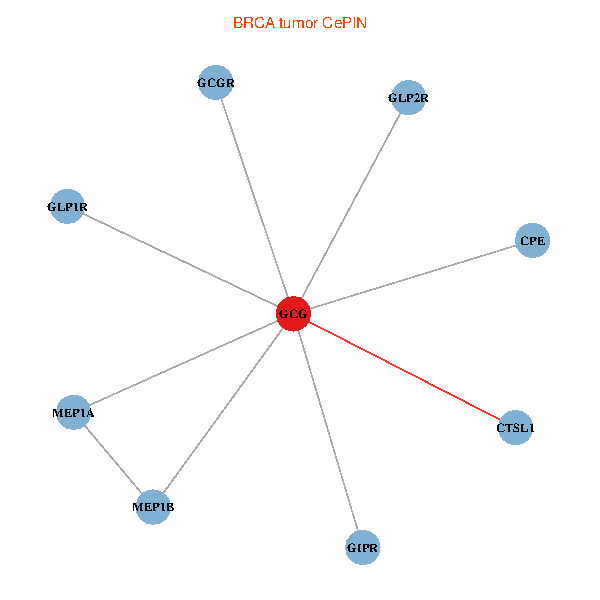
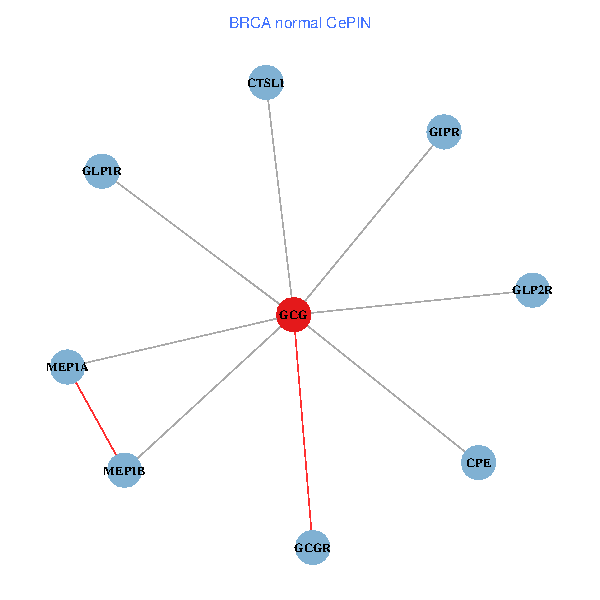
| BRCA (tumor) | BRCA (normal) |
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
 |  |
|
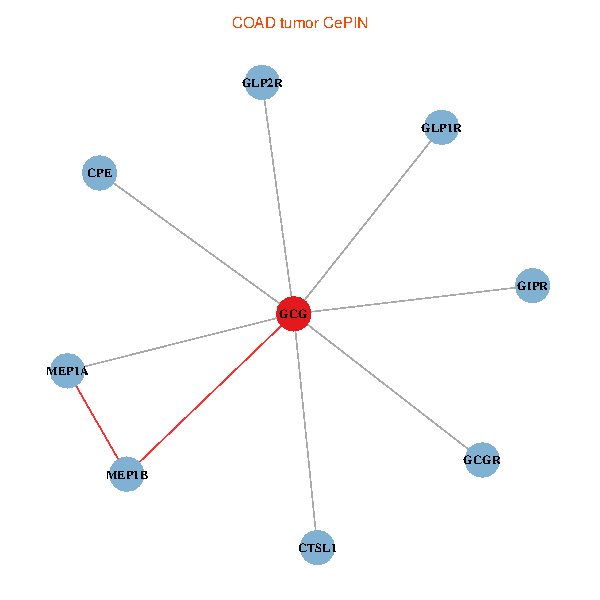
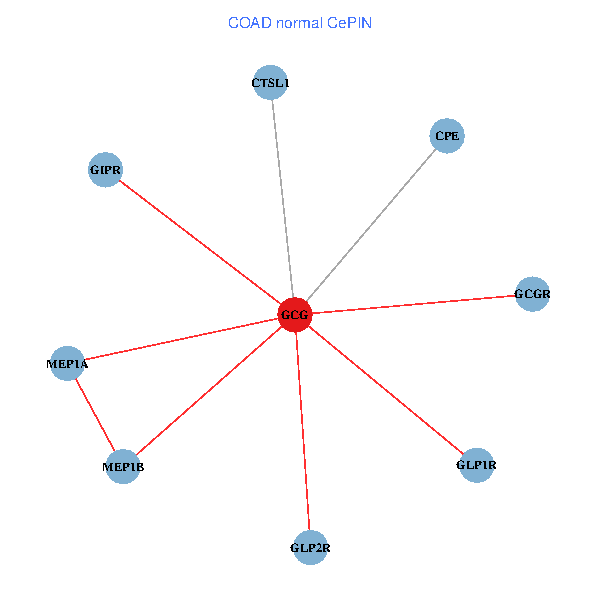
| COAD (tumor) | COAD (normal) |
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
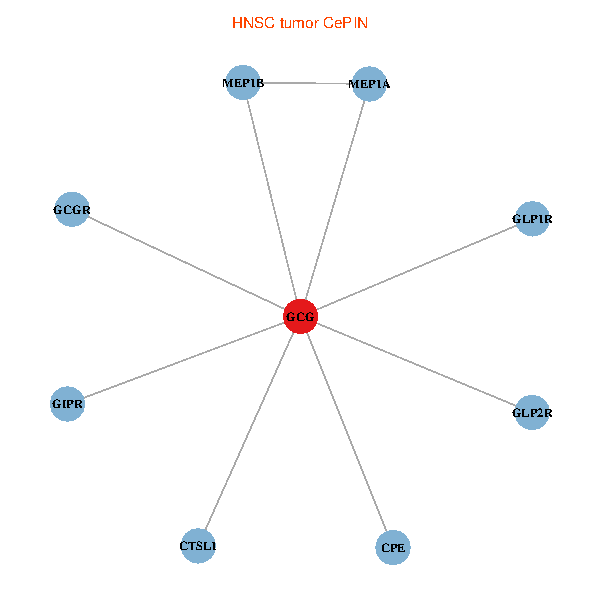
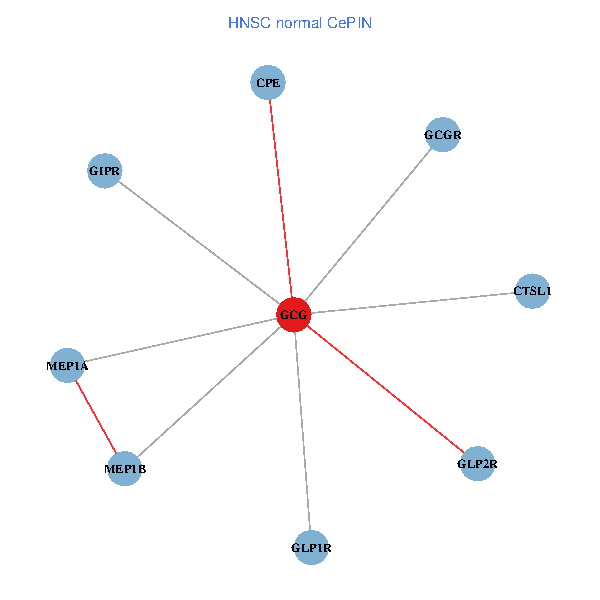
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
 |  |
|
| KICH (tumor) | KICH (normal) |
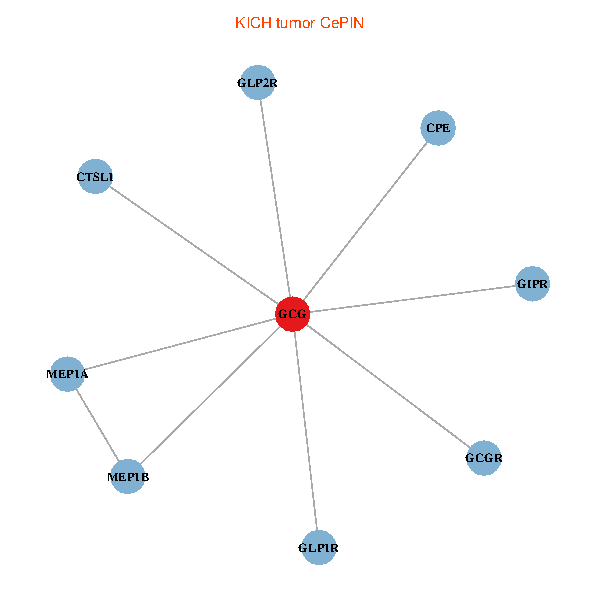
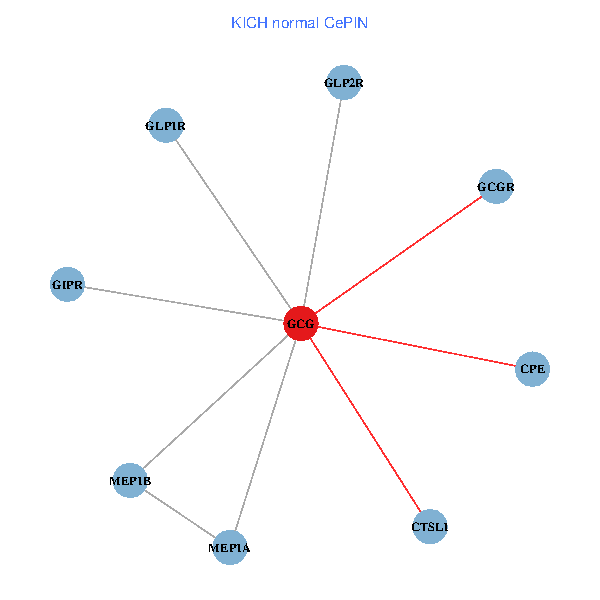
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
 |  |
|
| KIRC (tumor) | KIRC (normal) |
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
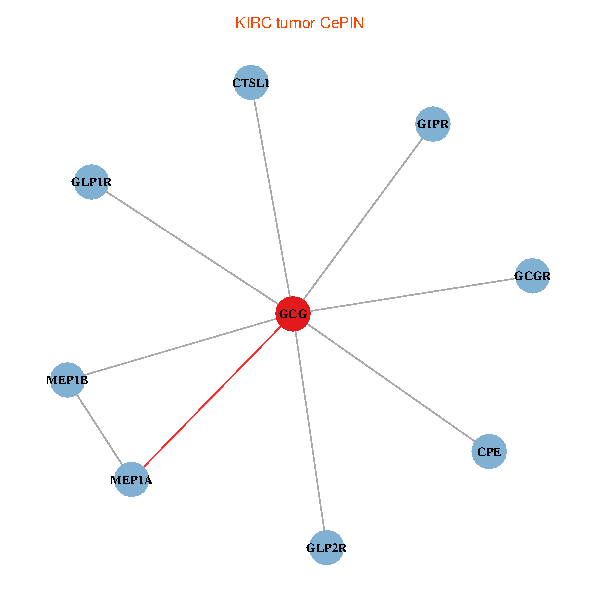 | 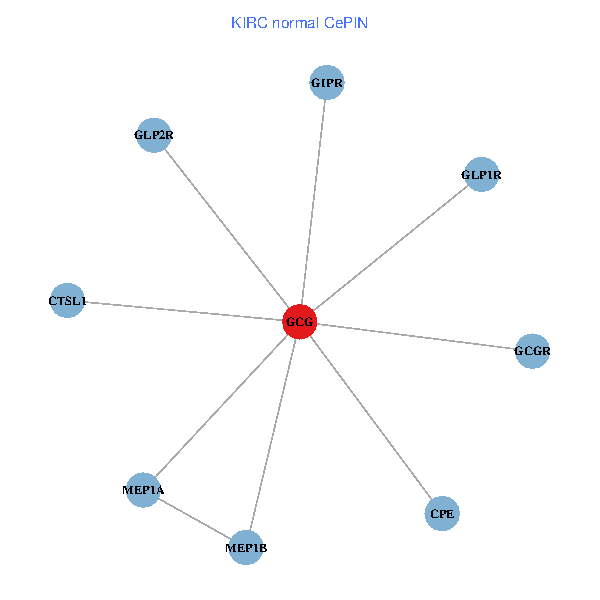 |
|
| KIRP (tumor) | KIRP (normal) |
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
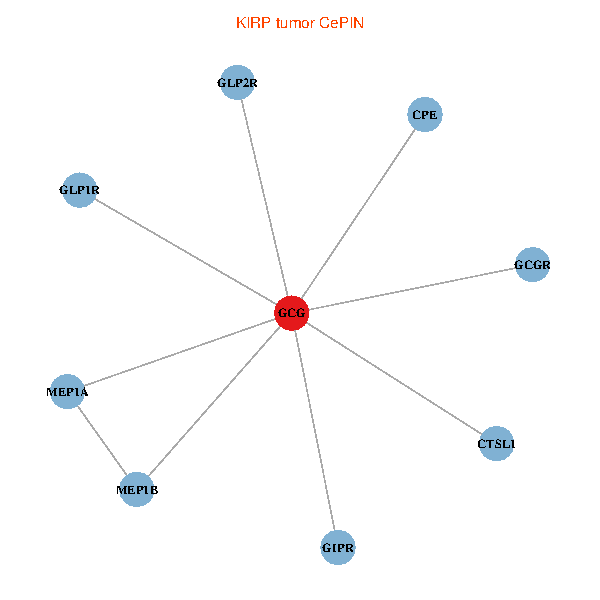 | 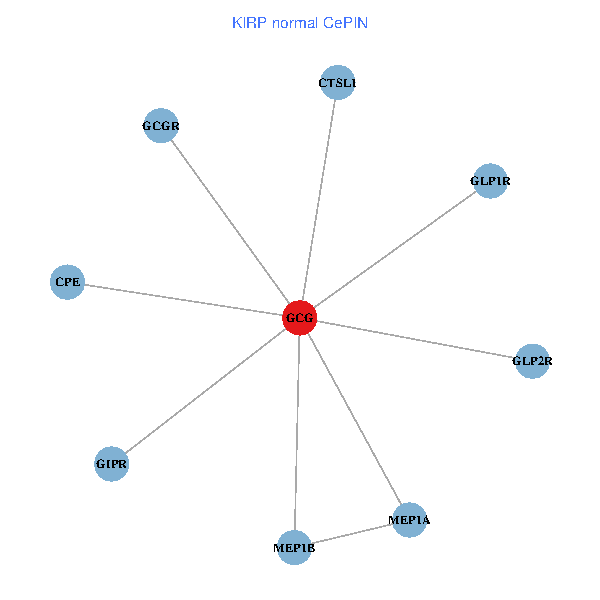 |
|
| LIHC (tumor) | LIHC (normal) |
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
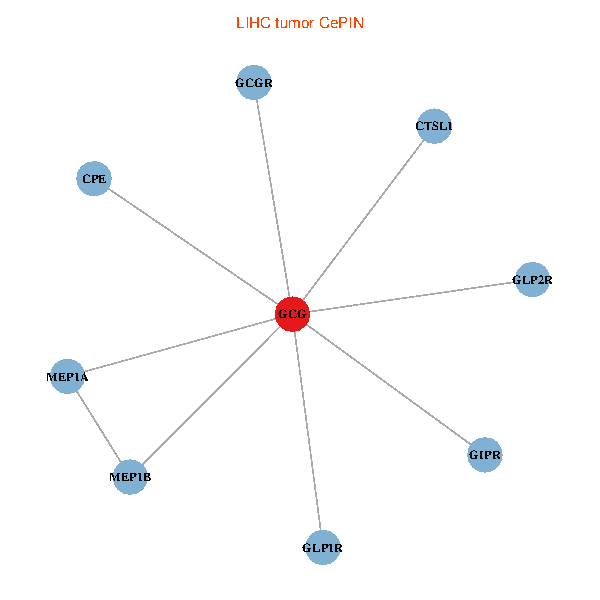 | 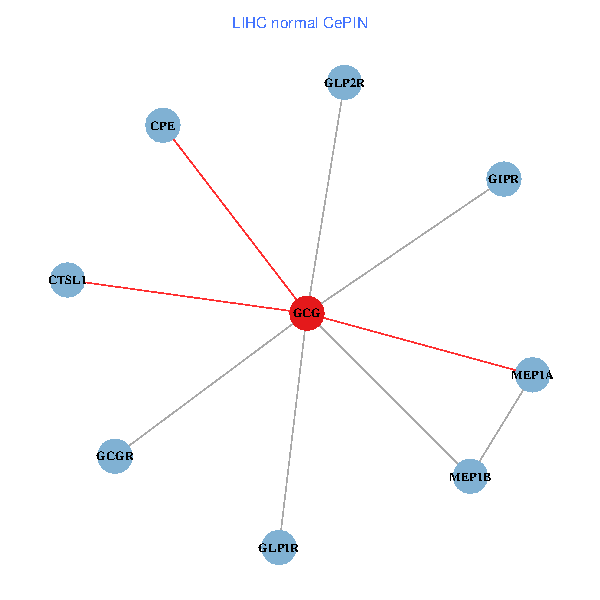 |
|
| LUAD (tumor) | LUAD (normal) |
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
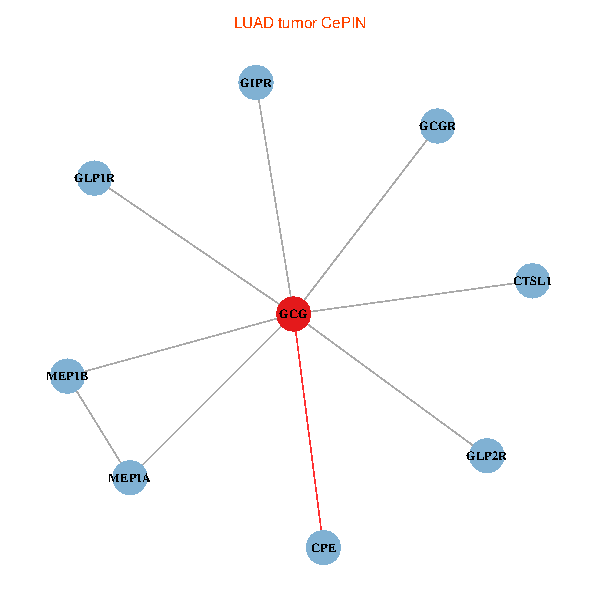 | 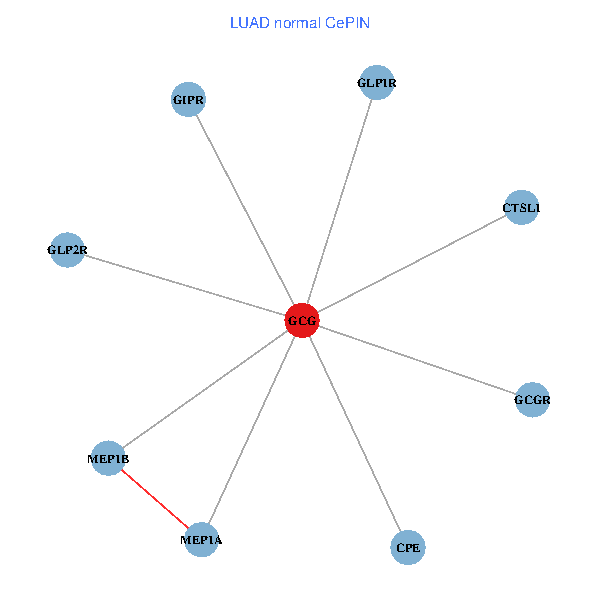 |
|
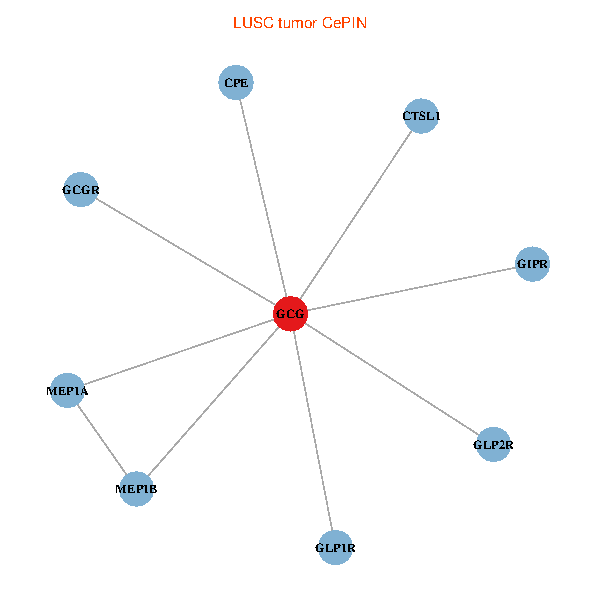
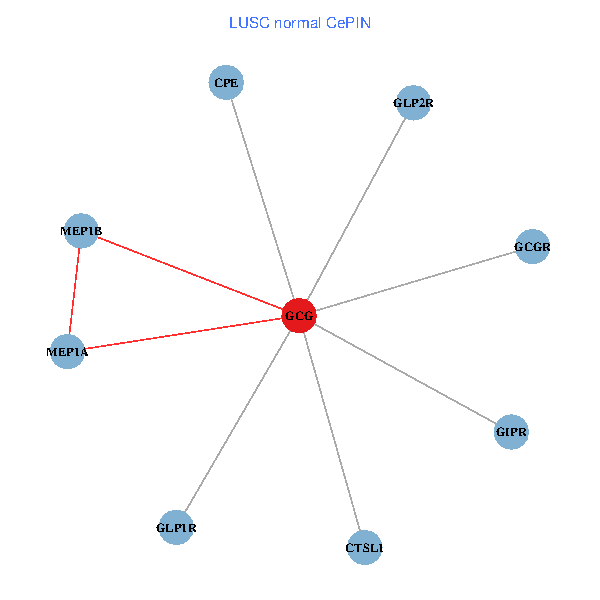
| LUSC (tumor) | LUSC (normal) |
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
 |  |
|
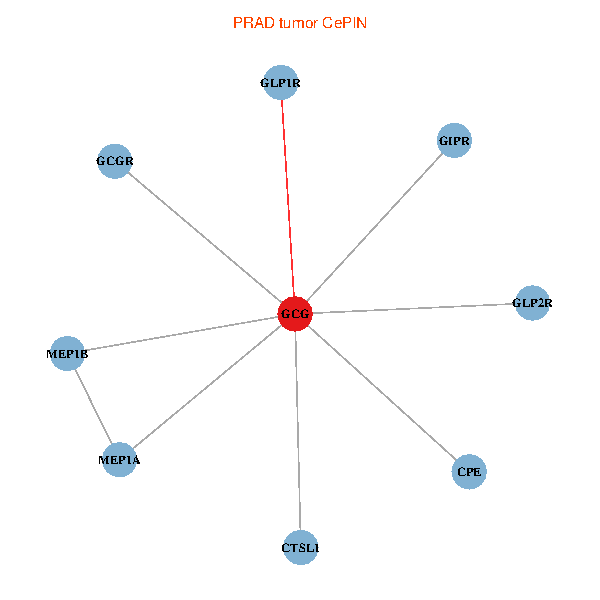
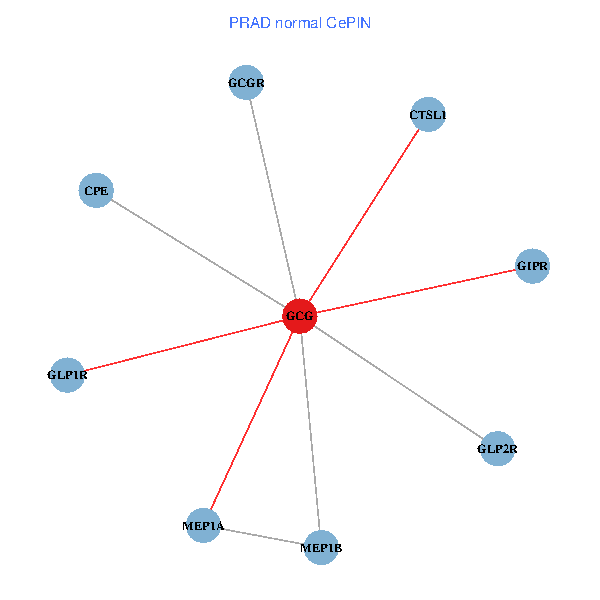
| PRAD (tumor) | PRAD (normal) |
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
 |  |
|
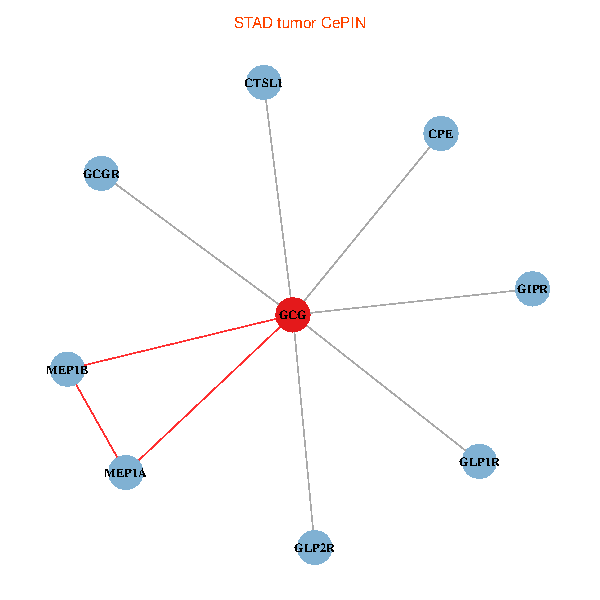
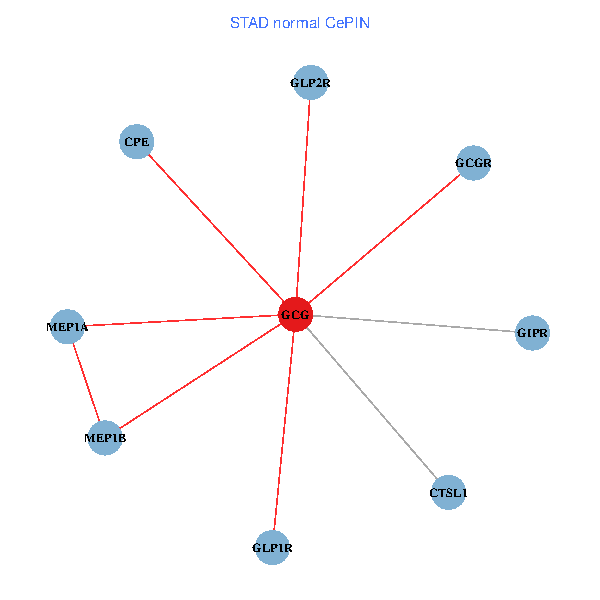
| STAD (tumor) | STAD (normal) |
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
 |  |
|
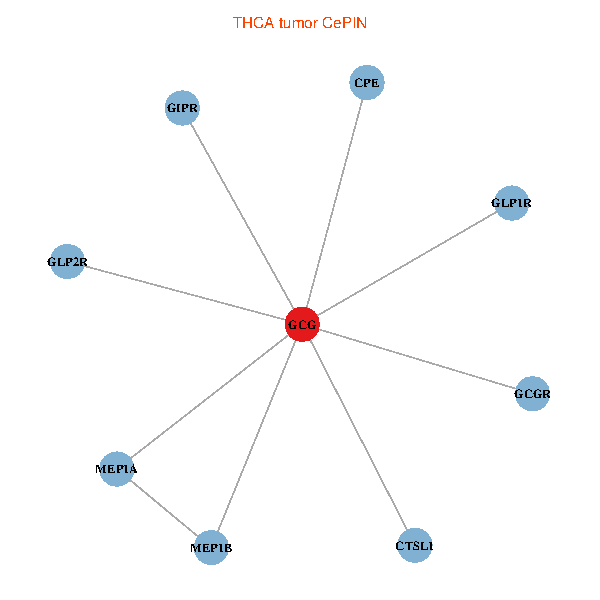
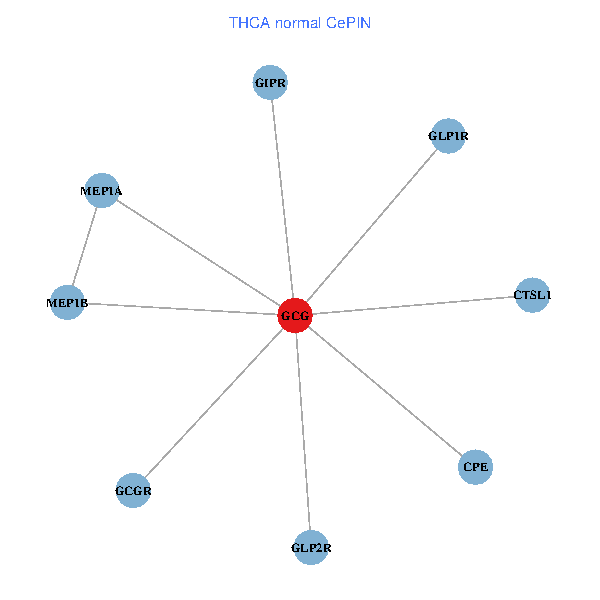
| THCA (tumor) | THCA (normal) |
| GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (tumor) | GCG, CTSL1, MEP1B, MEP1A, GLP1R, GIPR, CPE, GCGR, GLP2R (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 89 | BeFree,CTD_human,GAD |
| umls:C0011849 | Diabetes Mellitus | 50 | BeFree,GAD |
| umls:C0011847 | Diabetes | 41 | BeFree |
| umls:C0020456 | Hyperglycemia | 27 | BeFree,CTD_human |
| umls:C0028754 | Obesity | 27 | BeFree,CTD_human,GAD |
| umls:C0020615 | Hypoglycemia | 20 | BeFree |
| umls:C0011854 | Diabetes Mellitus, Insulin-Dependent | 12 | BeFree |
| umls:C0270952 | Muscular Dystrophy, Oculopharyngeal | 11 | BeFree |
| umls:C0020459 | Hyperinsulinism | 7 | BeFree,CTD_human |
| umls:C0271650 | Impaired glucose tolerance | 5 | BeFree,LHGDN |
| umls:C0428977 | Bradycardia | 5 | CTD_human |
| umls:C0017689 | Glucagonoma | 4 | BeFree |
| umls:C0021670 | insulinoma | 4 | BeFree |
| umls:C0004153 | Atherosclerosis | 3 | BeFree,GAD |
| umls:C0006142 | Malignant neoplasm of breast | 3 | BeFree |
| umls:C0149521 | Pancreatitis, Chronic | 3 | BeFree |
| umls:C0235974 | Pancreatic carcinoma | 3 | BeFree |
| umls:C0346647 | Malignant neoplasm of pancreas | 3 | BeFree |
| umls:C0524620 | Metabolic Syndrome X | 3 | BeFree |
| umls:C0678222 | Breast Carcinoma | 3 | BeFree |
| umls:C0003850 | Arteriosclerosis | 2 | BeFree |
| umls:C0007102 | Malignant tumor of colon | 2 | BeFree |
| umls:C0007222 | Cardiovascular Diseases | 2 | BeFree |
| umls:C0009319 | Colitis | 2 | BeFree,CTD_human |
| umls:C0020538 | Hypertensive disease | 2 | BeFree,CTD_human |
| umls:C0020649 | Hypotension | 2 | CTD_human |
| umls:C0022104 | Irritable Bowel Syndrome | 2 | BeFree,LHGDN |
| umls:C0027627 | Neoplasm Metastasis | 2 | BeFree,LHGDN |
| umls:C0031511 | Pheochromocytoma | 2 | BeFree |
| umls:C0032002 | Pituitary Diseases | 2 | BeFree |
| umls:C0036992 | Short Bowel Syndrome | 2 | BeFree,LHGDN |
| umls:C0155626 | Acute myocardial infarction | 2 | BeFree |
| umls:C0206754 | Neuroendocrine Tumors | 2 | BeFree |
| umls:C0242339 | Dyslipidemias | 2 | BeFree |
| umls:C0342276 | Maturity onset diabetes mellitus in young | 2 | BeFree |
| umls:C0376358 | Malignant neoplasm of prostate | 2 | BeFree |
| umls:C0400966 | Non-alcoholic Fatty Liver Disease | 2 | BeFree |
| umls:C1834014 | Oculopharyngodistal Myopathy | 2 | BeFree |
| umls:C1956346 | Coronary Artery Disease | 2 | BeFree |
| umls:C0001418 | Adenocarcinoma | 1 | LHGDN |
| umls:C0003125 | Anorexia Nervosa | 1 | LHGDN |
| umls:C0004509 | Azoospermia | 1 | BeFree |
| umls:C0007107 | Malignant neoplasm of larynx | 1 | BeFree |
| umls:C0007193 | Cardiomyopathy, Dilated | 1 | BeFree |
| umls:C0007370 | Catalepsy | 1 | CTD_human |
| umls:C0009375 | Colonic Neoplasms | 1 | CTD_human |
| umls:C0009421 | Comatose | 1 | CTD_human |
| umls:C0009806 | Constipation | 1 | CTD_human |
| umls:C0010054 | Coronary Arteriosclerosis | 1 | BeFree |
| umls:C0010068 | Coronary heart disease | 1 | BeFree |
| umls:C0010346 | Crohn Disease | 1 | BeFree |
| umls:C0011853 | Diabetes Mellitus, Experimental | 1 | RGD |
| umls:C0011881 | Diabetic Nephropathy | 1 | BeFree |
| umls:C0011991 | Diarrhea | 1 | BeFree |
| umls:C0016057 | Fibrosarcoma | 1 | BeFree |
| umls:C0016470 | Food Allergy | 1 | LHGDN |
| umls:C0018799 | Heart Diseases | 1 | CTD_human |
| umls:C0018801 | Heart failure | 1 | CTD_human |
| umls:C0019562 | Von Hippel-Lindau Syndrome | 1 | BeFree |
| umls:C0020473 | Hyperlipidemia | 1 | CTD_human |
| umls:C0020672 | Hypothermia, natural | 1 | CTD_human |
| umls:C0021831 | Intestinal Diseases | 1 | BeFree |
| umls:C0021841 | Intestinal Neoplasms | 1 | BeFree |
| umls:C0023890 | Liver Cirrhosis | 1 | BeFree |
| umls:C0023895 | Liver diseases | 1 | BeFree |
| umls:C0025267 | Multiple Endocrine Neoplasia Type 1 | 1 | BeFree |
| umls:C0025268 | Multiple Endocrine Neoplasia Type 2a | 1 | BeFree |
| umls:C0025286 | Meningioma | 1 | BeFree |
| umls:C0025517 | Metabolic Diseases | 1 | BeFree |
| umls:C0026847 | Spinal Muscular Atrophy | 1 | BeFree |
| umls:C0027051 | Myocardial Infarction | 1 | LHGDN |
| umls:C0027819 | Neuroblastoma | 1 | BeFree |
| umls:C0028259 | Nodule | 1 | BeFree |
| umls:C0032584 | polyps | 1 | BeFree |
| umls:C0033860 | Psoriasis | 1 | BeFree |
| umls:C0037763 | Spasm | 1 | CTD_human |
| umls:C0038354 | Stomach Diseases | 1 | CTD_human |
| umls:C0038522 | Subacute Sclerosing Panencephalitis | 1 | BeFree |
| umls:C0038580 | Substance Dependence | 1 | BeFree |
| umls:C0039231 | Tachycardia | 1 | CTD_human |
| umls:C0039239 | Sinus Tachycardia | 1 | CTD_human |
| umls:C0042373 | Vascular Diseases | 1 | CTD_human |
| umls:C0085110 | Severe Combined Immunodeficiency | 1 | BeFree |
| umls:C0085207 | Gestational Diabetes | 1 | BeFree |
| umls:C0162429 | Malnutrition | 1 | BeFree |
| umls:C0205734 | Diabetes, Autoimmune | 1 | BeFree |
| umls:C0206624 | Hepatoblastoma | 1 | BeFree |
| umls:C0238198 | Gastrointestinal Stromal Tumors | 1 | BeFree |
| umls:C0271708 | Fasting Hypoglycemia | 1 | BeFree |
| umls:C0271710 | Reactive hypoglycemia | 1 | BeFree |
| umls:C0281784 | Benign Meningioma | 1 | BeFree |
| umls:C0333297 | Chronic ulcer | 1 | BeFree |
| umls:C0339273 | Corneal dystrophy, Lattice type 3 | 1 | BeFree |
| umls:C0342277 | Diabetes mellitus autosomal dominant type II (disorder) | 1 | BeFree |
| umls:C0342289 | Diabetes-deafness syndrome maternally transmitted (disorder) | 1 | BeFree |
| umls:C0424295 | Hyperactive behavior | 1 | CTD_human |
| umls:C0524851 | Neurodegenerative Disorders | 1 | BeFree |
| umls:C0600139 | Prostate carcinoma | 1 | BeFree |
| umls:C0600260 | Lung Diseases, Obstructive | 1 | BeFree |
| umls:C0699790 | Colon Carcinoma | 1 | BeFree |
| umls:C0700095 | Central neuroblastoma | 1 | BeFree |
| umls:C0730345 | Microalbuminuria | 1 | BeFree |
| umls:C0858600 | Taste sweet | 1 | BeFree |
| umls:C0878500 | Intraepithelial Neoplasia | 1 | BeFree |
| umls:C1261473 | Sarcoma | 1 | BeFree |
| umls:C1318485 | Liver regeneration disorder | 1 | BeFree |
| umls:C1335302 | Pancreatic Ductal Adenocarcinoma | 1 | BeFree |
| umls:C1510472 | Drug Dependence | 1 | BeFree |
| umls:C1565489 | Renal Insufficiency | 1 | LHGDN |
| umls:C1623038 | Cirrhosis | 1 | BeFree |
| umls:C1859486 | BIETTI CRYSTALLINE CORNEORETINAL DYSTROPHY | 1 | BeFree |
| umls:C1879526 | Aberrant Crypt Foci | 1 | CTD_human |
| umls:C2239176 | Liver carcinoma | 1 | BeFree |
| umls:C3241937 | Nonalcoholic Steatohepatitis | 1 | BeFree |
| umls:C3714619 | Insulin resistance syndrome | 1 | BeFree |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))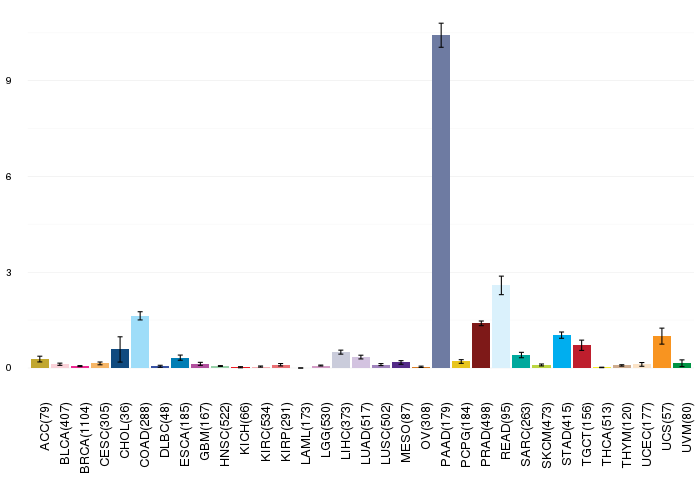
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))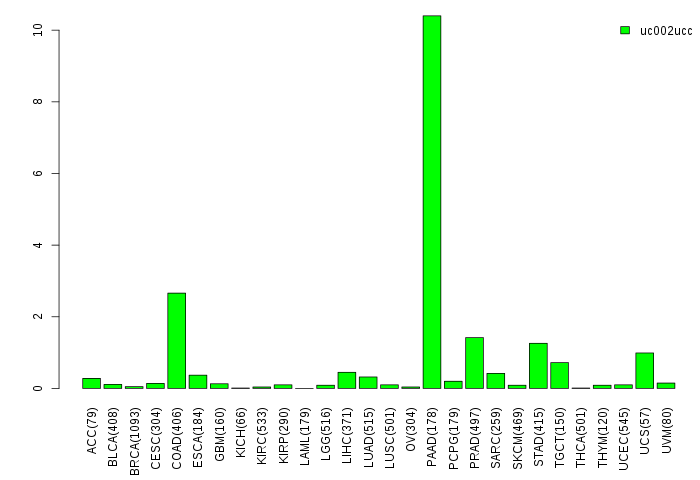
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data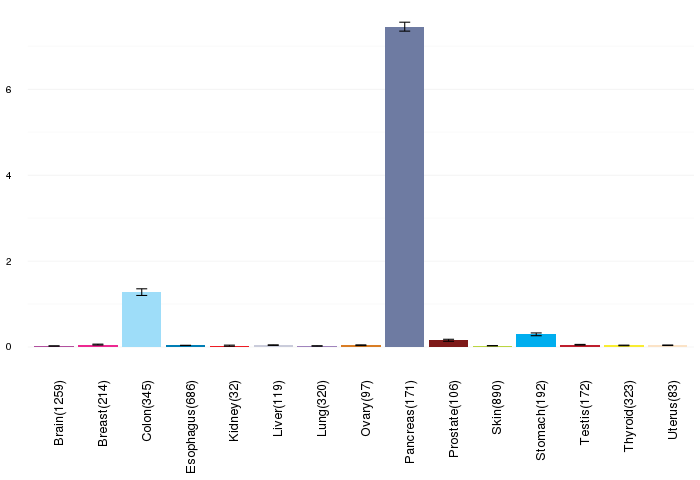
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))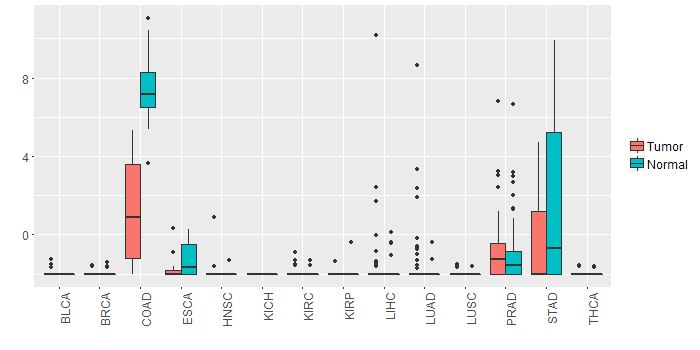
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.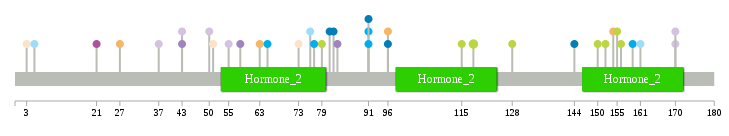

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 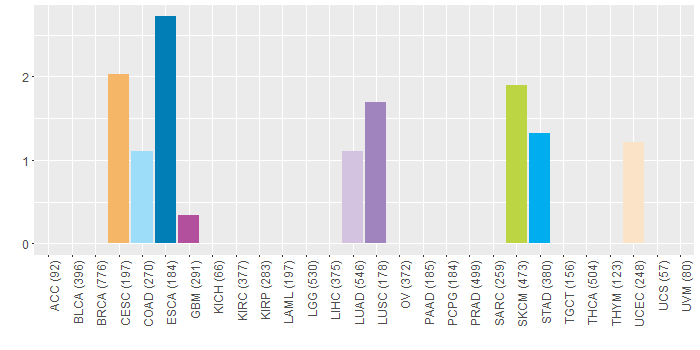
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)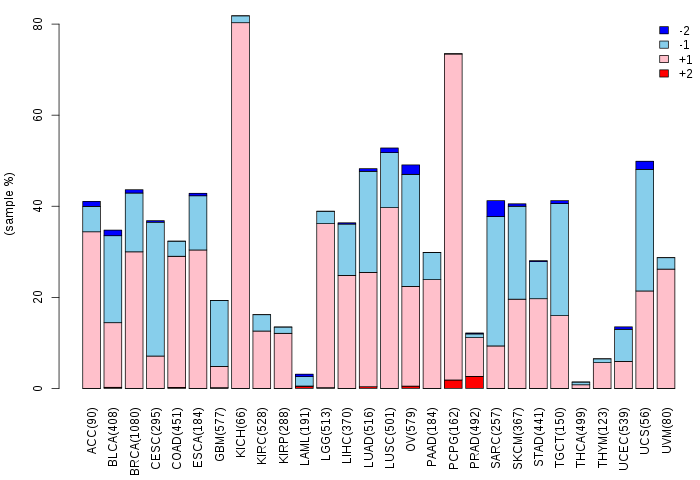
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 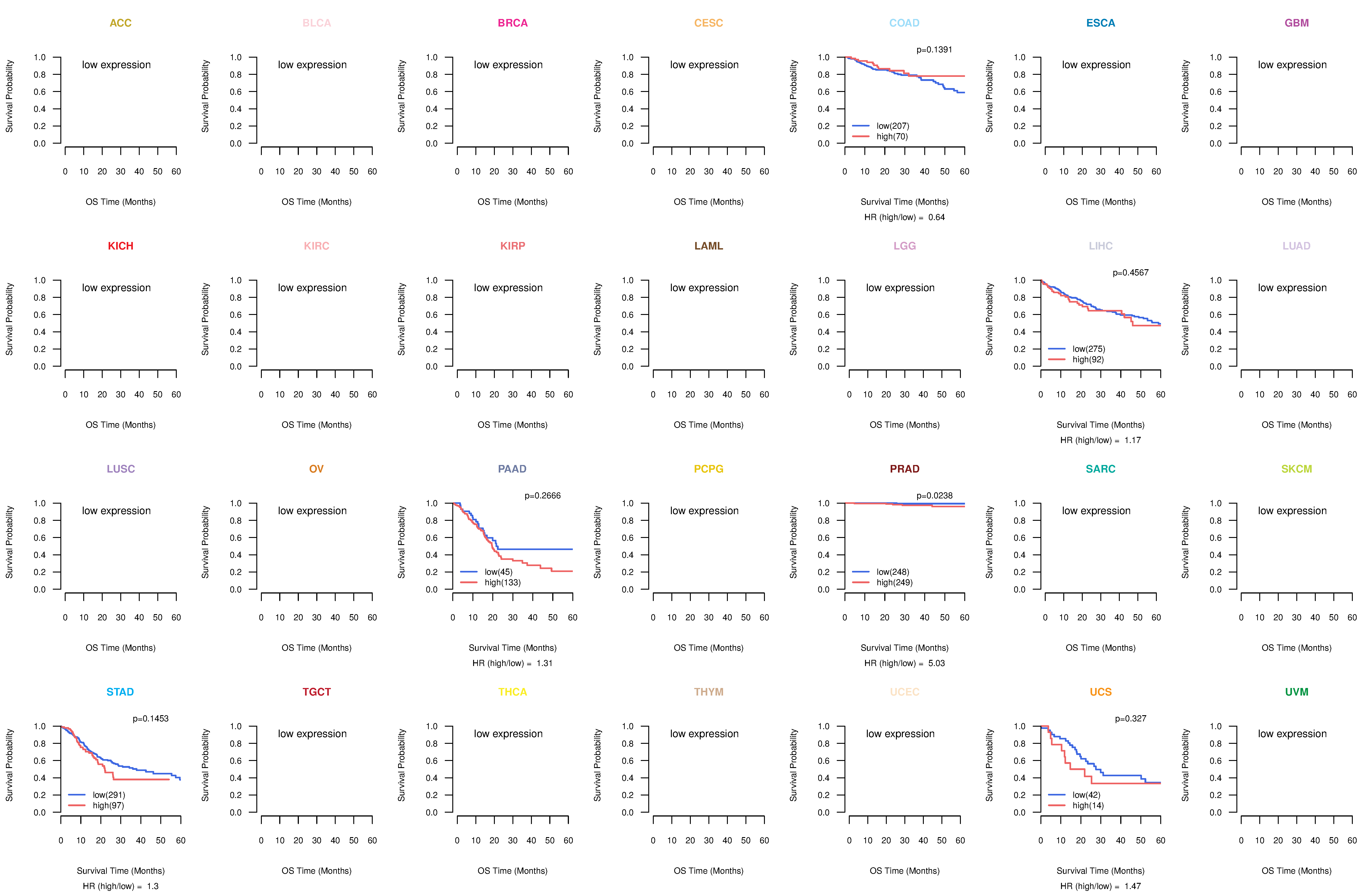
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 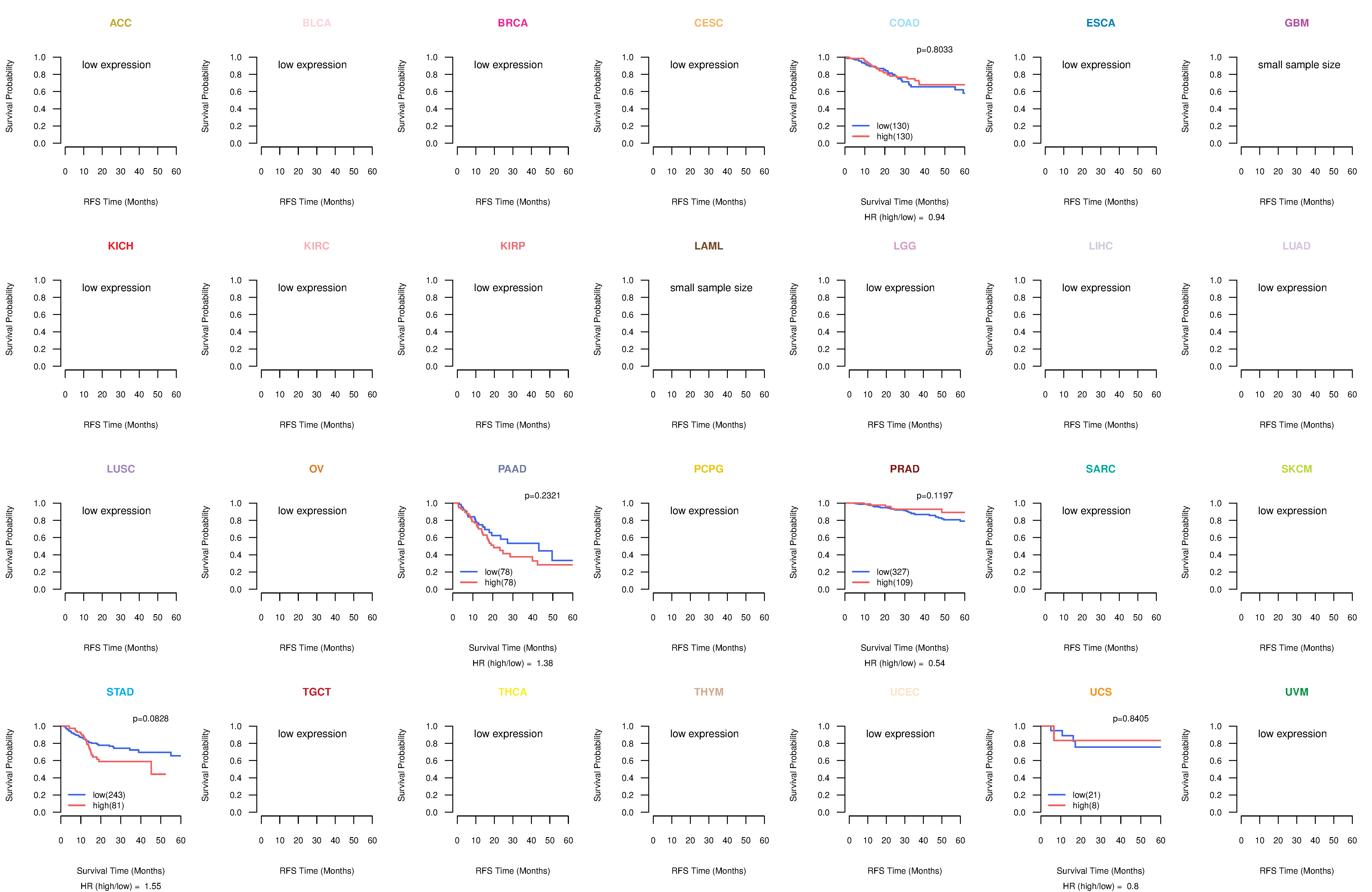
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 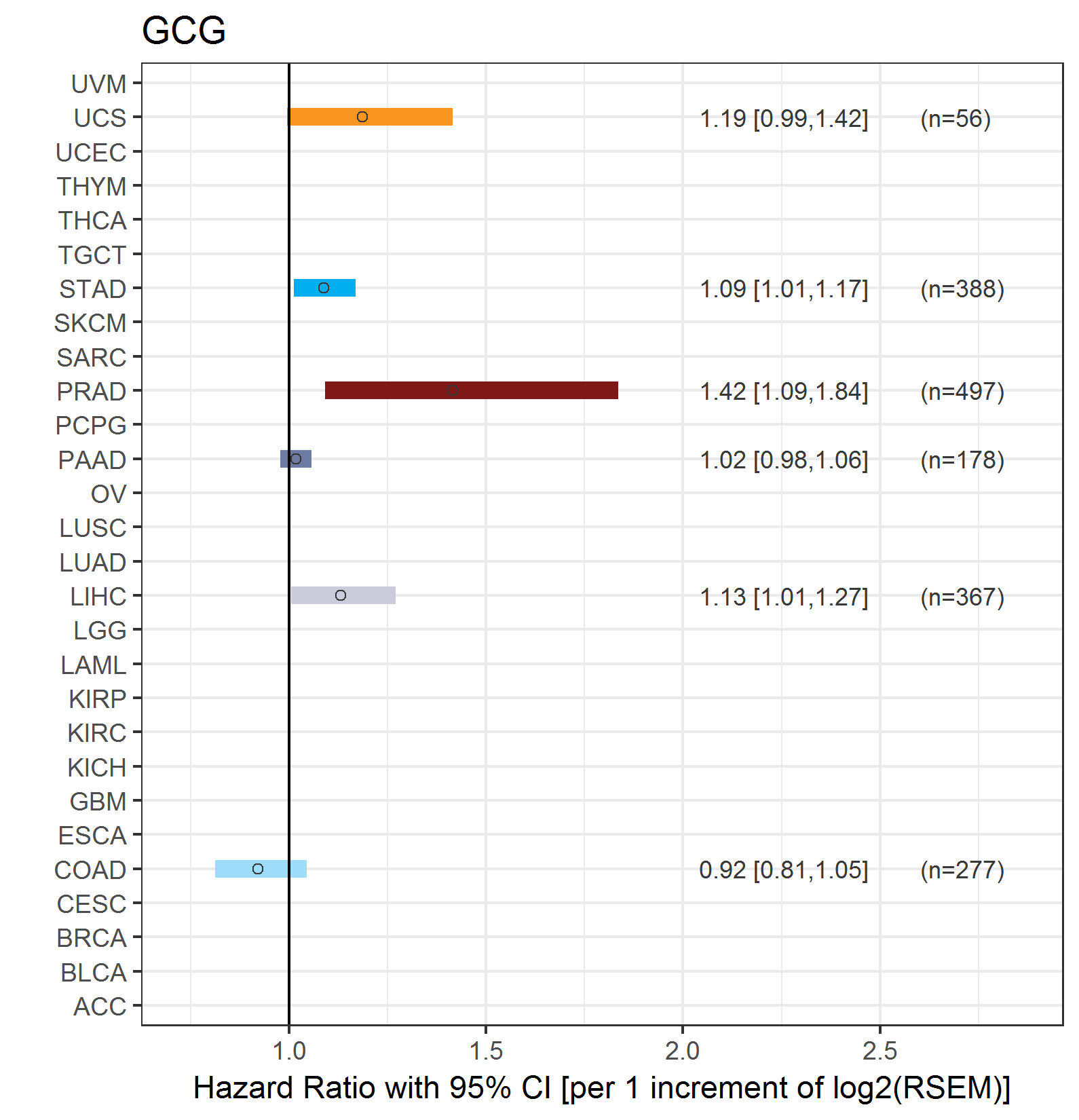
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 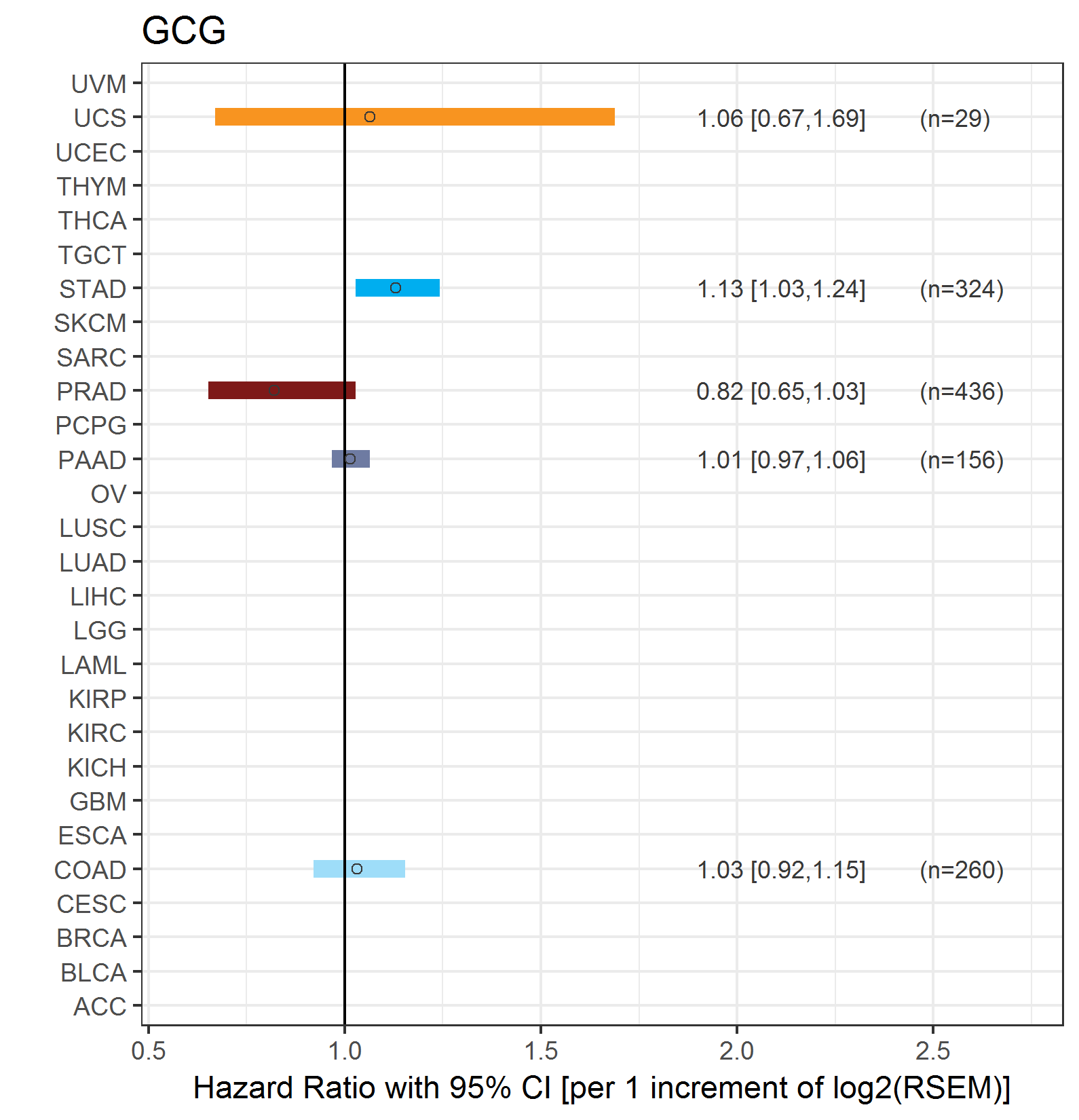
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























