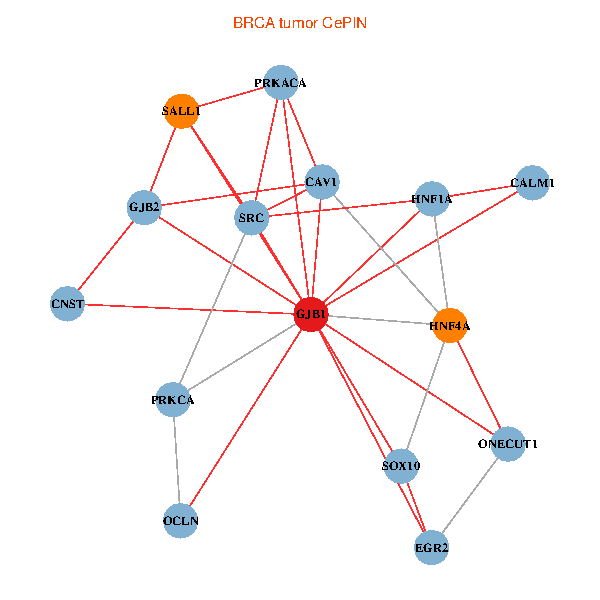
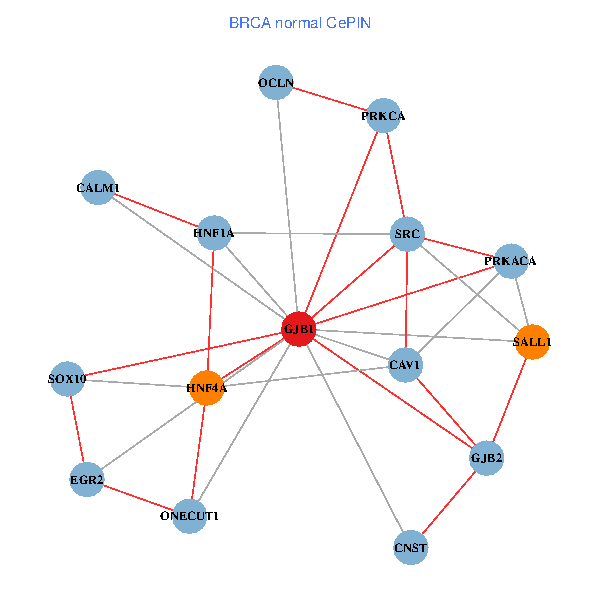
| BRCA (tumor) | BRCA (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
 |  |
|
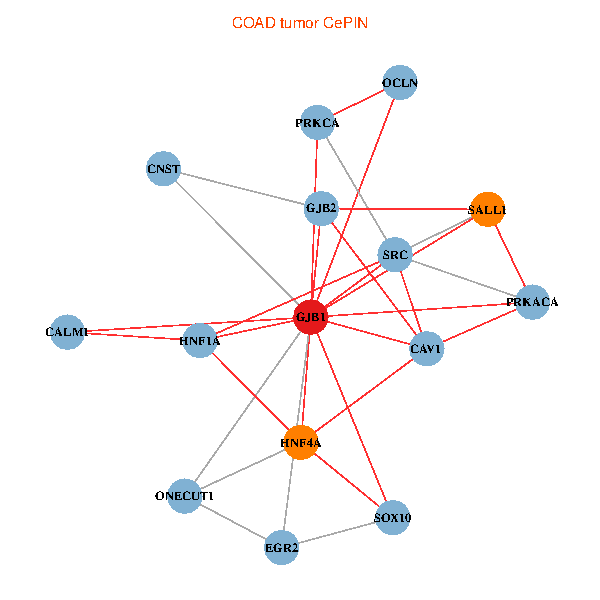
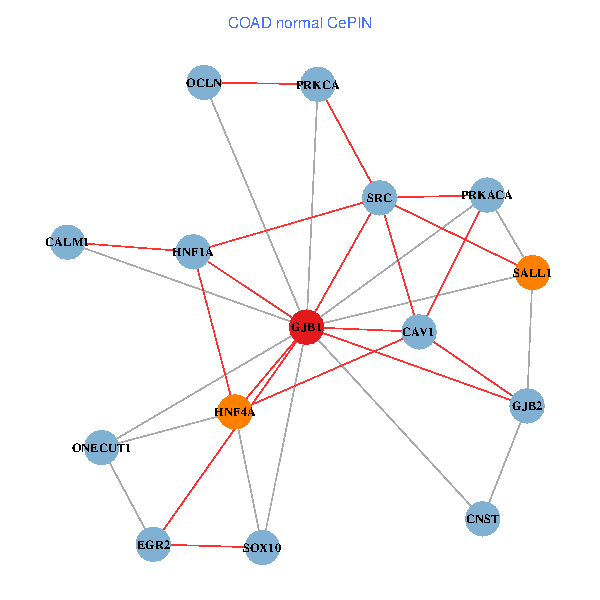
| COAD (tumor) | COAD (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
 |  |
|
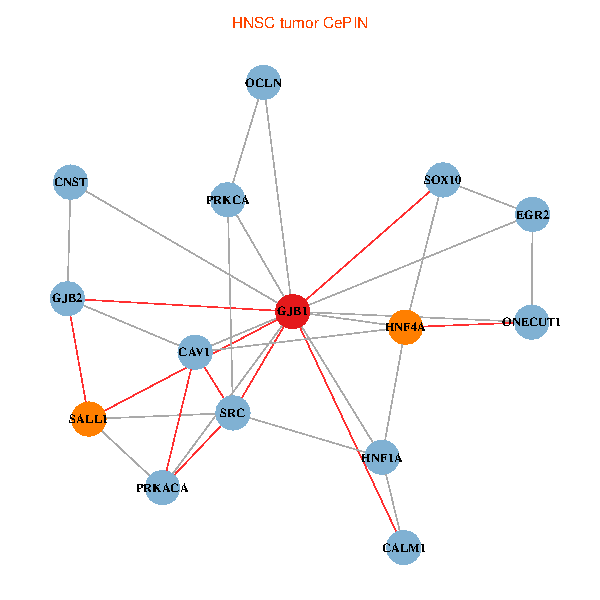
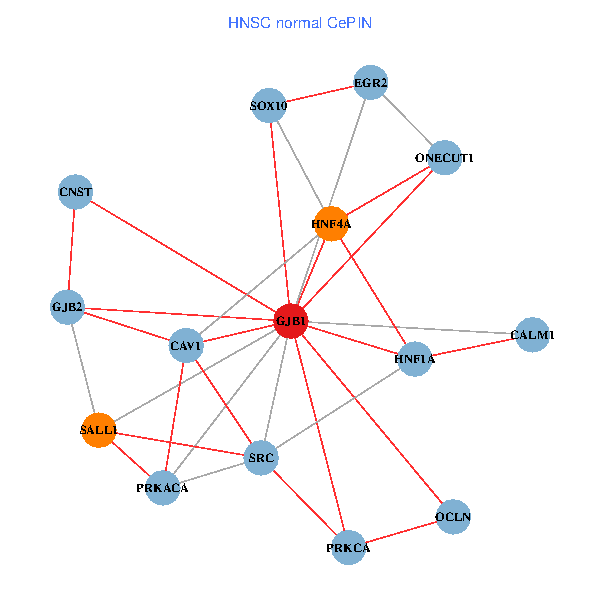
| HNSC (tumor) | HNSC (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
 |  |
|
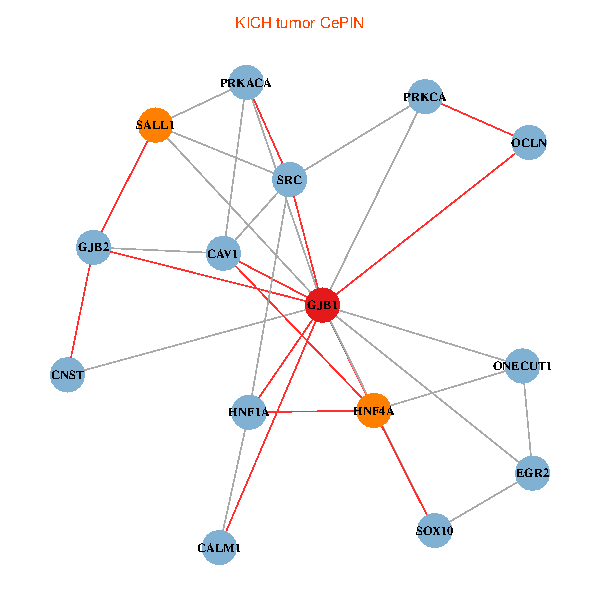
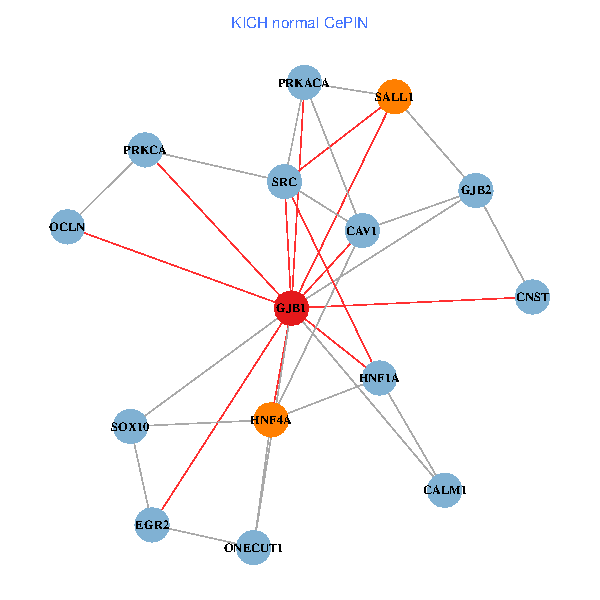
| KICH (tumor) | KICH (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
 |  |
|
| KIRC (tumor) | KIRC (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
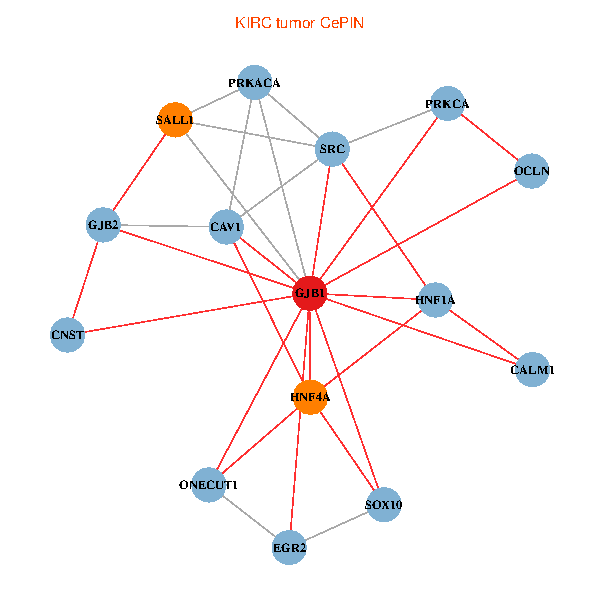 | 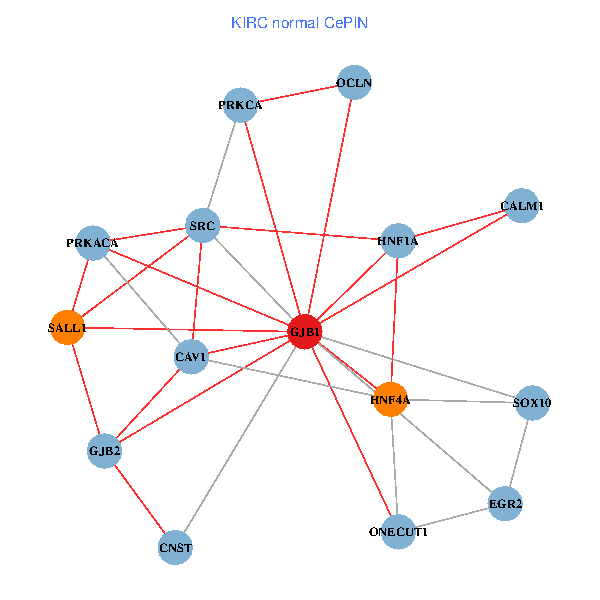 |
|
| KIRP (tumor) | KIRP (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
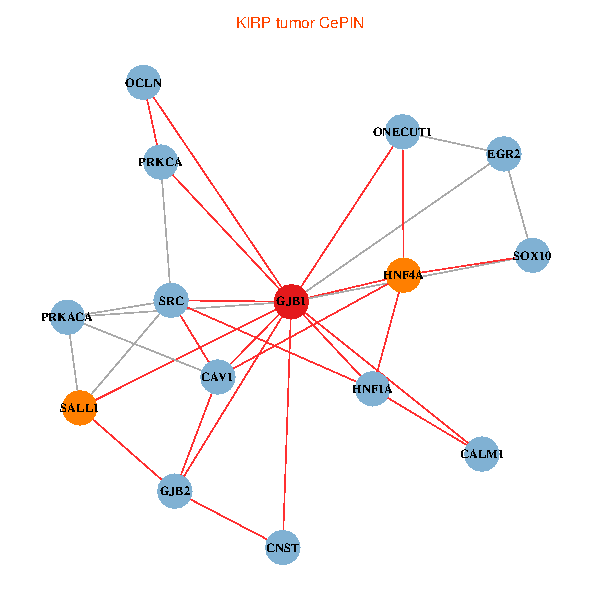 | 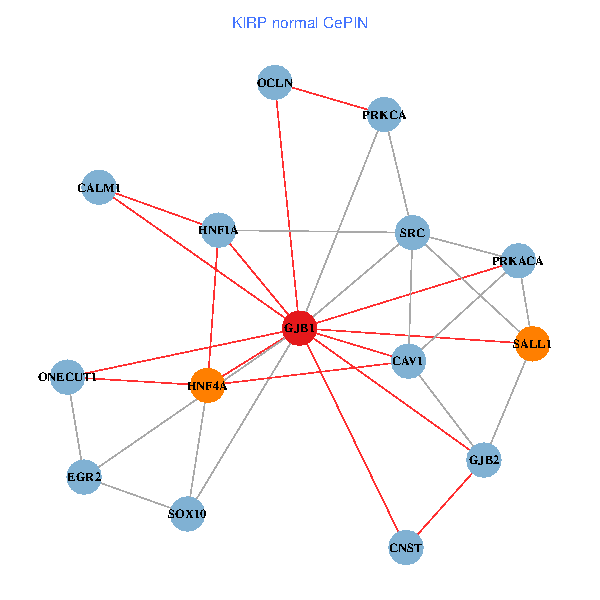 |
|
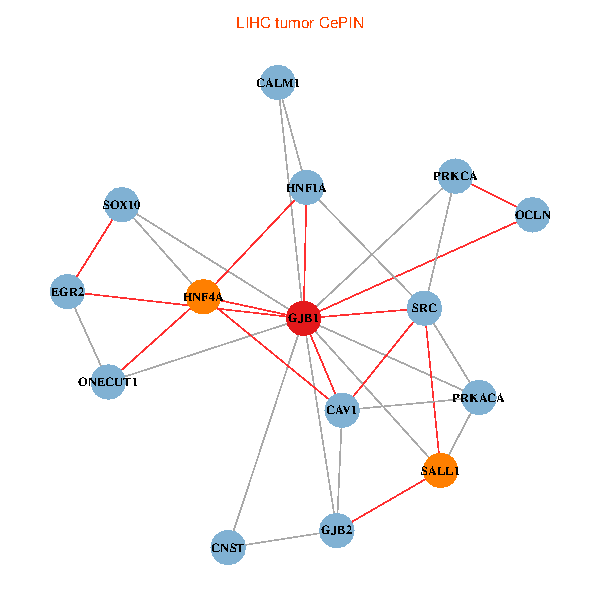
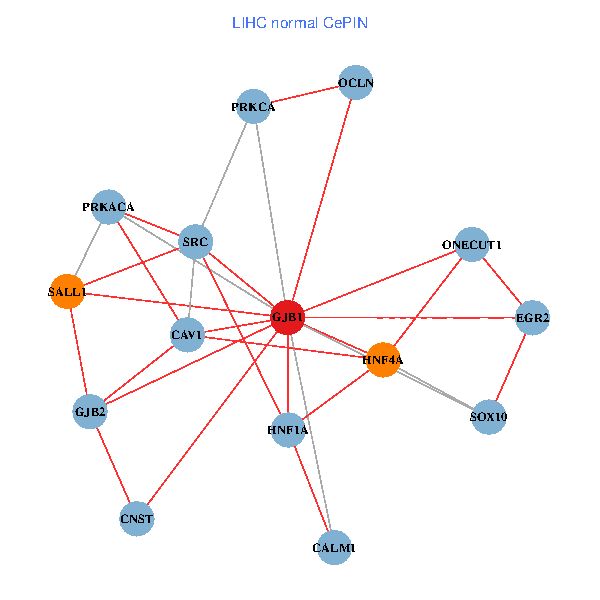
| LIHC (tumor) | LIHC (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
 |  |
|
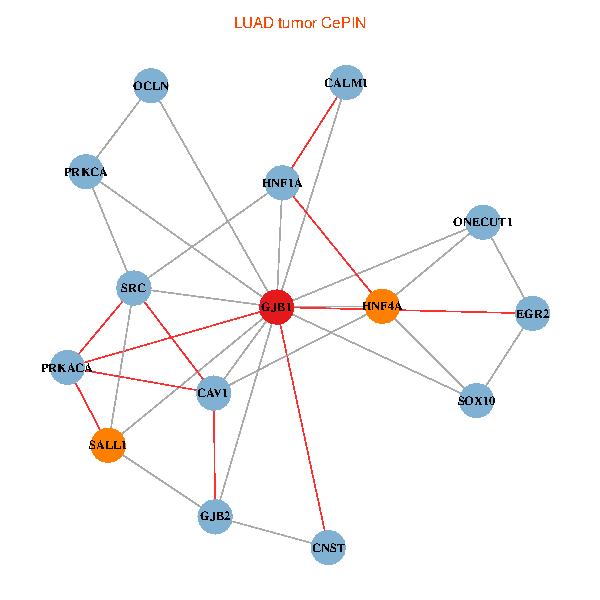
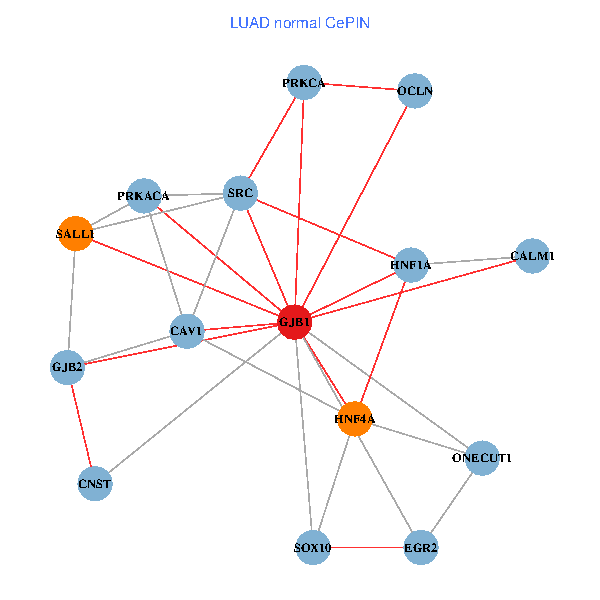
| LUAD (tumor) | LUAD (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
 |  |
|
| LUSC (tumor) | LUSC (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
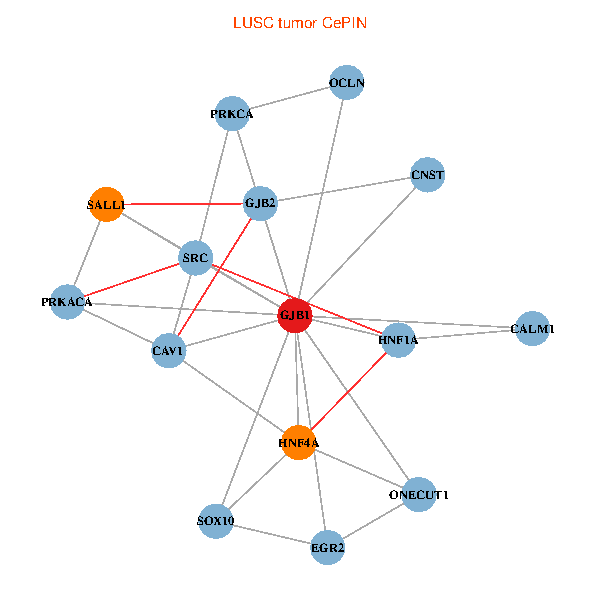 | 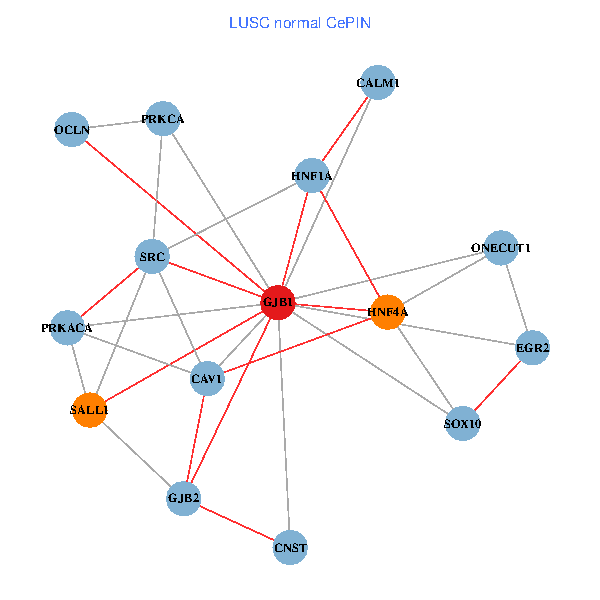 |
|
| PRAD (tumor) | PRAD (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
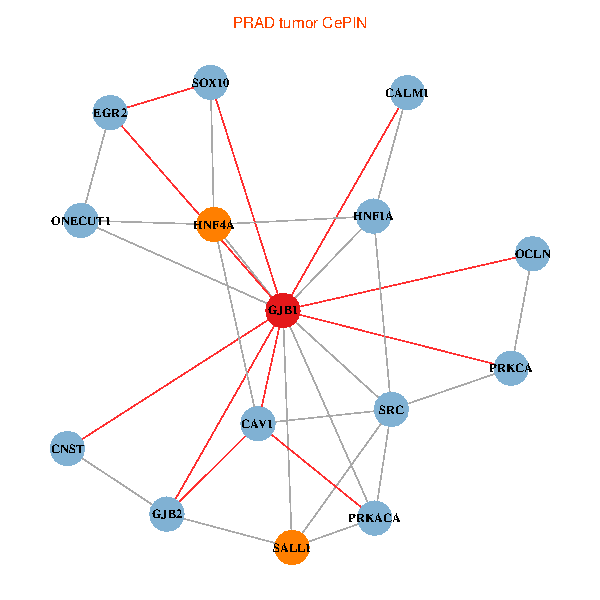 | 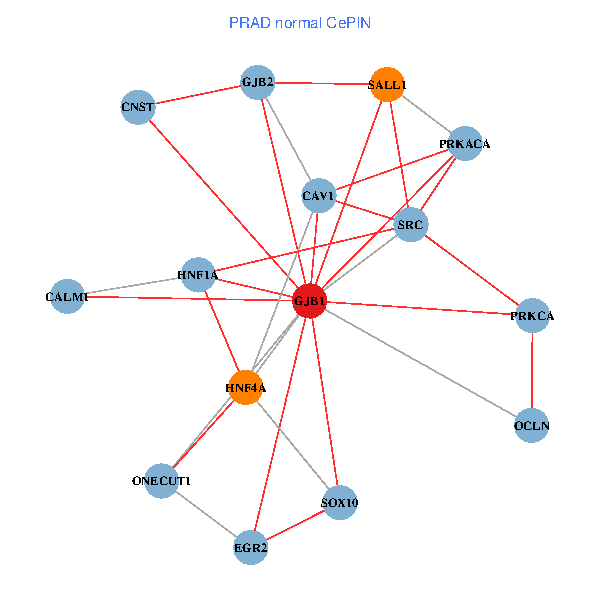 |
|
| STAD (tumor) | STAD (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
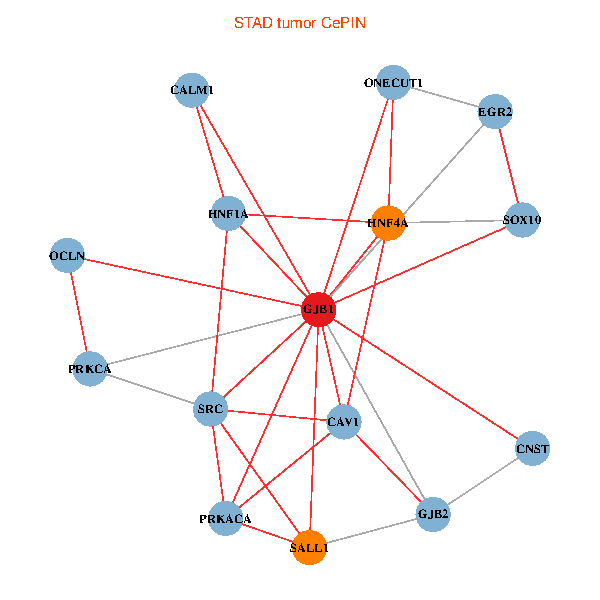 | 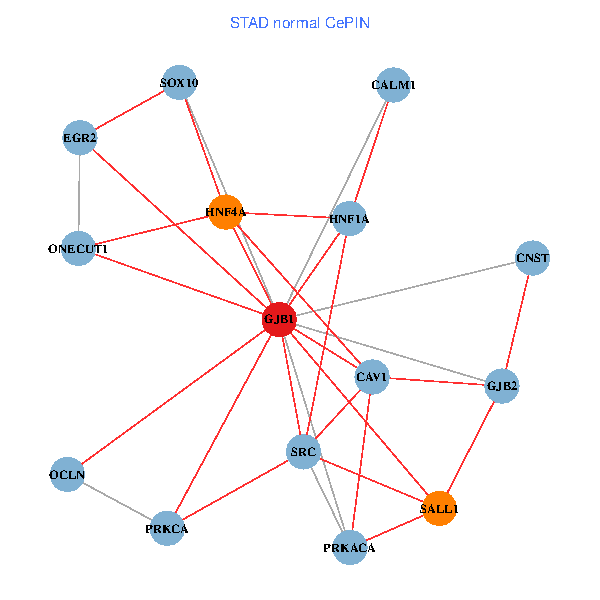 |
|
| THCA (tumor) | THCA (normal) |
| GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (tumor) | GJB1, HNF4A, ONECUT1, SRC, PRKCA, SOX10, PRKACA, CALM1, HNF1A, CAV1, EGR2, SALL1, CNST, OCLN, GJB2 (normal) |
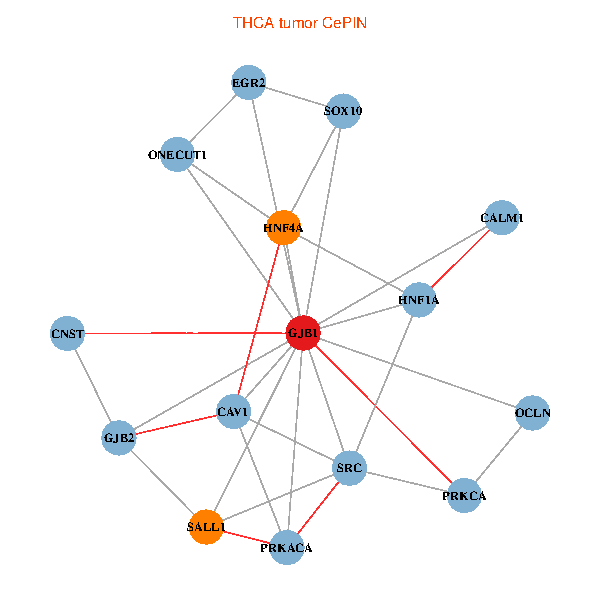 | 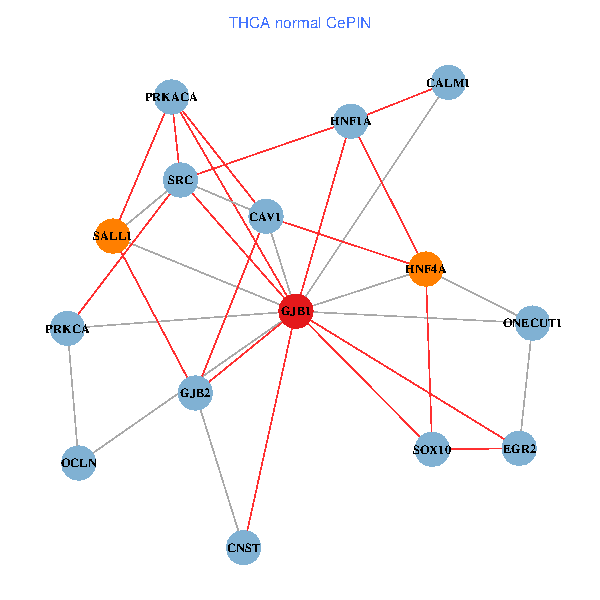 |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0007959 | Charcot-Marie-Tooth Disease | 146 | BeFree,CLINVAR,GAD,LHGDN |
| umls:C0393808 | Charcot-Marie-Tooth disease, X-linked, 1 | 83 | BeFree,CLINVAR,CTD_human,MGD,ORPHANET,UNIPROT |
| umls:C0442874 | Neuropathy | 21 | BeFree |
| umls:C0270922 | Peripheral demyelinating neuropathy | 16 | BeFree |
| umls:C0031117 | Peripheral Neuropathy | 12 | BeFree,LHGDN |
| umls:C2239176 | Liver carcinoma | 11 | BeFree,CTD_human,LHGDN |
| umls:C0007134 | Renal Cell Carcinoma | 10 | BeFree,CTD_human,LHGDN |
| umls:C0023903 | Liver neoplasms | 8 | BeFree,CTD_human |
| umls:C0027888 | Hereditary Motor and Sensory Neuropathies | 6 | BeFree |
| umls:C0598589 | Inherited neuropathies | 6 | BeFree |
| umls:C0011195 | Dejerine-Sottas Disease (disorder) | 5 | BeFree,CLINVAR,UNIPROT |
| umls:C0751036 | Hereditary Motor and Sensory Neuropathy Type I | 5 | BeFree |
| umls:C0001418 | Adenocarcinoma | 4 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 4 | BeFree,CTD_human,LHGDN |
| umls:C0393814 | Hereditary liability to pressure palsies | 4 | BeFree |
| umls:C0596263 | Carcinogenesis | 4 | BeFree |
| umls:C1512409 | Hepatocarcinogenesis | 4 | BeFree |
| umls:C0018784 | Sensorineural Hearing Loss (disorder) | 3 | BeFree |
| umls:C0024121 | Lung Neoplasms | 3 | CTD_human,LHGDN |
| umls:C0235031 | Neurologic Symptoms | 3 | BeFree |
| umls:C0278678 | Metastatic Renal Cell Cancer | 3 | BeFree |
| umls:C0376358 | Malignant neoplasm of prostate | 3 | BeFree |
| umls:C2932678 | Inherited Peripheral Neuropathy | 3 | BeFree |
| umls:C0004134 | Ataxia | 2 | BeFree,LHGDN |
| umls:C0023904 | Liver Neoplasms, Experimental | 2 | CTD_human,RGD |
| umls:C0024623 | Malignant neoplasm of stomach | 2 | BeFree |
| umls:C0026269 | Mitral Valve Stenosis | 2 | BeFree |
| umls:C0026769 | Multiple Sclerosis | 2 | BeFree |
| umls:C0027126 | Myotonic Dystrophy | 2 | BeFree |
| umls:C0151313 | Sensory neuropathy | 2 | BeFree |
| umls:C0152013 | Adenocarcinoma of lung (disorder) | 2 | BeFree |
| umls:C0152025 | Polyneuropathy | 2 | BeFree |
| umls:C0152027 | Sensory Disorders | 2 | BeFree |
| umls:C0338656 | Impaired cognition | 2 | BeFree |
| umls:C0392553 | Hereditary peripheral neuropathy | 2 | BeFree |
| umls:C0422879 | CNS symptom | 2 | BeFree |
| umls:C0600139 | Prostate carcinoma | 2 | BeFree |
| umls:C0684249 | Carcinoma of lung | 2 | BeFree |
| umls:C0686377 | CNS metastases | 2 | BeFree |
| umls:C0699791 | Stomach Carcinoma | 2 | BeFree |
| umls:C0740457 | Malignant neoplasm of kidney | 2 | BeFree |
| umls:C0947751 | Vascular inflammations | 2 | BeFree |
| umls:C1378703 | Renal carcinoma | 2 | BeFree |
| umls:C1520166 | Xenograft Model | 2 | BeFree |
| umls:C0001430 | Adenoma | 1 | CTD_human |
| umls:C0003130 | Anoxia | 1 | LHGDN |
| umls:C0005398 | Cholestasis, Extrahepatic | 1 | RGD |
| umls:C0006142 | Malignant neoplasm of breast | 1 | BeFree |
| umls:C0013362 | Dysarthria | 1 | LHGDN |
| umls:C0015230 | Exanthema | 1 | BeFree |
| umls:C0017638 | Glioma | 1 | BeFree |
| umls:C0018781 | Noise-induced hearing loss | 1 | GAD |
| umls:C0021051 | Immunologic Deficiency Syndromes | 1 | BeFree |
| umls:C0023520 | Leukodystrophy | 1 | BeFree |
| umls:C0030554 | Paresthesia | 1 | BeFree |
| umls:C0031511 | Pheochromocytoma | 1 | BeFree |
| umls:C0032927 | Precancerous Conditions | 1 | CTD_human |
| umls:C0035126 | Reperfusion Injury | 1 | CTD_human |
| umls:C0087012 | Ataxia, Spinocerebellar | 1 | BeFree |
| umls:C0162309 | Adrenoleukodystrophy | 1 | BeFree |
| umls:C0235974 | Pancreatic carcinoma | 1 | BeFree |
| umls:C0242379 | Malignant neoplasm of lung | 1 | BeFree |
| umls:C0270914 | Hereditary Motor and Sensory-Neuropathy Type II | 1 | BeFree |
| umls:C0270921 | Axonal neuropathy | 1 | BeFree |
| umls:C0270933 | Inflammatory neuropathy | 1 | BeFree |
| umls:C0332853 | Anastomosis | 1 | BeFree |
| umls:C0346647 | Malignant neoplasm of pancreas | 1 | BeFree |
| umls:C0374997 | Helicobacter pylori (H. pylori) infection in conditions classified elsewhere and of unspecified site | 1 | BeFree |
| umls:C0393819 | Polyradiculoneuropathy, Chronic Inflammatory Demyelinating | 1 | BeFree |
| umls:C0393907 | Axonal sensorimotor neuropathy | 1 | BeFree |
| umls:C0410226 | Congenital Myotonic Dystrophy | 1 | BeFree |
| umls:C0587246 | Weakness of limb | 1 | BeFree |
| umls:C0678222 | Breast Carcinoma | 1 | BeFree |
| umls:C0848332 | Spots on skin | 1 | BeFree |
| umls:C0850666 | Helicobacter pylori infection | 1 | BeFree |
| umls:C0920350 | Autoimmune thyroiditis | 1 | RGD |
| umls:C1140680 | Malignant neoplasm of ovary | 1 | GAD |
| umls:C1527231 | Adrenomyeloneuropathy | 1 | BeFree |
| umls:C1720983 | Channelopathies | 1 | BeFree |
| umls:C0796205 | SPINOCEREBELLAR ATAXIA, X-LINKED 1 | 0 | ORPHANET |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))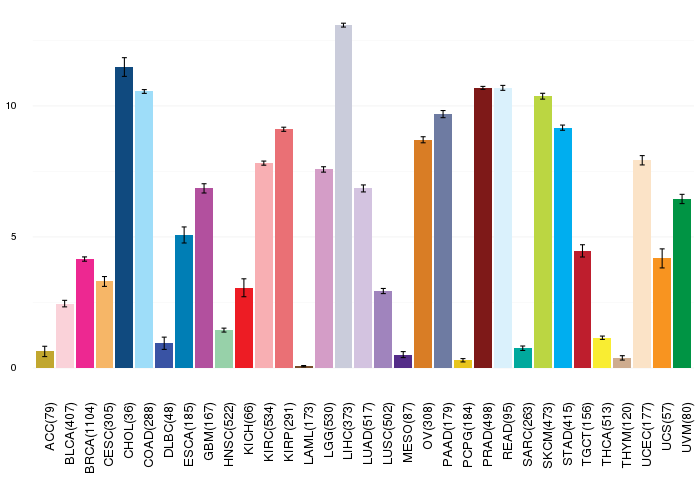
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))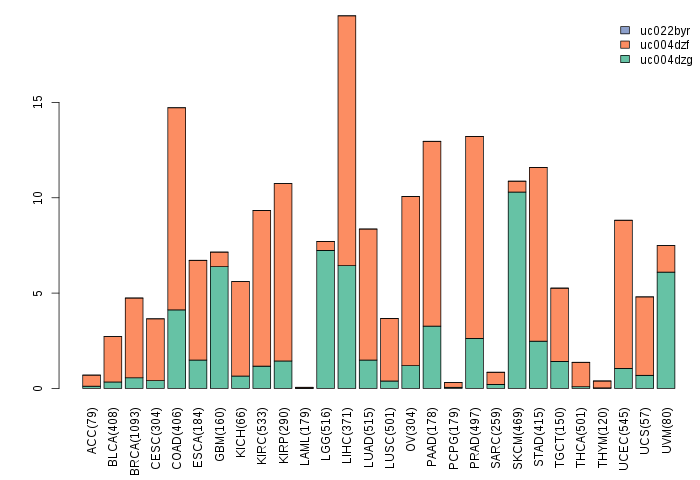
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data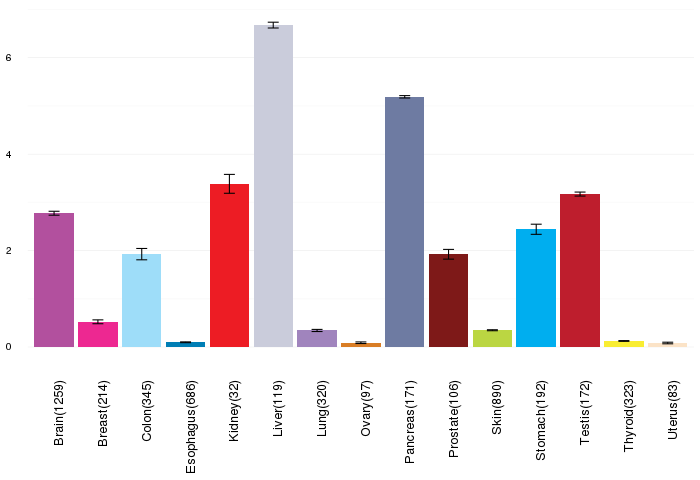
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))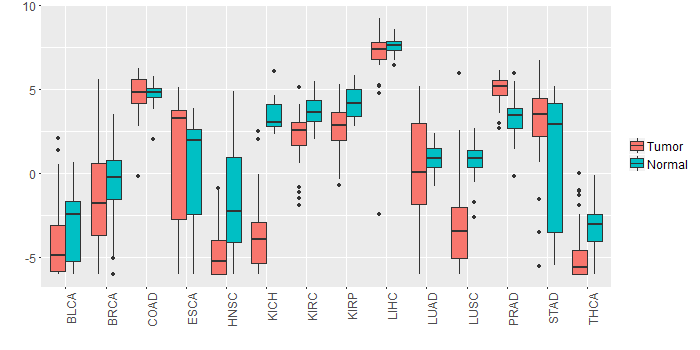
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.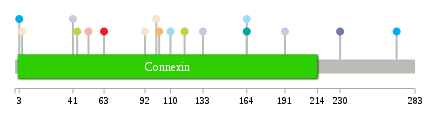

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 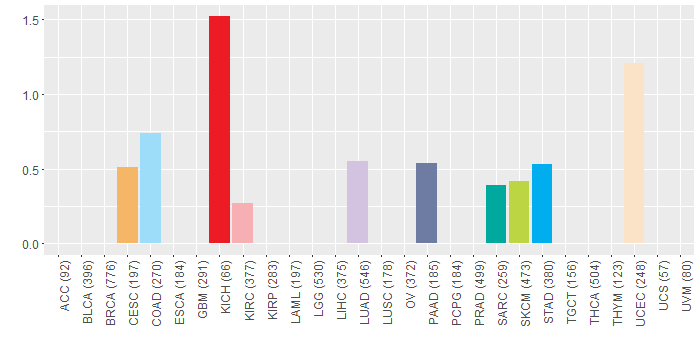
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)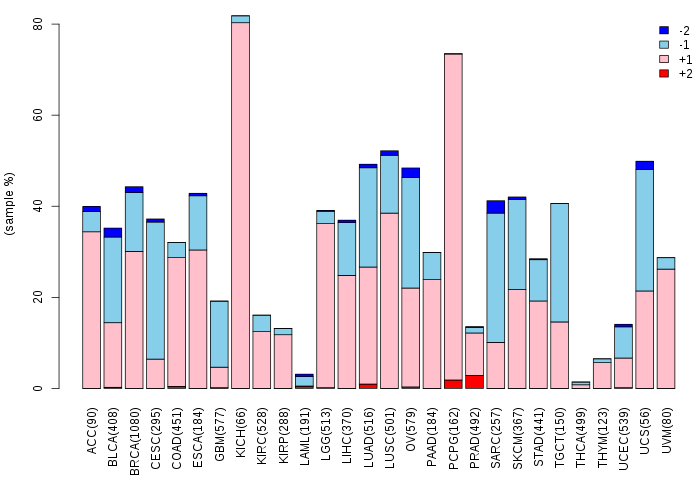
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 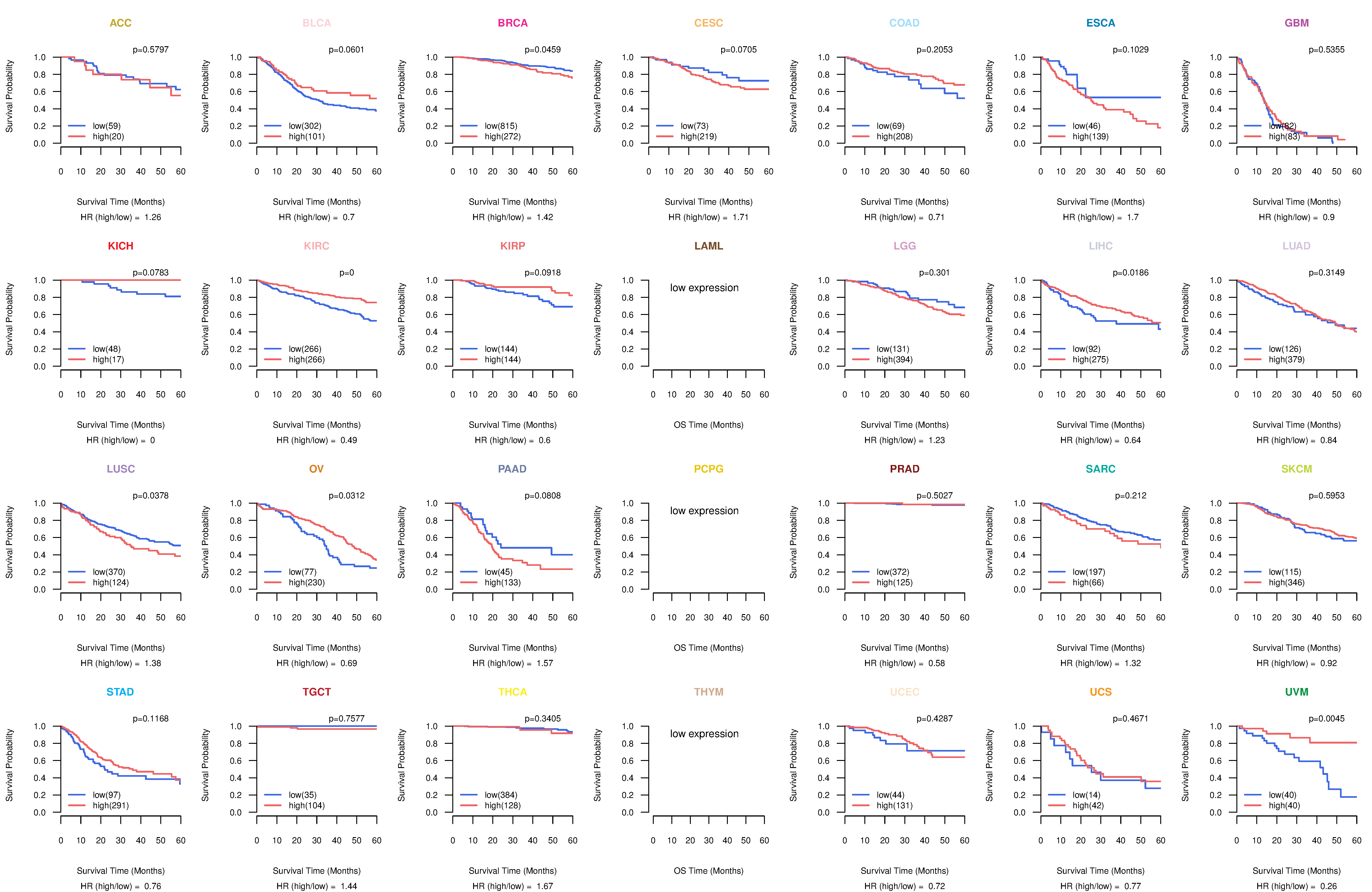
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 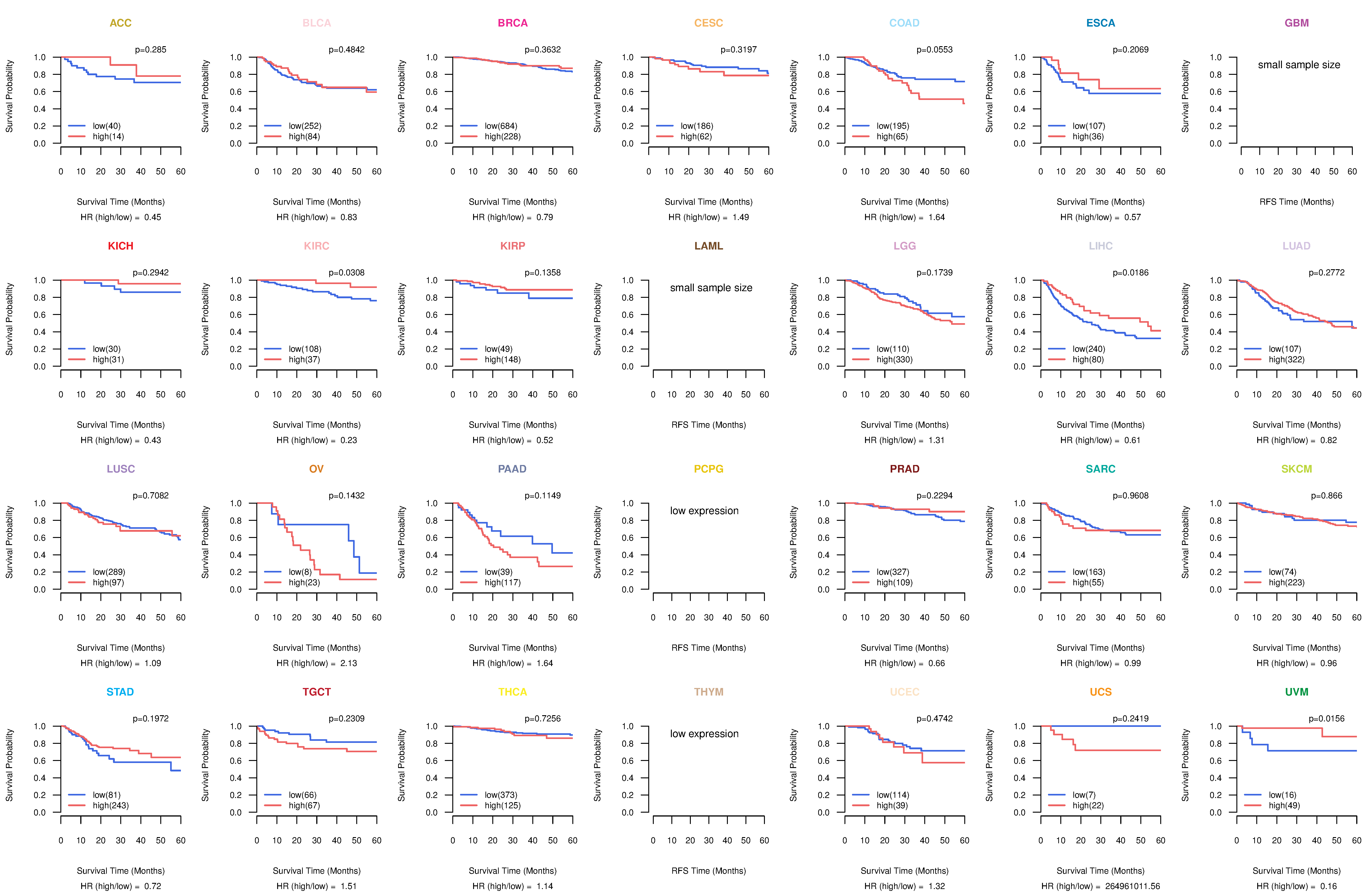
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 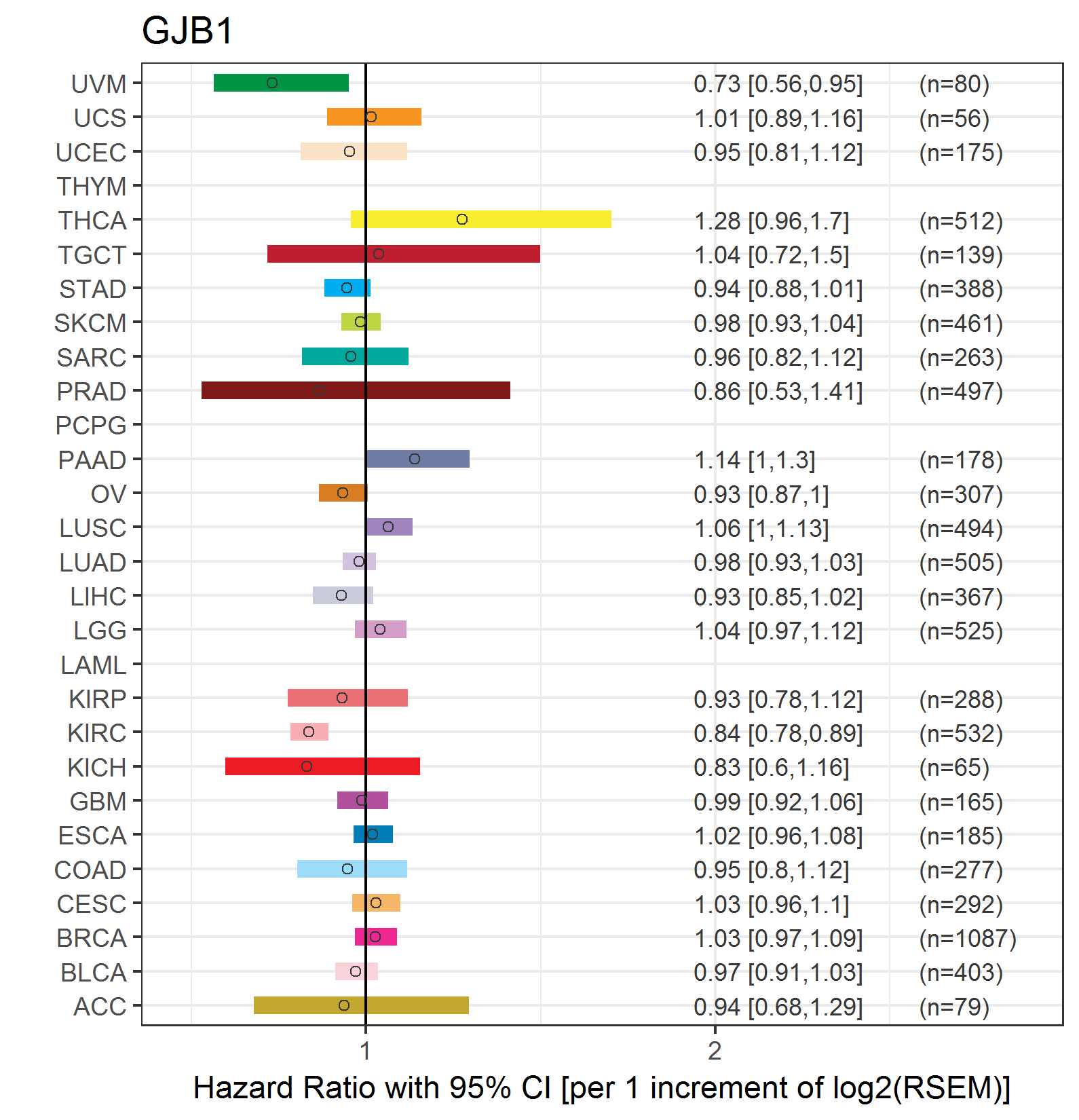
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 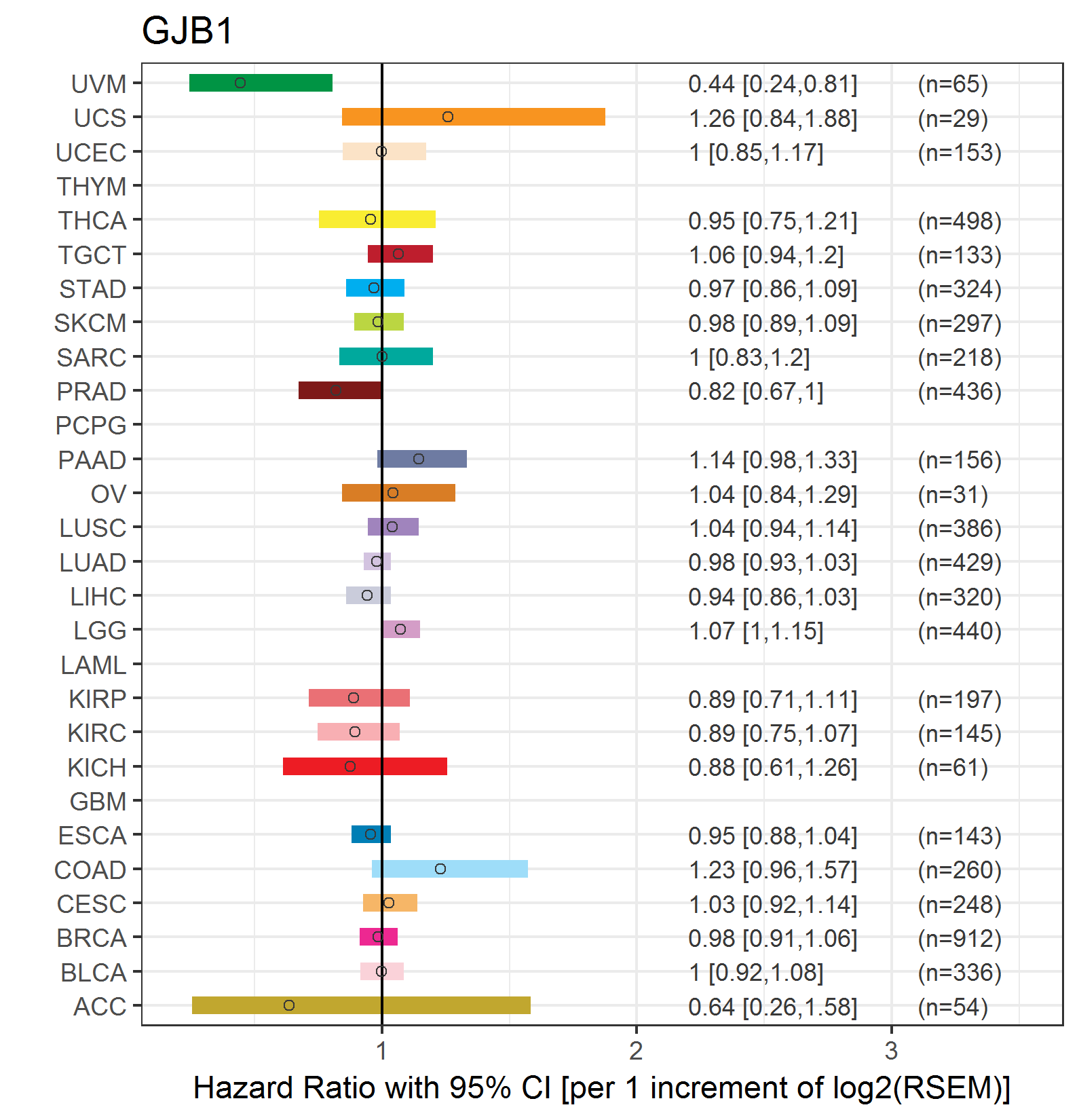
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























