FBN2, MFAP2, MYOC, CALR, FBLN5, HSPG2, ATXN7, CXCL11, VCAN, ELN, EFEMP2, LTBP1, DCN, FBLN2, CXCL13, RHOB, CXCL9, CXCL10, LOX, MFAP5 (normal) |
 |  |
|
| THCA (tumor) | THCA (normal) |
| FBN1, FBN2, MFAP2, MYOC, CALR, FBLN5, HSPG2, ATXN7, CXCL11, SPRY2, VCAN, ELN, LTBP1, DCN, FBLN2, CXCL13, RHOB, CXCL9, CXCL10, LOX, MFAP5 (tumor) | FBN1, FBN2, MFAP2, SERPINB2, MYOC, FBLN5, HSPG2, ATXN7, CXCL11, SPRY2, VCAN, ELN, EFEMP2, LTBP1, DCN, FBLN2, CXCL13, RHOB, CXCL9, LOX, MFAP5 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0024796 | Marfan Syndrome | 310 | BeFree,CLINVAR,CTD_human,GAD,LHGDN,MGD,ORPHANET,UNIPROT |
| umls:C0009782 | Connective Tissue Diseases | 40 | BeFree,GAD |
| umls:C0013581 | Ectopia Lentis | 37 | BeFree,CTD_human,GAD,LHGDN,ORPHANET |
| umls:C0036421 | Systemic Scleroderma | 13 | BeFree,GAD,LHGDN |
| umls:C0162872 | Aortic Aneurysm, Thoracic | 11 | BeFree,CTD_human,GAD |
| umls:C0265313 | Weill-Marchesani syndrome | 10 | BeFree,CTD_human,ORPHANET |
| umls:C0220668 | Congenital contractural arachnodactyly | 9 | BeFree |
| umls:C0410787 | Hereditary Connective Tissue Disorder | 9 | BeFree |
| umls:C0003486 | Aortic Aneurysm | 8 | BeFree,GAD |
| umls:C0340643 | Dissection of aorta | 8 | BeFree |
| umls:C1851286 | Ectopia lentis isolated | 8 | BeFree,CLINVAR |
| umls:C0011644 | Scleroderma | 7 | BeFree,GAD |
| umls:C1956346 | Coronary Artery Disease | 7 | BeFree,GAD |
| umls:C0002940 | Aneurysm | 6 | BeFree |
| umls:C0026267 | Mitral Valve Prolapse Syndrome | 6 | BeFree,GAD,LHGDN |
| umls:C2697932 | Loeys-Dietz Syndrome | 6 | BeFree |
| umls:C0003493 | Aortic Diseases | 5 | BeFree |
| umls:C0003706 | Arachnodactyly | 5 | BeFree,CTD_human |
| umls:C0007222 | Cardiovascular Diseases | 5 | BeFree,LHGDN |
| umls:C0020538 | Hypertensive disease | 5 | BeFree,RGD |
| umls:C0265004 | Dilatation of aorta | 5 | BeFree |
| umls:C0729233 | Dissecting aneurysm of the thoracic aorta | 5 | BeFree |
| umls:C1812607 | Aortic aneurysm and dissection | 5 | BeFree |
| umls:C3489726 | Geleophysic dysplasia | 5 | BeFree,ORPHANET |
| umls:C0010068 | Coronary heart disease | 4 | BeFree |
| umls:C0017601 | Glaucoma | 4 | BeFree,LHGDN |
| umls:C0023787 | Lipodystrophy | 4 | BeFree |
| umls:C0221032 | Familial generalized lipodystrophy | 4 | BeFree |
| umls:C0265287 | Acromicric Dysplasia | 4 | BeFree,ORPHANET,UNIPROT |
| umls:C0340629 | Aortic aneurysm without mention of rupture NOS | 4 | BeFree |
| umls:C0010054 | Coronary Arteriosclerosis | 3 | BeFree |
| umls:C0019880 | Homocystinuria | 3 | BeFree,LHGDN |
| umls:C0023316 | Lens Subluxation | 3 | BeFree,LHGDN |
| umls:C0029463 | Osteosarcoma | 3 | BeFree |
| umls:C0031099 | Periodontitis | 3 | BeFree |
| umls:C0034067 | Pulmonary Emphysema | 3 | BeFree |
| umls:C0238669 | Aortic root dilatation | 3 | BeFree |
| umls:C0243050 | Cardiovascular Abnormalities | 3 | BeFree |
| umls:C0410702 | Adolescent idiopathic scoliosis | 3 | BeFree |
| umls:C0585442 | Osteosarcoma of bone | 3 | BeFree |
| umls:C1321551 | Shprintzen-Goldberg syndrome | 3 | BeFree,CTD_human,ORPHANET |
| umls:C1858556 | OVERLAP CONNECTIVE TISSUE DISEASE | 3 | BeFree,CLINVAR,CTD_human |
| umls:C3541518 | ECTOPIA LENTIS 1, ISOLATED, AUTOSOMAL DOMINANT | 3 | CTD_human,UNIPROT |
| umls:C0000768 | Congenital Abnormality | 2 | BeFree |
| umls:C0003496 | Aortic Rupture | 2 | CTD_human,GAD |
| umls:C0003499 | Supravalvular aortic stenosis | 2 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 2 | BeFree,GAD |
| umls:C0007193 | Cardiomyopathy, Dilated | 2 | BeFree |
| umls:C0009402 | Colorectal Carcinoma | 2 | BeFree |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 2 | BeFree |
| umls:C0013336 | Dwarfism | 2 | BeFree |
| umls:C0017612 | Glaucoma, Open-Angle | 2 | BeFree,GAD |
| umls:C0017658 | Glomerulonephritis | 2 | RGD |
| umls:C0027092 | Myopia | 2 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 2 | BeFree |
| umls:C0033377 | Ptosis | 2 | BeFree |
| umls:C0036341 | Schizophrenia | 2 | BeFree |
| umls:C0042025 | Urinary Stress Incontinence | 2 | BeFree |
| umls:C0085413 | Polycystic Kidney, Autosomal Dominant | 2 | BeFree |
| umls:C0085580 | Essential Hypertension | 2 | BeFree |
| umls:C0152459 | Linear atrophy | 2 | BeFree |
| umls:C0206368 | Exfoliation Syndrome | 2 | BeFree,GAD |
| umls:C0221357 | Brachydactyly | 2 | BeFree |
| umls:C0241165 | Thick skin | 2 | BeFree |
| umls:C0265385 | Autosomal dominant hereditary disorder | 2 | BeFree |
| umls:C0268365 | Marfanoid hypermobility syndrome | 2 | BeFree |
| umls:C0345050 | Congenital aneurysm of ascending aorta | 2 | BeFree,CLINVAR |
| umls:C0345392 | Congenital kyphoscoliosis | 2 | BeFree |
| umls:C0406586 | Wiedemann-Rautenstrauch syndrome | 2 | BeFree |
| umls:C0575158 | Kyphoscoliosis deformity of spine | 2 | BeFree |
| umls:C0595936 | Aqueous Humor Disorders | 2 | BeFree |
| umls:C0600033 | Acquired Kyphoscoliosis | 2 | BeFree |
| umls:C0856747 | Aneurysm of ascending aorta | 2 | BeFree |
| umls:C1527249 | Colorectal Cancer | 2 | BeFree |
| umls:C2746069 | Familial ectopia lentis | 2 | BeFree |
| umls:C3178782 | Aortic Stiffness | 2 | GAD |
| umls:C0003507 | Aortic Valve Stenosis | 1 | GAD |
| umls:C0004138 | Ataxias, Hereditary | 1 | BeFree |
| umls:C0005586 | Bipolar Disorder | 1 | GAD |
| umls:C0005911 | Body Weight Changes | 1 | GAD |
| umls:C0009319 | Colitis | 1 | BeFree |
| umls:C0010278 | Craniosynostosis | 1 | BeFree |
| umls:C0010674 | Cystic Fibrosis | 1 | BeFree |
| umls:C0011206 | Delirium | 1 | BeFree |
| umls:C0011570 | Mental Depression | 1 | BeFree |
| umls:C0011581 | Depressive disorder | 1 | BeFree |
| umls:C0011633 | Dermatomyositis | 1 | BeFree |
| umls:C0011853 | Diabetes Mellitus, Experimental | 1 | RGD |
| umls:C0012359 | Pathological Dilatation | 1 | GAD |
| umls:C0013720 | Ehlers-Danlos Syndrome | 1 | BeFree |
| umls:C0014118 | Endocarditis | 1 | BeFree |
| umls:C0014175 | Endometriosis | 1 | CTD_human |
| umls:C0015310 | Exotropia | 1 | BeFree |
| umls:C0018801 | Heart failure | 1 | BeFree |
| umls:C0018802 | Congestive heart failure | 1 | BeFree |
| umls:C0019555 | Hip Dislocation, Congenital | 1 | BeFree,GAD |
| umls:C0020456 | Hyperglycemia | 1 | BeFree,CTD_human |
| umls:C0020459 | Hyperinsulinism | 1 | BeFree,CTD_human |
| umls:C0022661 | Kidney Failure, Chronic | 1 | GAD |
| umls:C0022821 | Kyphosis deformity of spine | 1 | BeFree |
| umls:C0023267 | Fibroid Tumor | 1 | BeFree |
| umls:C0024299 | Lymphoma | 1 | BeFree |
| umls:C0026265 | Diseases of mitral valve | 1 | BeFree |
| umls:C0026266 | Mitral Valve Insufficiency | 1 | BeFree |
| umls:C0026848 | Myopathy | 1 | BeFree |
| umls:C0029925 | Ovarian Carcinoma | 1 | BeFree |
| umls:C0030552 | Paresis | 1 | BeFree |
| umls:C0030567 | Parkinson Disease | 1 | GAD |
| umls:C0033975 | Psychotic Disorders | 1 | BeFree |
| umls:C0033999 | Pterygium | 1 | BeFree |
| umls:C0036439 | Scoliosis, unspecified | 1 | BeFree |
| umls:C0038013 | Ankylosing spondylitis | 1 | BeFree |
| umls:C0039685 | Tetralogy of Fallot | 1 | BeFree,GAD,LHGDN |
| umls:C0040336 | Tobacco Use Disorder | 1 | GAD |
| umls:C0042133 | Uterine Fibroids | 1 | BeFree |
| umls:C0042373 | Vascular Diseases | 1 | BeFree |
| umls:C0043194 | Wiskott-Aldrich Syndrome | 1 | BeFree |
| umls:C0086437 | Joint laxity | 1 | BeFree |
| umls:C0151786 | Muscle Weakness | 1 | BeFree |
| umls:C0206062 | Lung Diseases, Interstitial | 1 | BeFree |
| umls:C0206682 | Follicular thyroid carcinoma | 1 | BeFree |
| umls:C0232197 | Fibrillation | 1 | BeFree |
| umls:C0235833 | Congenital diaphragmatic hernia | 1 | BeFree |
| umls:C0265227 | Schinzel-Giedion syndrome | 1 | BeFree |
| umls:C0265673 | Congenital kyphosis | 1 | BeFree |
| umls:C0267725 | Paraesophageal hernia | 1 | BeFree |
| umls:C0268407 | Senile cardiac amyloidosis | 1 | BeFree |
| umls:C0338484 | Familial Hemiplegic Migraine | 1 | BeFree |
| umls:C0410528 | Skeletal dysplasia | 1 | BeFree |
| umls:C0549473 | Thyroid carcinoma | 1 | BeFree |
| umls:C0559260 | Congenital scoliosis | 1 | BeFree |
| umls:C0596263 | Carcinogenesis | 1 | BeFree |
| umls:C0677932 | Progressive Neoplastic Disease | 1 | BeFree |
| umls:C0678222 | Breast Carcinoma | 1 | BeFree |
| umls:C0679408 | Lesion of stomach | 1 | BeFree |
| umls:C0700208 | Acquired scoliosis | 1 | BeFree |
| umls:C0746102 | Chronic lung disease | 1 | BeFree |
| umls:C0751587 | CADASIL Syndrome | 1 | BeFree |
| umls:C1140680 | Malignant neoplasm of ovary | 1 | BeFree |
| umls:C1145670 | Respiratory Failure | 1 | BeFree |
| umls:C1299432 | Multi vessel coronary artery disease | 1 | BeFree |
| umls:C1301700 | Cardiovascular morbidity | 1 | BeFree |
| umls:C1800706 | Idiopathic Pulmonary Fibrosis | 1 | BeFree |
| umls:C1836635 | Loeys-Dietz Aortic Aneurysm Syndrome | 1 | BeFree |
| umls:C1861456 | Stiff Skin Syndrome | 1 | BeFree,CLINVAR,CTD_human,UNIPROT |
| umls:C1869115 | Weill-Marchesani Syndrome, Autosomal Dominant | 1 | BeFree,CLINVAR,MGD |
| umls:C2751492 | AMYLOIDOSIS, HEREDITARY, TRANSTHYRETIN-RELATED | 1 | BeFree |
| umls:C3280054 | GELEOPHYSIC DYSPLASIA 2 | 1 | CLINVAR,UNIPROT |
| umls:C3539781 | Progressive cGVHD | 1 | BeFree |
| umls:C1851718 | Emphysema, Hereditary Pulmonary | 0 | MGD |
| umls:C1866983 | SCLERODERMA, FAMILIAL PROGRESSIVE | 0 | MGD |
| umls:C2931588 | GEMSS syndrome | 0 | ORPHANET |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))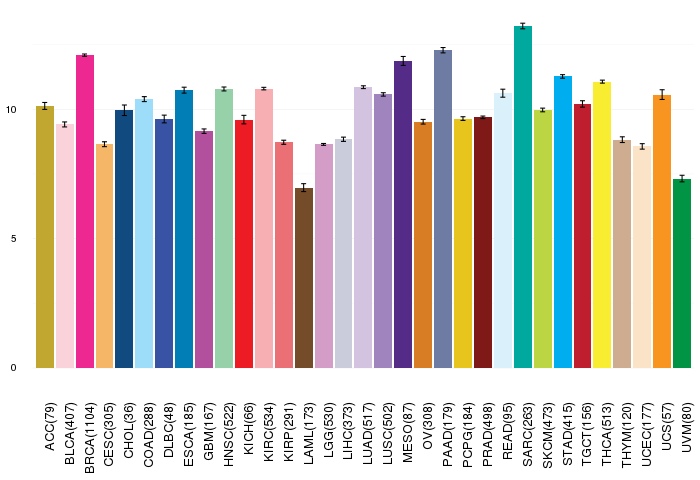
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))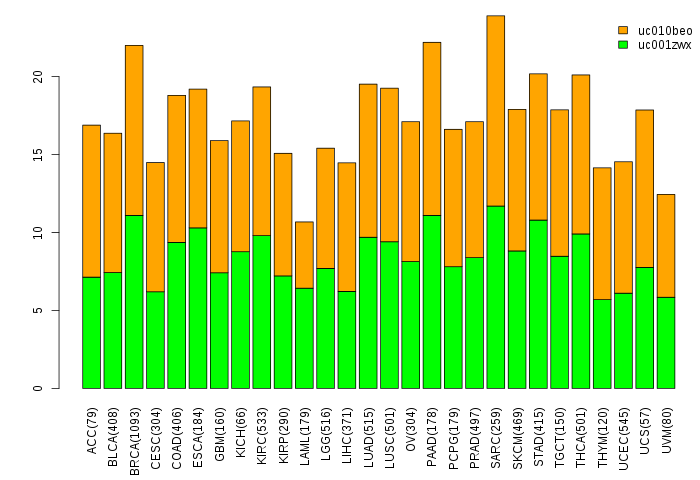
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data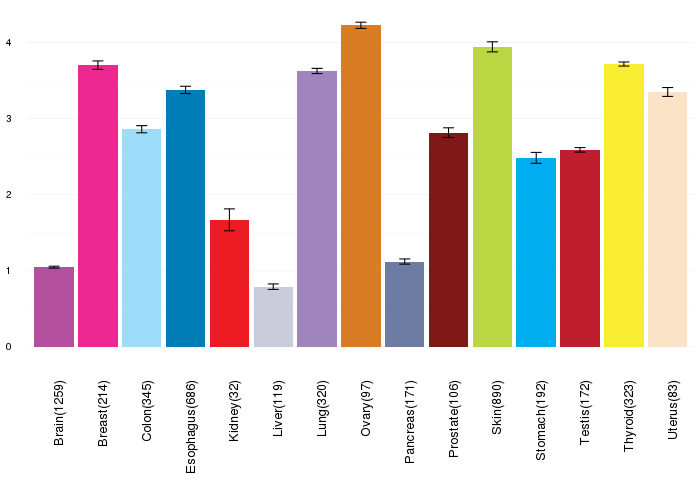
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))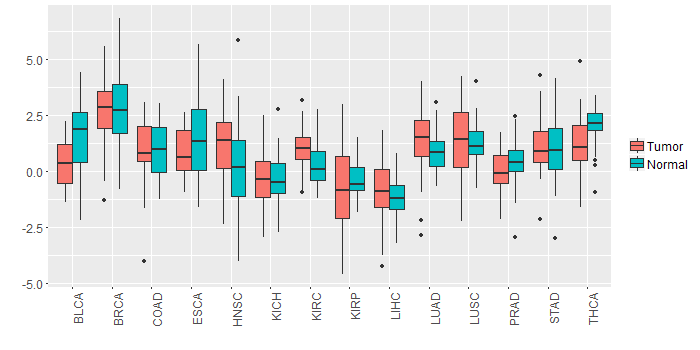
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 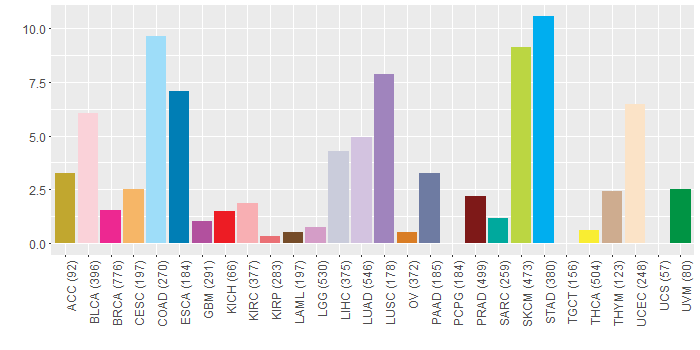
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)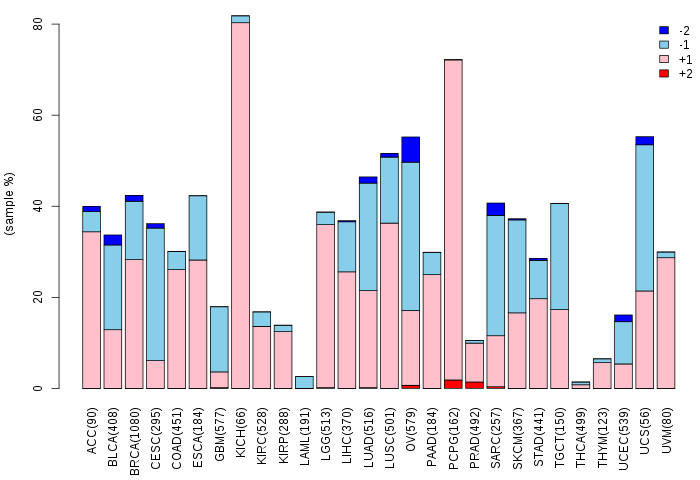
 Fusion genes including TissGene
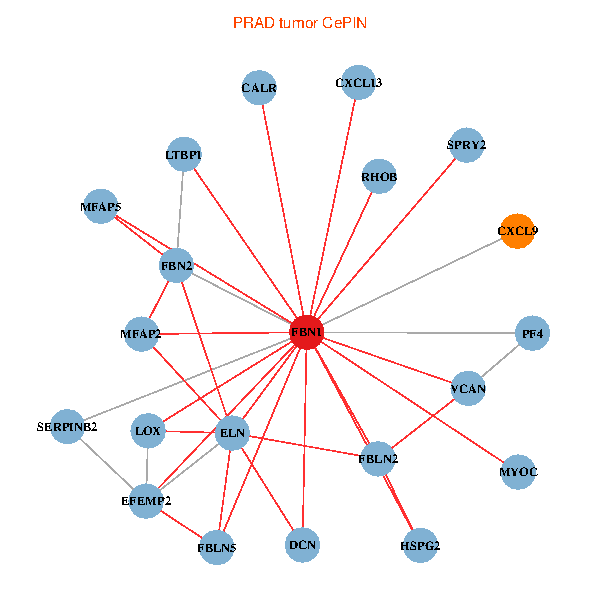
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























