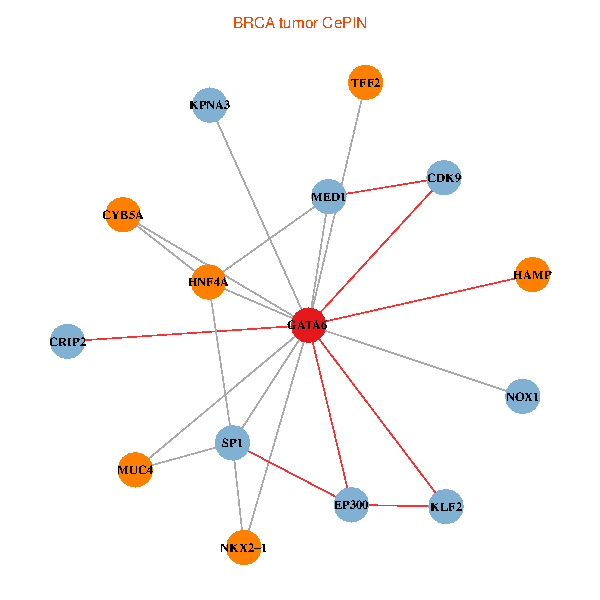
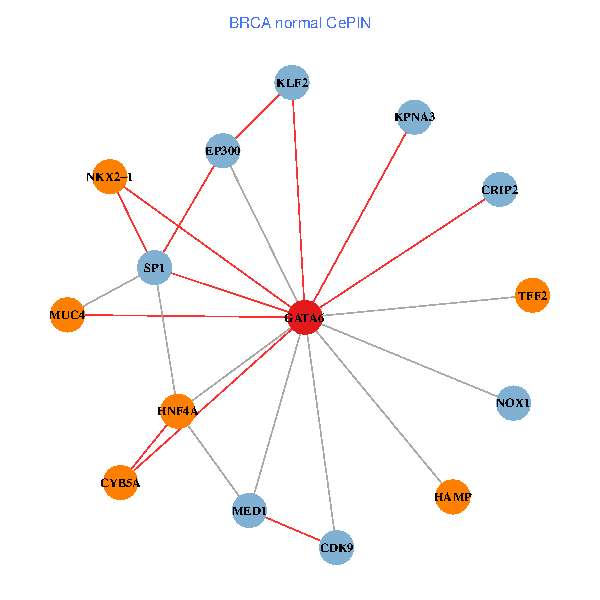
| BRCA (tumor) | BRCA (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
 |  |
|
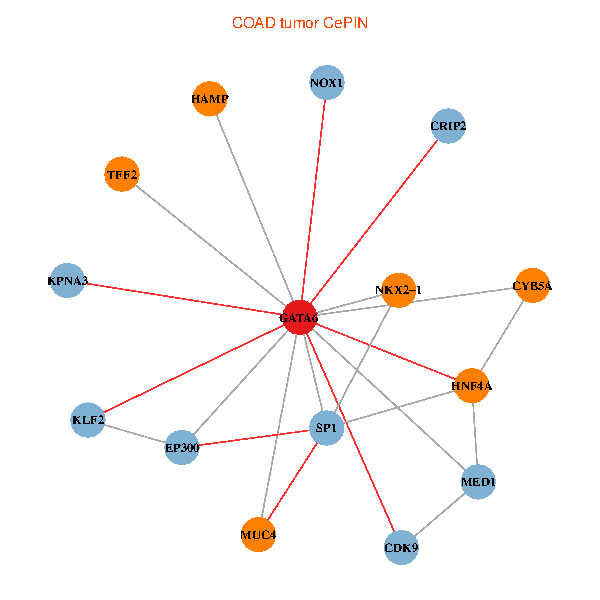
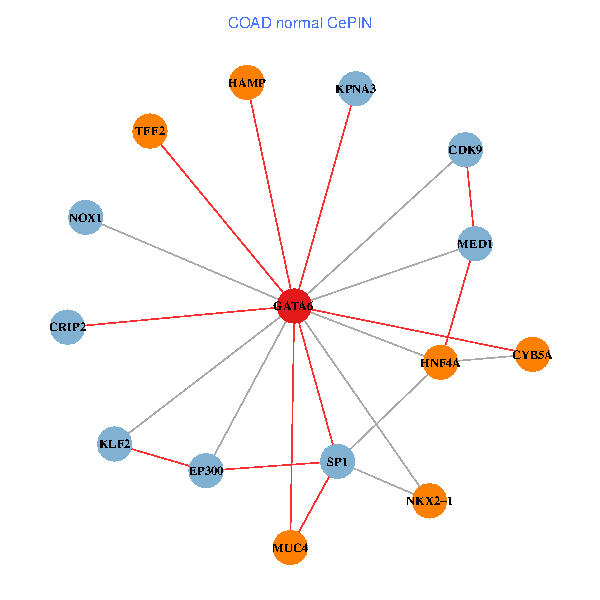
| COAD (tumor) | COAD (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
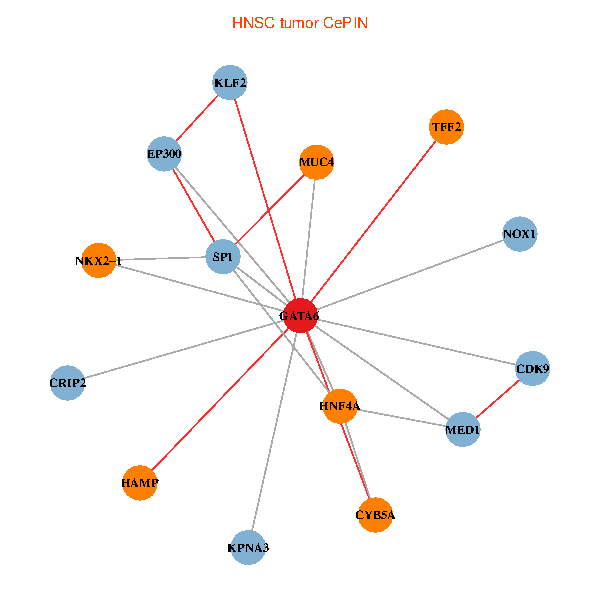 | 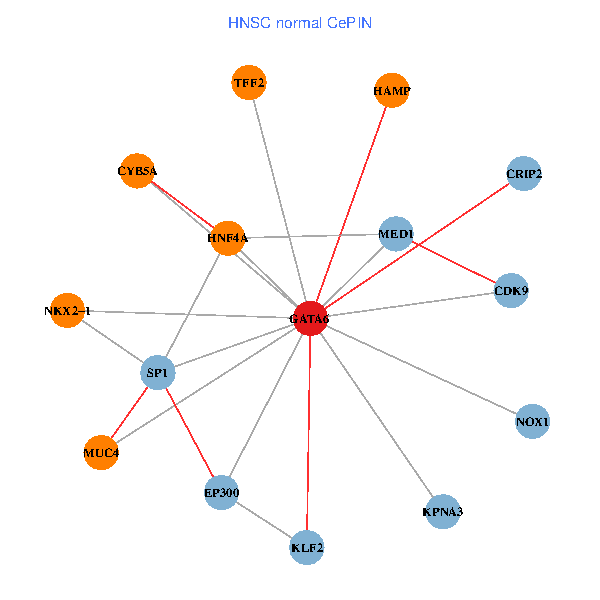 |
|
| KICH (tumor) | KICH (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
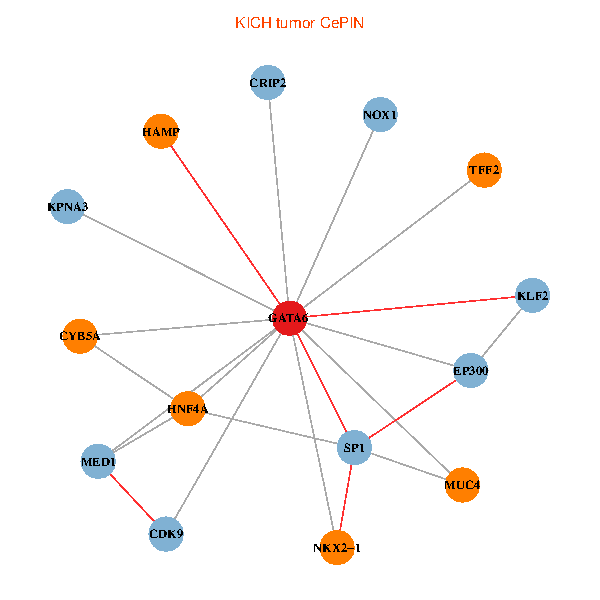 | 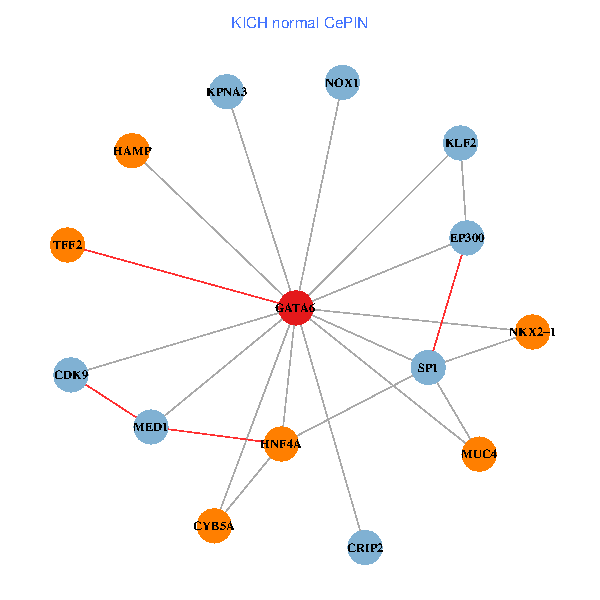 |
|
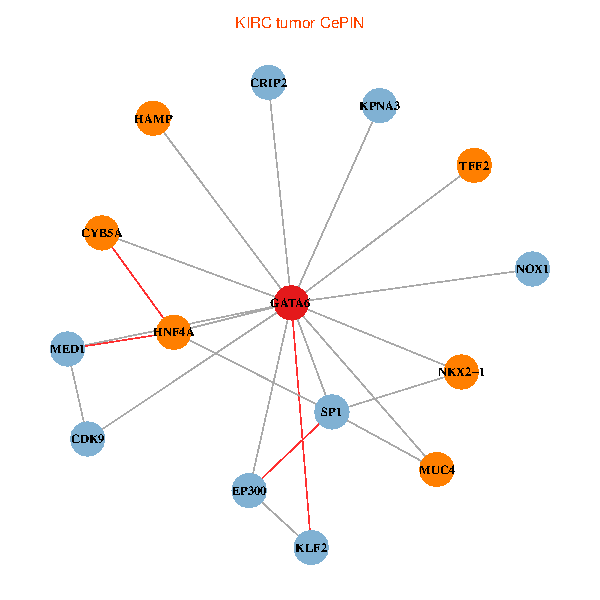
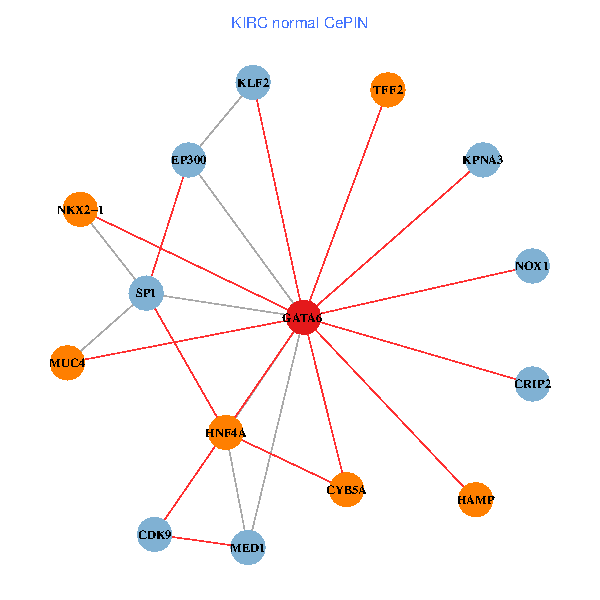
| KIRC (tumor) | KIRC (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
 |  |
|
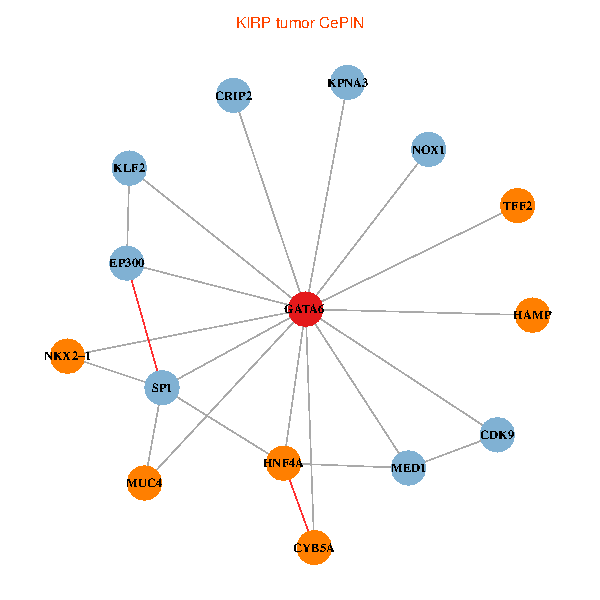
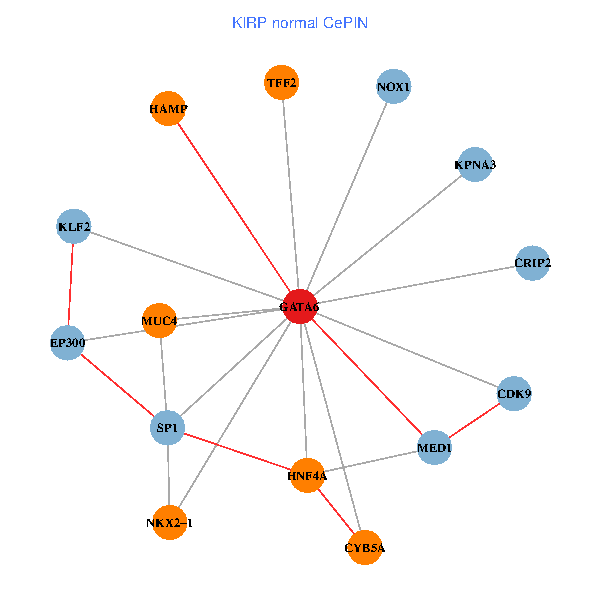
| KIRP (tumor) | KIRP (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
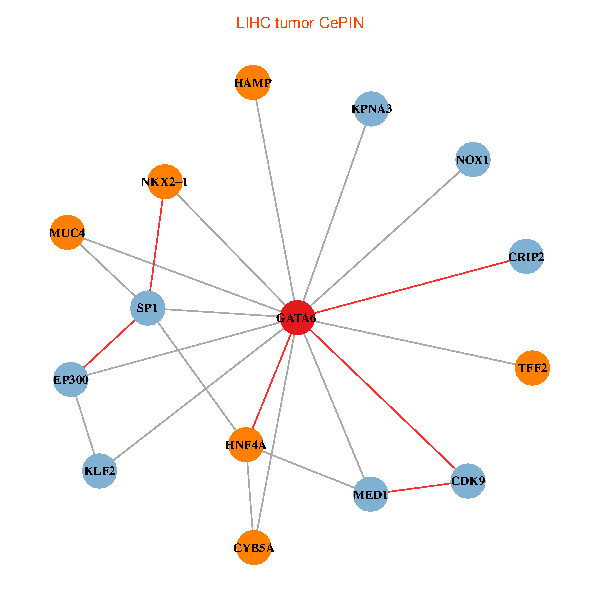 | 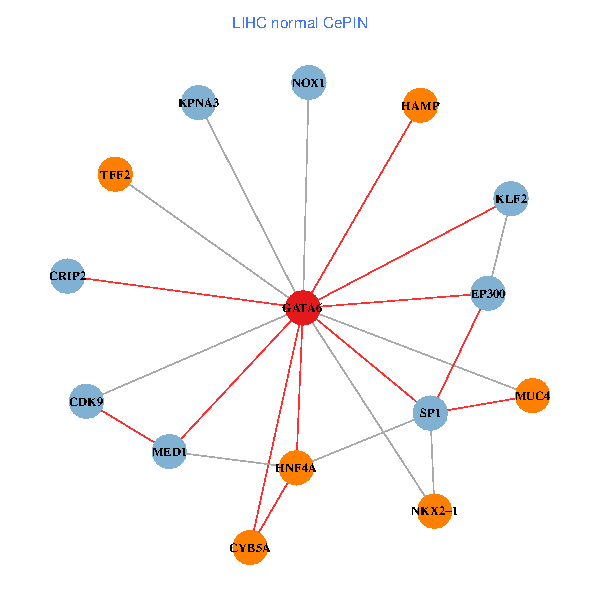 |
|
| LUAD (tumor) | LUAD (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
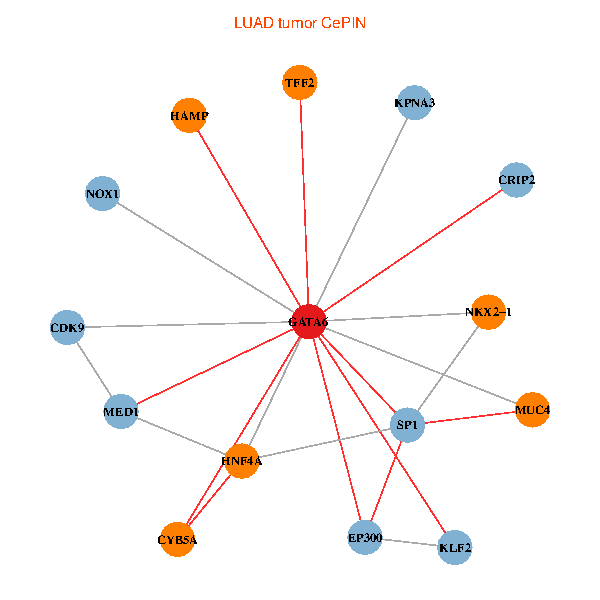 | 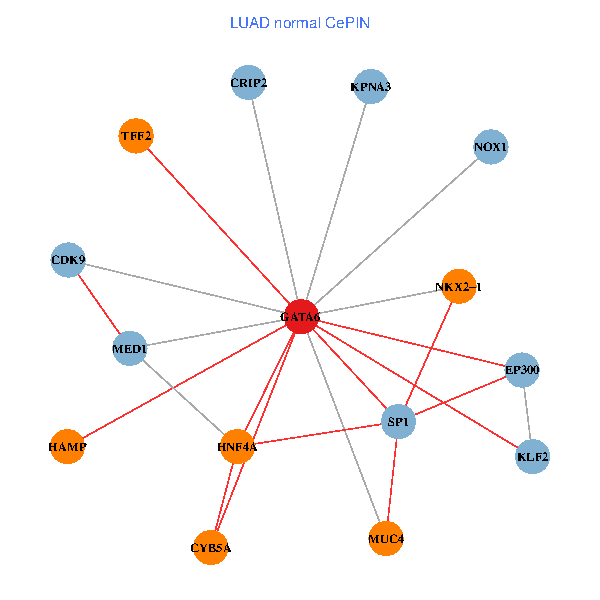 |
|
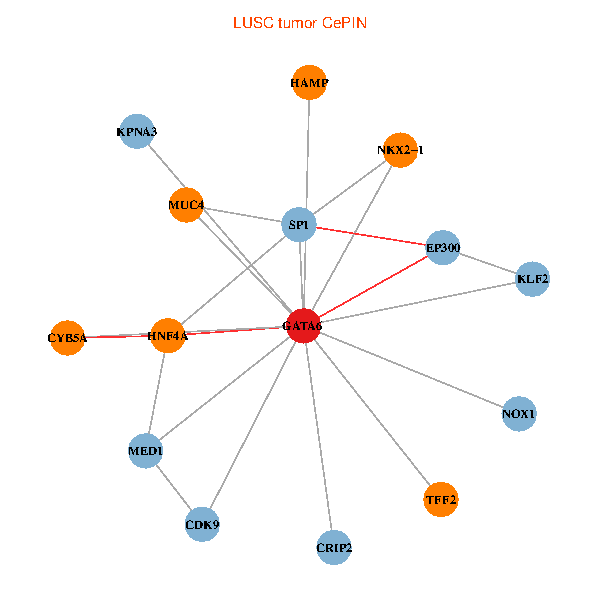
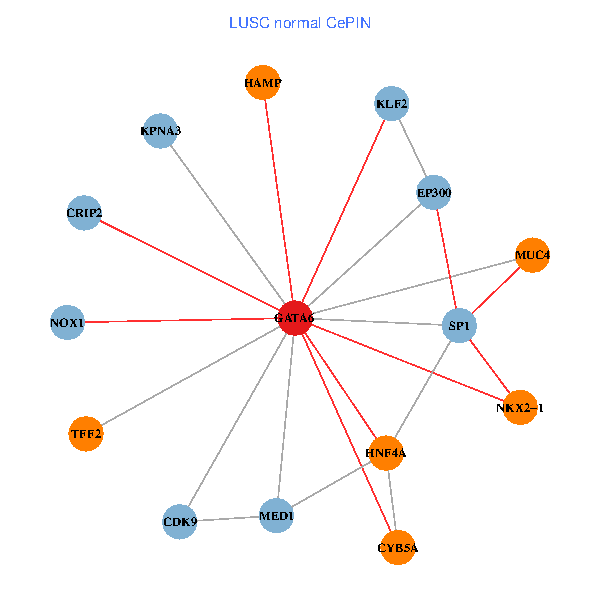
| LUSC (tumor) | LUSC (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
 |  |
|
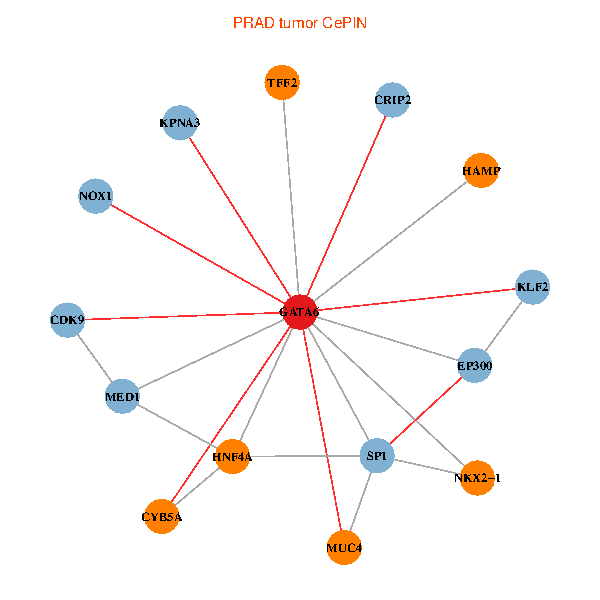
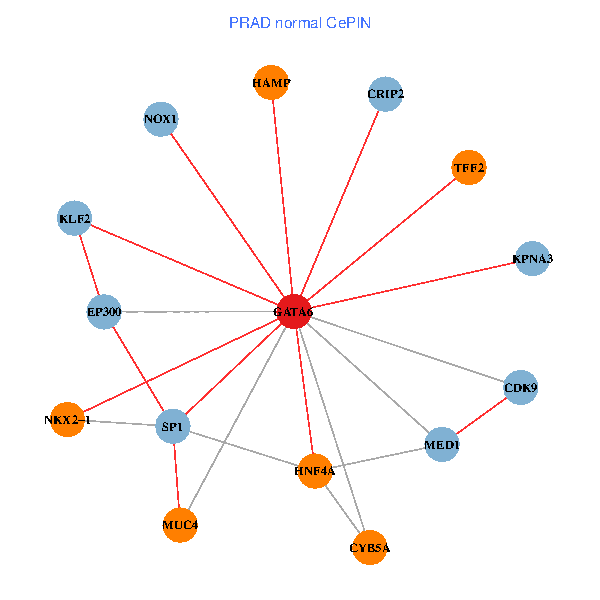
| PRAD (tumor) | PRAD (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
 |  |
|
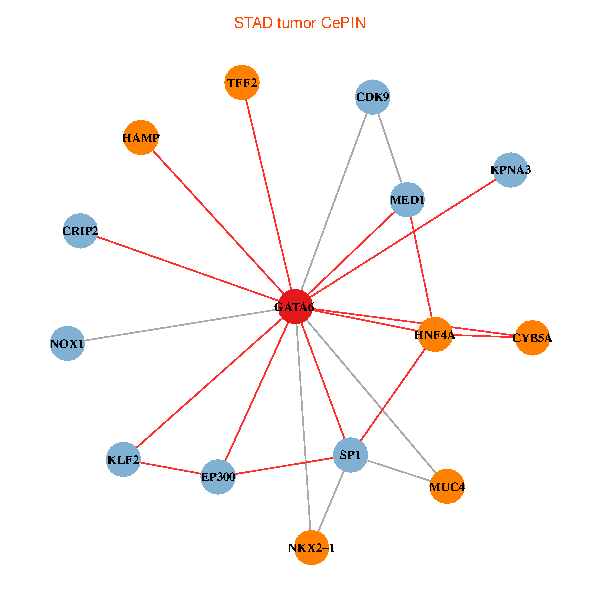
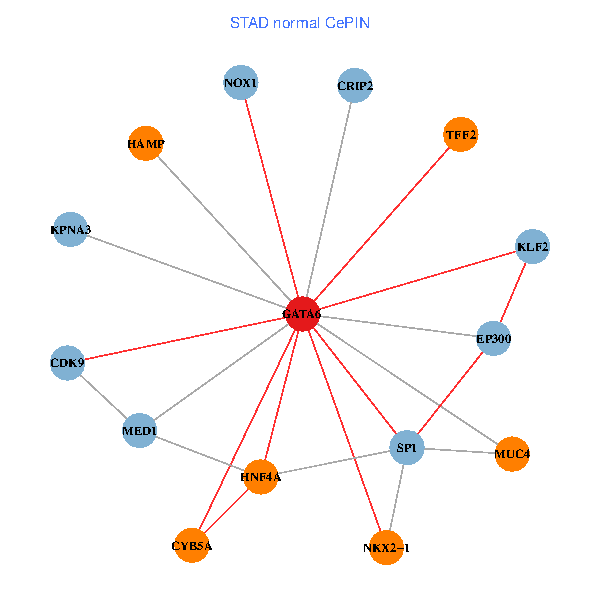
| STAD (tumor) | STAD (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
 |  |
|
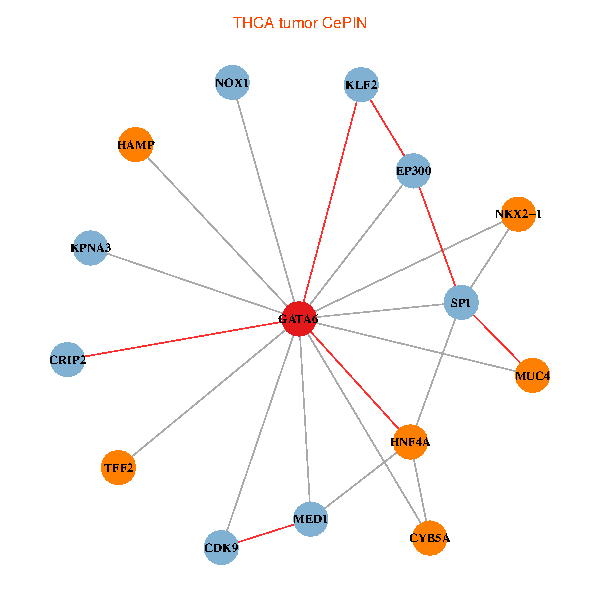
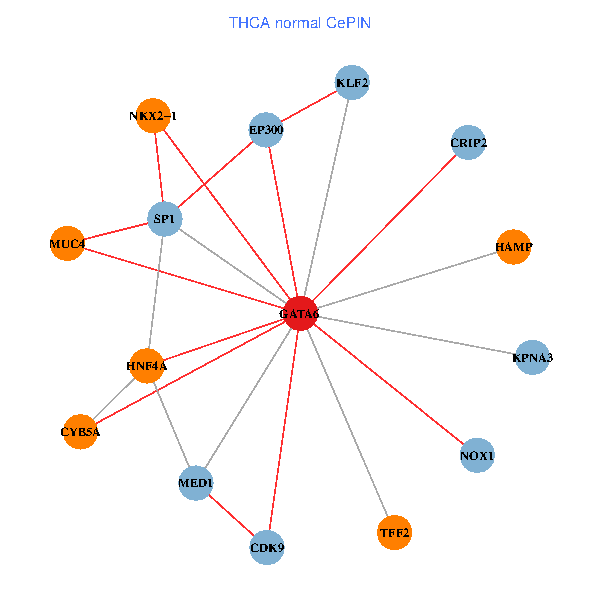
| THCA (tumor) | THCA (normal) |
| GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (tumor) | GATA6, HNF4A, SP1, CDK9, EP300, NKX2-1, CRIP2, MED1, CYB5A, MUC4, NOX1, HAMP, TFF2, KPNA3, KLF2 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0596263 | Carcinogenesis | 8 | BeFree |
| umls:C0018798 | Congenital Heart Defects | 7 | BeFree,GAD |
| umls:C0024623 | Malignant neoplasm of stomach | 5 | BeFree |
| umls:C0152021 | Congenital heart disease | 5 | BeFree |
| umls:C0699791 | Stomach Carcinoma | 5 | BeFree |
| umls:C0000768 | Congenital Abnormality | 4 | BeFree |
| umls:C0009402 | Colorectal Carcinoma | 4 | BeFree |
| umls:C0010068 | Coronary heart disease | 4 | BeFree |
| umls:C0011847 | Diabetes | 4 | BeFree |
| umls:C0011849 | Diabetes Mellitus | 4 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 4 | BeFree |
| umls:C0039685 | Tetralogy of Fallot | 4 | BeFree,CLINVAR,GAD,ORPHANET,UNIPROT |
| umls:C0235974 | Pancreatic carcinoma | 4 | BeFree |
| umls:C0346647 | Malignant neoplasm of pancreas | 4 | BeFree |
| umls:C0699790 | Colon Carcinoma | 4 | BeFree |
| umls:C1527249 | Colorectal Cancer | 4 | BeFree |
| umls:C0001418 | Adenocarcinoma | 3 | BeFree |
| umls:C0001430 | Adenoma | 3 | BeFree |
| umls:C0007102 | Malignant tumor of colon | 3 | BeFree |
| umls:C0018818 | Ventricular Septal Defects | 3 | BeFree |
| umls:C0029928 | Ovarian Diseases | 3 | BeFree |
| umls:C0041207 | Truncus Arteriosus, Persistent | 3 | BeFree,CLINVAR |
| umls:C0158981 | Neonatal diabetes mellitus | 3 | BeFree |
| umls:C0206686 | Adrenocortical carcinoma | 3 | BeFree |
| umls:C1140680 | Malignant neoplasm of ovary | 3 | BeFree,GAD |
| umls:C0002793 | Anaplasia | 2 | BeFree |
| umls:C0029925 | Ovarian Carcinoma | 2 | BeFree |
| umls:C0032460 | Polycystic Ovary Syndrome | 2 | BeFree,LHGDN |
| umls:C0205851 | Germ cell tumor | 2 | BeFree |
| umls:C0232197 | Fibrillation | 2 | BeFree |
| umls:C0267963 | Exocrine pancreatic insufficiency | 2 | BeFree |
| umls:C0919267 | ovarian neoplasm | 2 | BeFree |
| umls:C1704272 | Benign Prostatic Hyperplasia | 2 | BeFree |
| umls:C0001624 | Adrenal Gland Neoplasms | 1 | BeFree |
| umls:C0004114 | Astrocytoma | 1 | BeFree |
| umls:C0005694 | Bladder neck obstruction | 1 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 1 | BeFree |
| umls:C0007138 | Carcinoma, Transitional Cell | 1 | BeFree |
| umls:C0007193 | Cardiomyopathy, Dilated | 1 | BeFree |
| umls:C0010346 | Crohn Disease | 1 | GAD |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | BeFree |
| umls:C0014145 | Yolk Sac Tumor | 1 | BeFree,LHGDN |
| umls:C0014175 | Endometriosis | 1 | BeFree |
| umls:C0017152 | Gastritis | 1 | BeFree |
| umls:C0017636 | Glioblastoma | 1 | BeFree |
| umls:C0017638 | Glioma | 1 | BeFree |
| umls:C0018816 | Heart Septal Defects | 1 | BeFree |
| umls:C0018817 | Atrial Septal Defects | 1 | BeFree,GAD |
| umls:C0019284 | Diaphragmatic Hernia | 1 | CTD_human |
| umls:C0020542 | Pulmonary Hypertension | 1 | RGD |
| umls:C0021390 | Inflammatory Bowel Diseases | 1 | GAD |
| umls:C0023903 | Liver neoplasms | 1 | BeFree |
| umls:C0024121 | Lung Neoplasms | 1 | CTD_human |
| umls:C0030297 | Pancreatic Neoplasm | 1 | LHGDN |
| umls:C0036421 | Systemic Scleroderma | 1 | BeFree |
| umls:C0036920 | Sezary Syndrome | 1 | BeFree |
| umls:C0038356 | Stomach Neoplasms | 1 | LHGDN |
| umls:C0039538 | Teratoma | 1 | BeFree,LHGDN |
| umls:C0039747 | Thecoma | 1 | BeFree |
| umls:C0085786 | Hamman-Rich syndrome | 1 | BeFree |
| umls:C0149782 | Squamous cell carcinoma of lung | 1 | BeFree |
| umls:C0152013 | Adenocarcinoma of lung (disorder) | 1 | BeFree |
| umls:C0152171 | Idiopathic pulmonary hypertension | 1 | BeFree |
| umls:C0206667 | Adrenal Cortical Adenoma | 1 | BeFree |
| umls:C0206698 | Cholangiocarcinoma | 1 | BeFree |
| umls:C0235833 | Congenital diaphragmatic hernia | 1 | BeFree,ORPHANET |
| umls:C0242379 | Malignant neoplasm of lung | 1 | BeFree |
| umls:C0268855 | Hypertrophy of bladder | 1 | BeFree |
| umls:C0281361 | Adenocarcinoma of pancreas | 1 | BeFree |
| umls:C0334520 | Teratoma, Malignant | 1 | BeFree |
| umls:C0334579 | Anaplastic astrocytoma | 1 | BeFree |
| umls:C0340427 | Familial dilated cardiomyopathy | 1 | BeFree |
| umls:C0574785 | Lower Urinary Tract Symptoms | 1 | BeFree |
| umls:C0678222 | Breast Carcinoma | 1 | BeFree |
| umls:C0686619 | Secondary malignant neoplasm of lymph node | 1 | BeFree |
| umls:C0850572 | Adenomatous polyp of colon | 1 | BeFree |
| umls:C0949059 | Polyp of large intestine | 1 | BeFree |
| umls:C1258085 | Barrett Epithelium | 1 | BeFree |
| umls:C1301034 | Pancreatic intraepithelial neoplasia | 1 | BeFree |
| umls:C1335302 | Pancreatic Ductal Adenocarcinoma | 1 | BeFree |
| umls:C1800706 | Idiopathic Pulmonary Fibrosis | 1 | BeFree |
| umls:C1838780 | Pancreatic Hypoplasia, Congenital, with Diabetes Mellitus and Congenital Heart Disease | 1 | BeFree,CLINVAR |
| umls:C1857586 | CONOTRUNCAL HEART MALFORMATIONS (disorder) | 1 | UNIPROT |
| umls:C2973725 | Pulmonary arterial hypertension | 1 | BeFree |
| umls:C3280939 | ATRIOVENTRICULAR SEPTAL DEFECT 5 | 1 | CLINVAR,UNIPROT |
| umls:C3280943 | ATRIAL SEPTAL DEFECT 9 | 1 | CLINVAR,UNIPROT |
| umls:C3714844 | Pulmonary Hypertension, Primary, 1, With Hereditary Hemorrhagic Telangiectasia | 1 | BeFree |
| umls:C4012454 | HEART DEFECTS, CONGENITAL, AND OTHER CONGENITAL ANOMALIES | 1 | UNIPROT |
| umls:C0344724 | Ostium secundum atrial septal defect | 0 | ORPHANET |
| umls:C0344735 | Partial atrioventricular canal | 0 | ORPHANET |
| umls:C2931296 | Yorifuji Okuno syndrome | 0 | ORPHANET |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))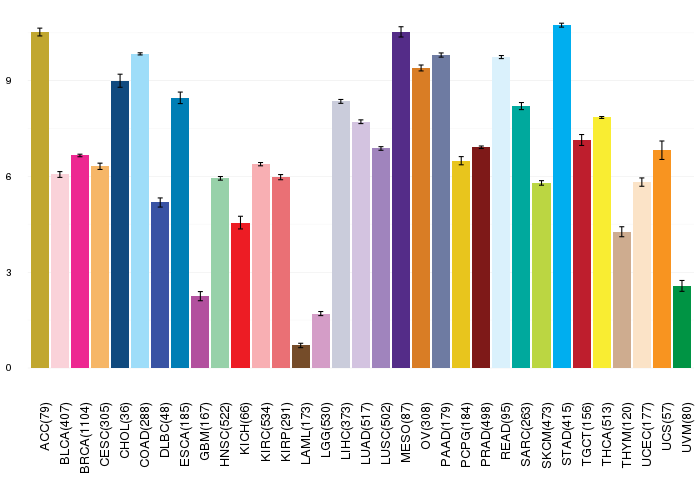
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))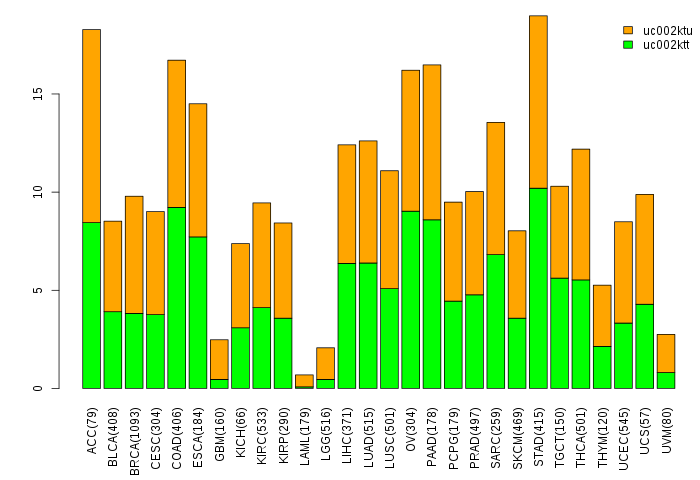
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data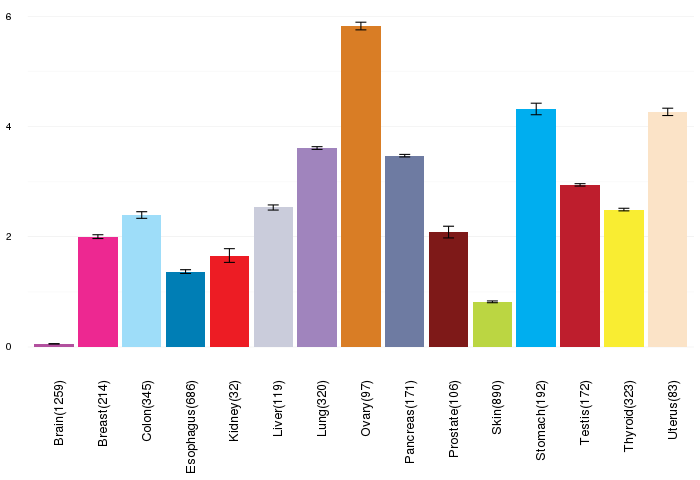
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))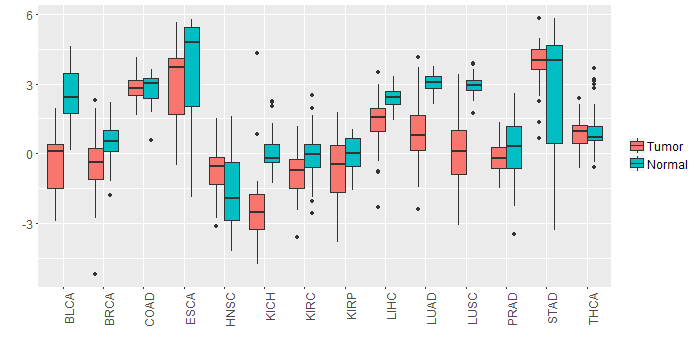
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 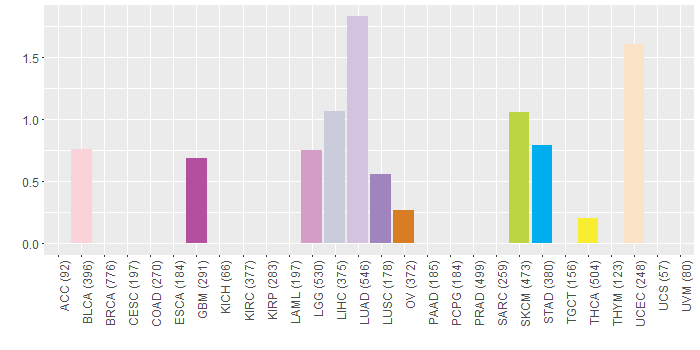
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)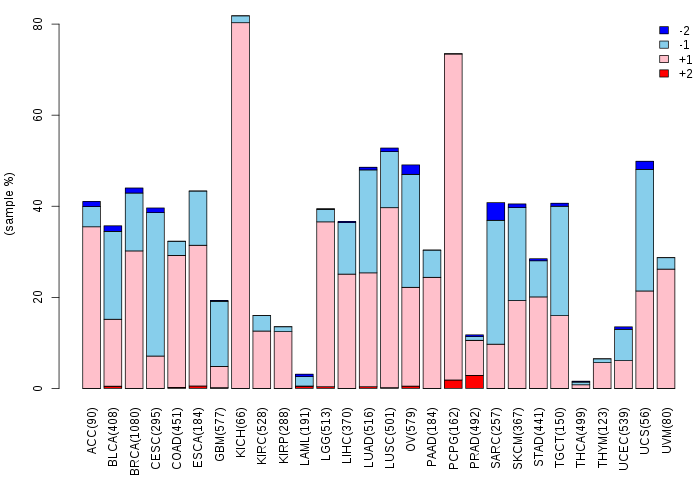
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 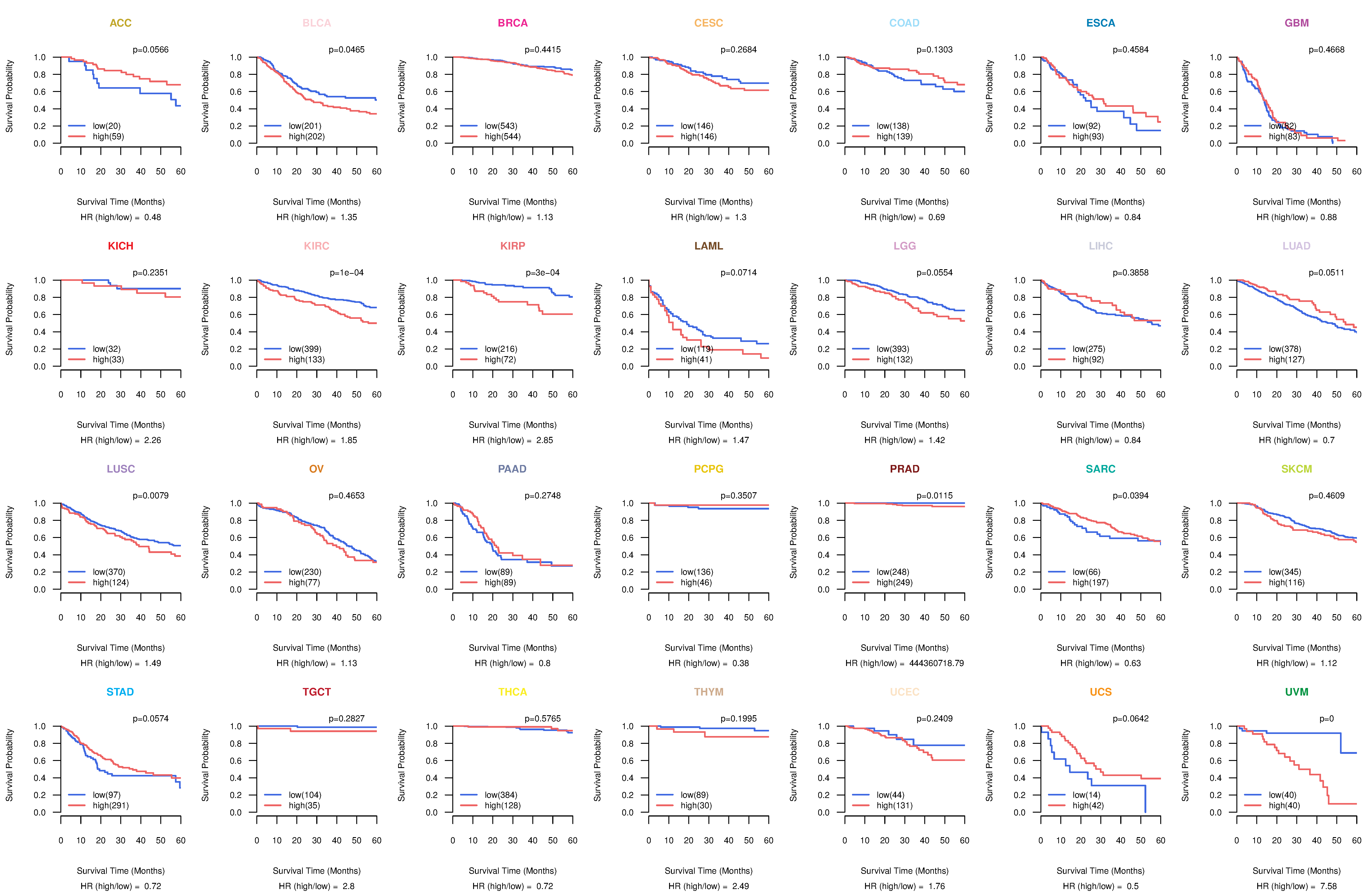
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 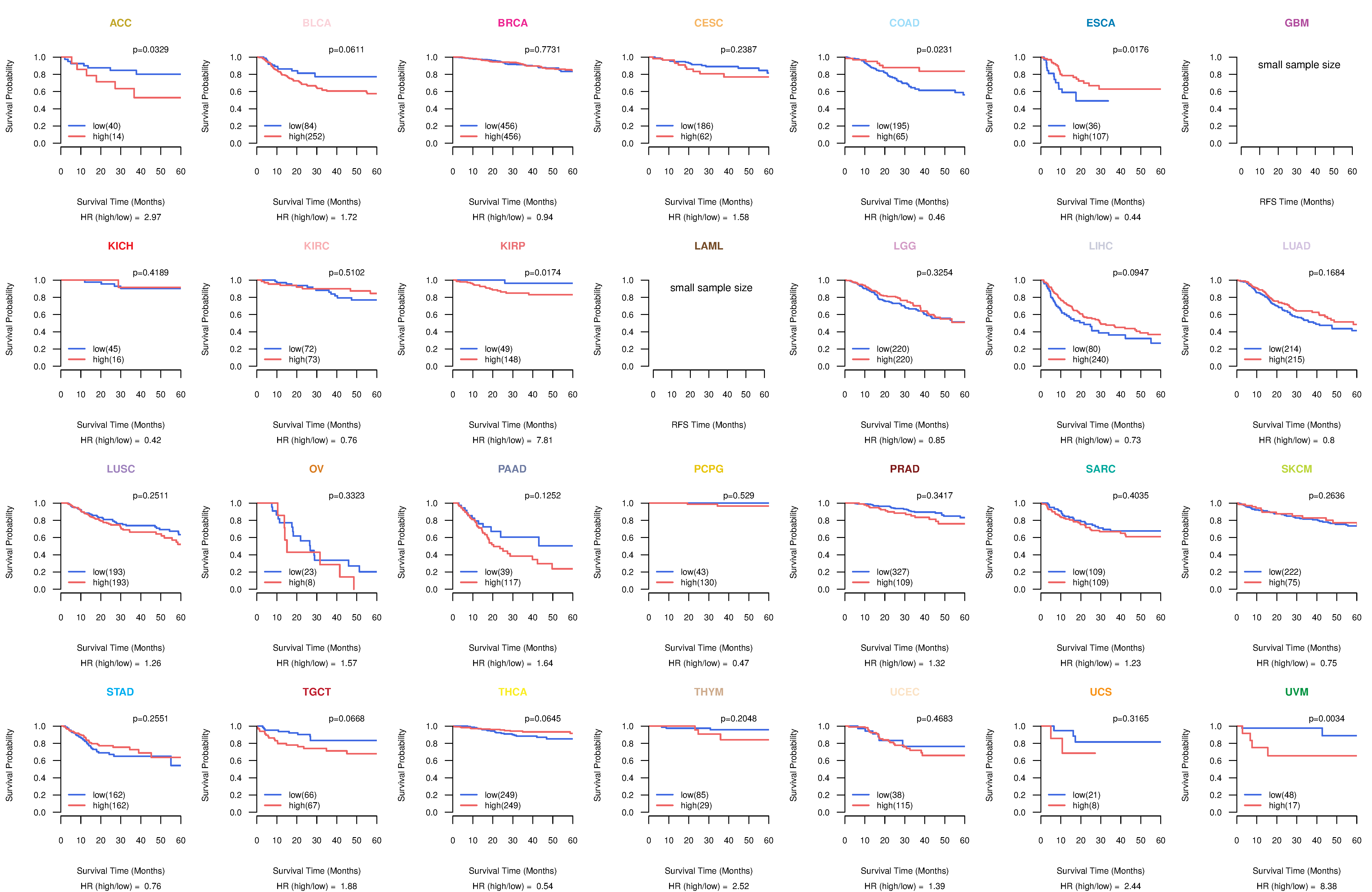
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 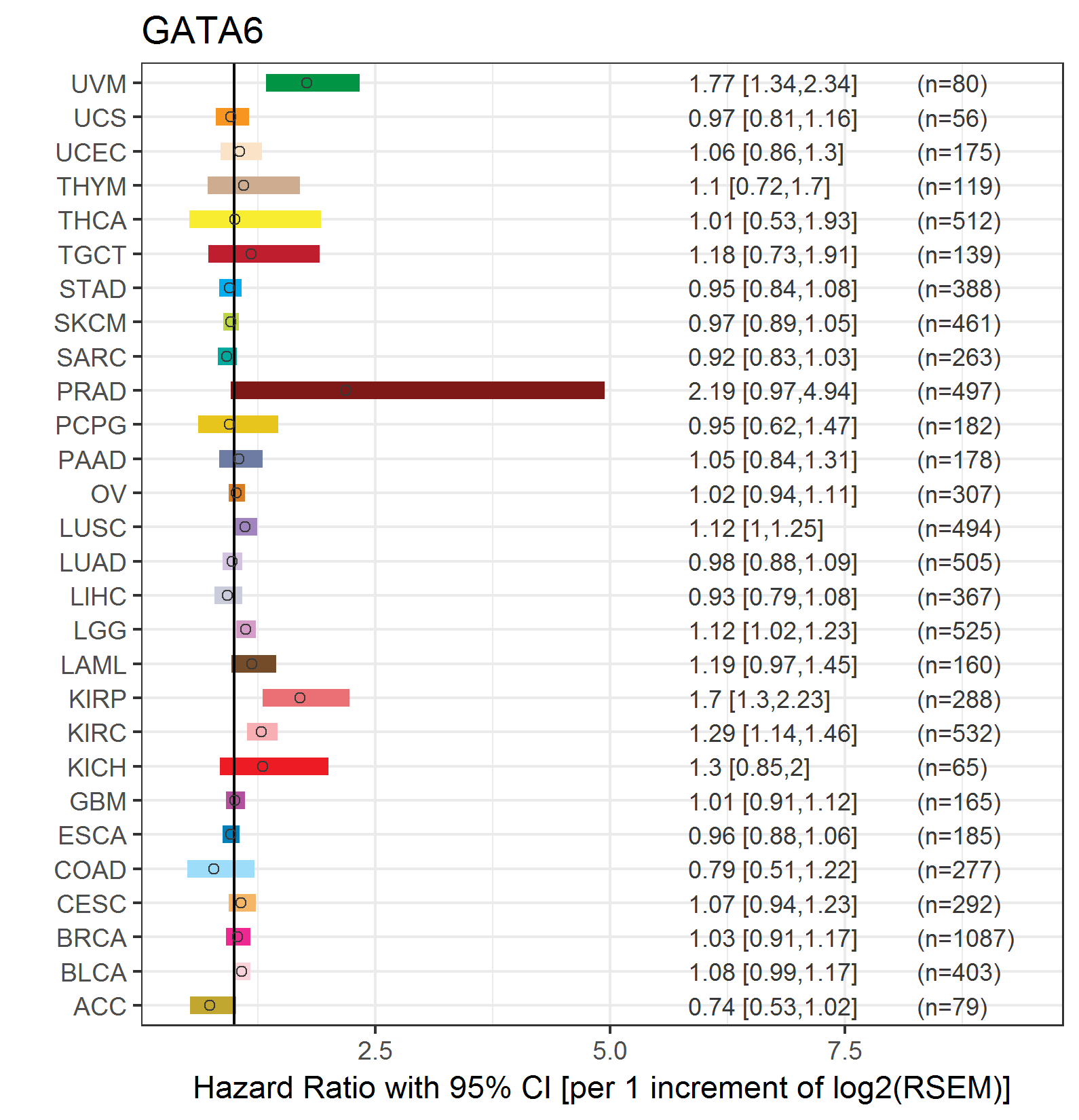
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 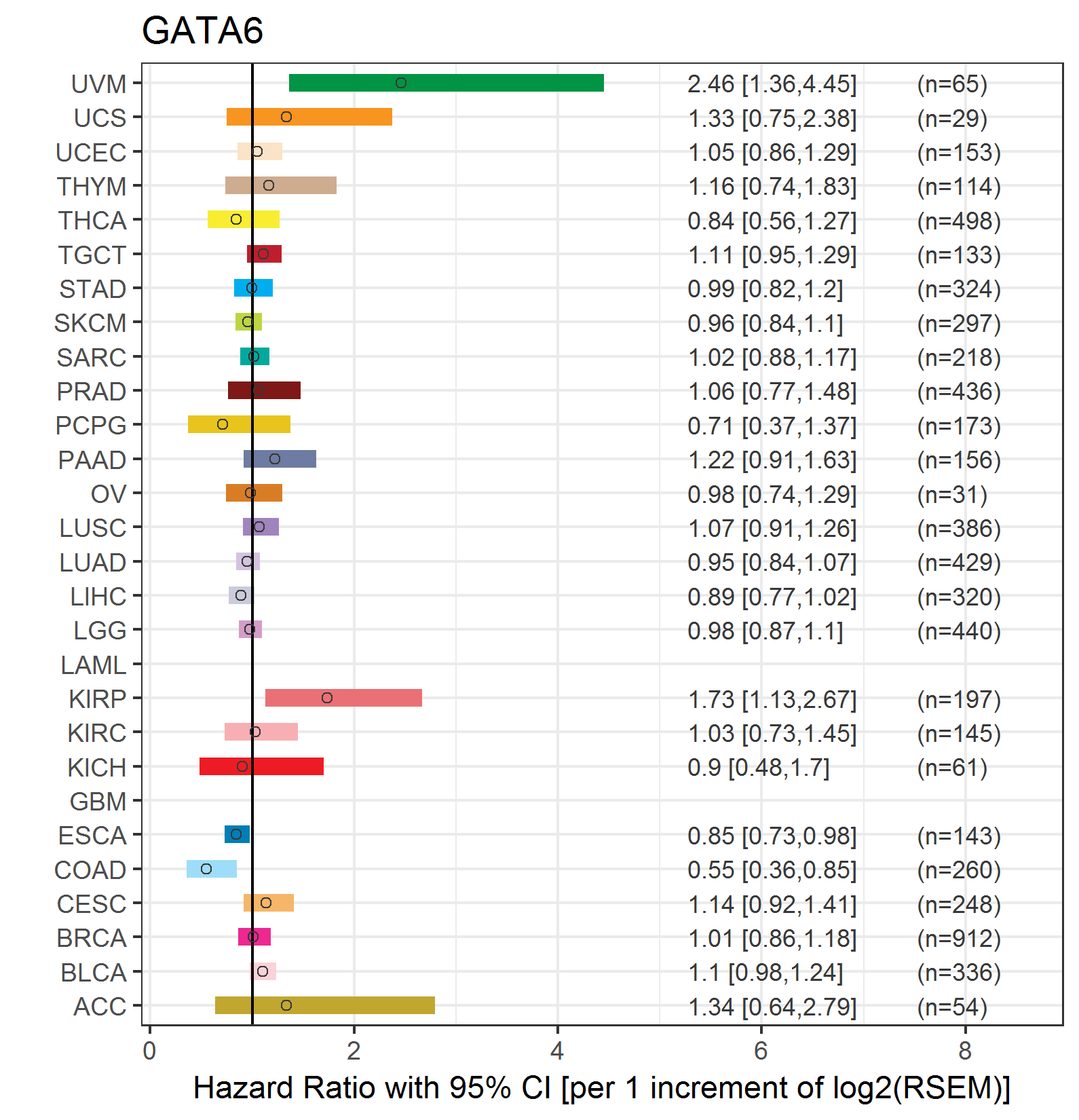
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























