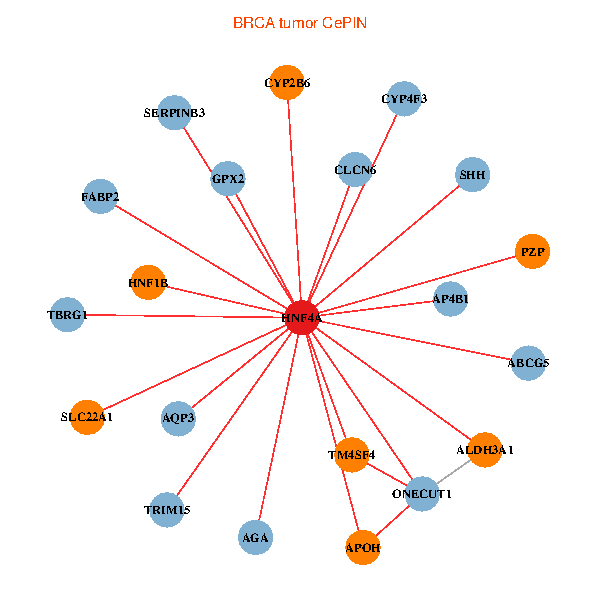
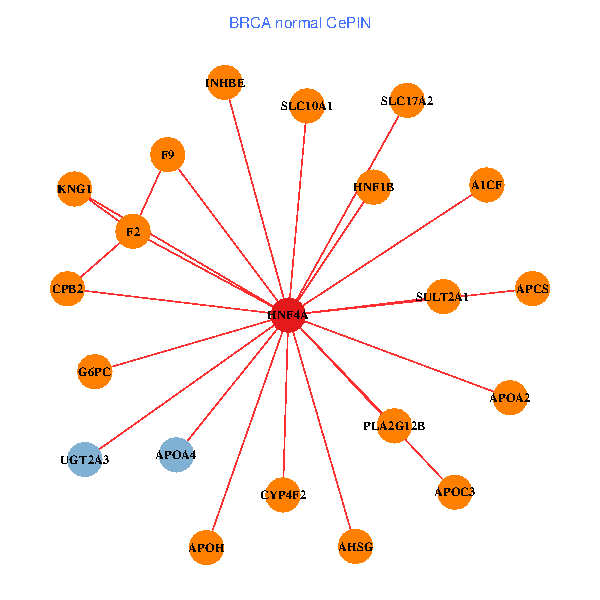
| BRCA (tumor) | BRCA (normal) |
| HNF4A, ONECUT1, GPX2, ALDH3A1, TRIM15, SLC22A1, ABCG5, SERPINB3, PZP, APOH, SHH, AQP3, AGA, HNF1B, AP4B1, CYP2B6, TBRG1, TM4SF4, CYP4F3, FABP2, CLCN6 (tumor) | HNF4A, APCS, CPB2, F2, KNG1, APOA2, AHSG, UGT2A3, APOH, F9, A1CF, APOC3, G6PC, CYP4F2, HNF1B, SULT2A1, INHBE, SLC10A1, SLC17A2, APOA4, PLA2G12B (normal) |
 |  |
|
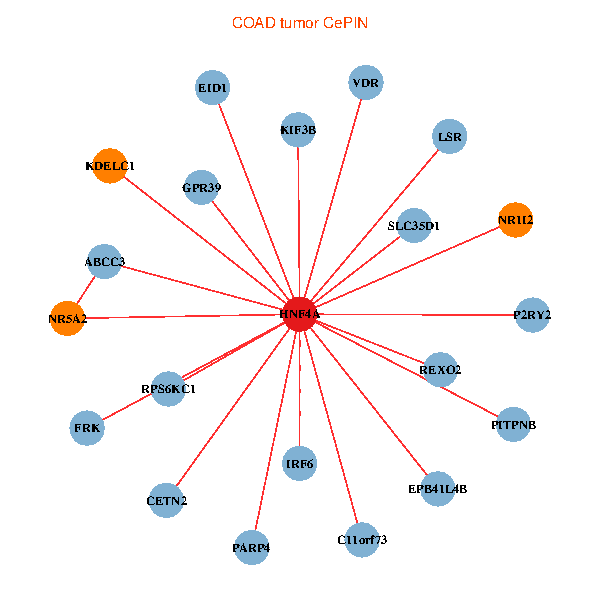
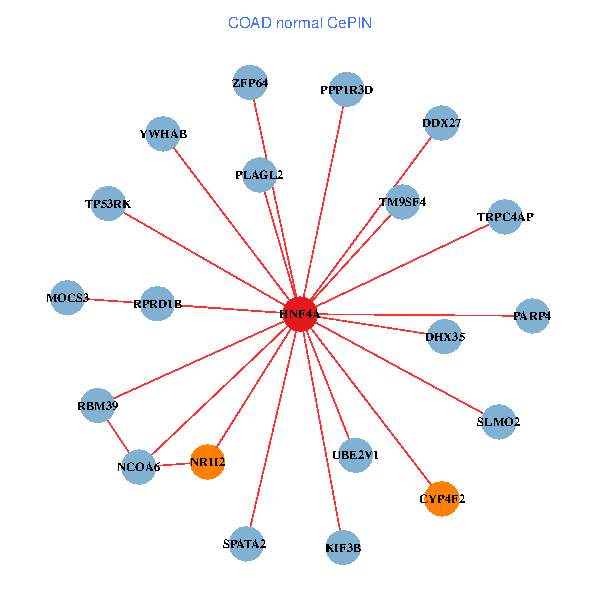
| COAD (tumor) | COAD (normal) |
| HNF4A, EID1, NR5A2, NR1I2, EPB41L4B, VDR, LSR, CETN2, IRF6, P2RY2, SLC35D1, KIF3B, PARP4, PITPNB, C11orf73, KDELC1, ABCC3, FRK, REXO2, RPS6KC1, GPR39 (tumor) | HNF4A, YWHAB, TRPC4AP, UBE2V1, SPATA2, NR1I2, NCOA6, PPP1R3D, MOCS3, ZFP64, RBM39, DDX27, SLMO2, DHX35, PLAGL2, RPRD1B, CYP4F2, KIF3B, PARP4, TP53RK, TM9SF4 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
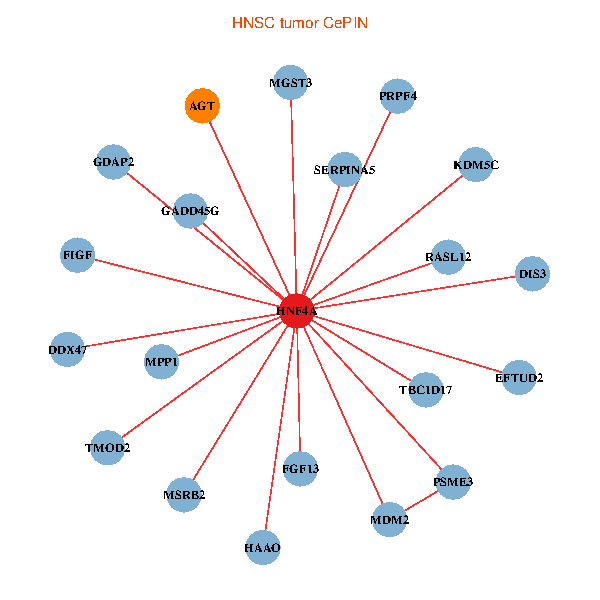
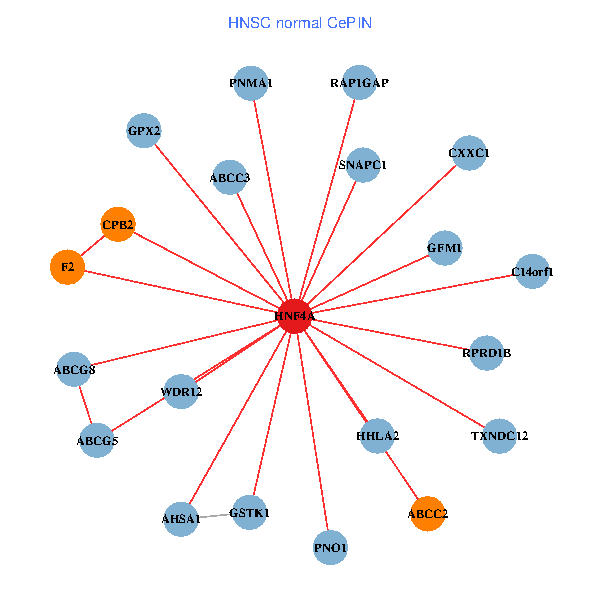
| HNF4A, MDM2, TBC1D17, EFTUD2, GADD45G, SERPINA5, PSME3, MPP1, FGF13, AGT, PRPF4, KDM5C, DIS3, TMOD2, DDX47, RASL12, MGST3, HAAO, MSRB2, GDAP2, FIGF (tumor) | HNF4A, GSTK1, RAP1GAP, AHSA1, C14orf1, CPB2, F2, GPX2, HHLA2, ABCC2, SNAPC1, TXNDC12, PNO1, ABCG8, PNMA1, ABCG5, CXXC1, RPRD1B, WDR12, GFM1, ABCC3 (normal) |
 |  |
|
| KICH (tumor) | KICH (normal) |
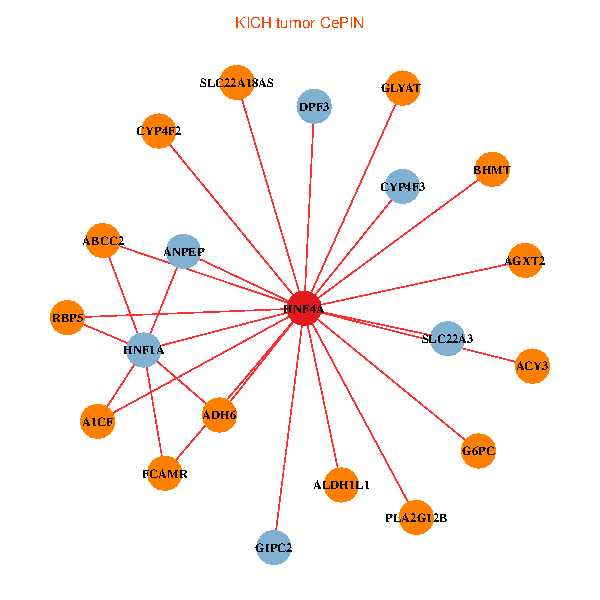
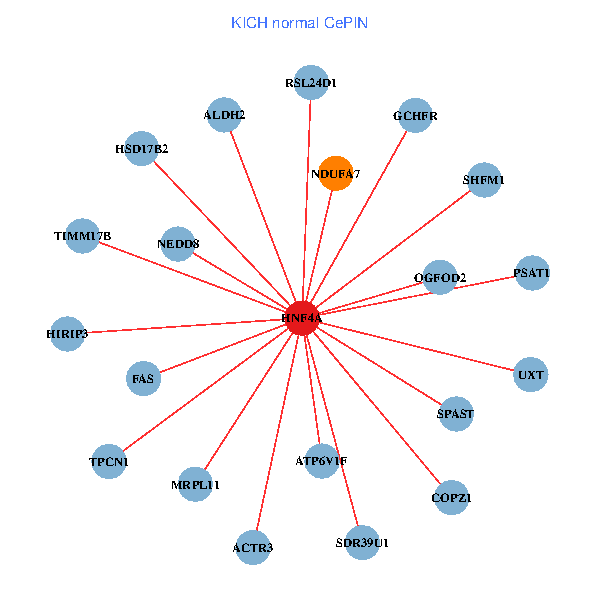
| HNF4A, GIPC2, DPF3, ADH6, HNF1A, ACY3, ANPEP, ABCC2, GLYAT, SLC22A3, AGXT2, BHMT, SLC22A18AS, A1CF, ALDH1L1, G6PC, CYP4F2, FCAMR, RBP5, CYP4F3, PLA2G12B (tumor) | HNF4A, PSAT1, FAS, NEDD8, SHFM1, COPZ1, ACTR3, SPAST, SDR39U1, UXT, NDUFA7, GCHFR, HIRIP3, HSD17B2, MRPL11, RSL24D1, ALDH2, TIMM17B, ATP6V1F, OGFOD2, TPCN1 (normal) |
 |  |
|
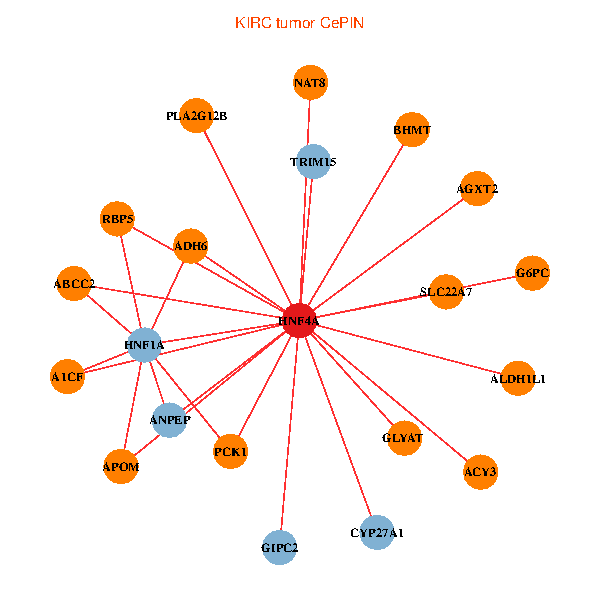
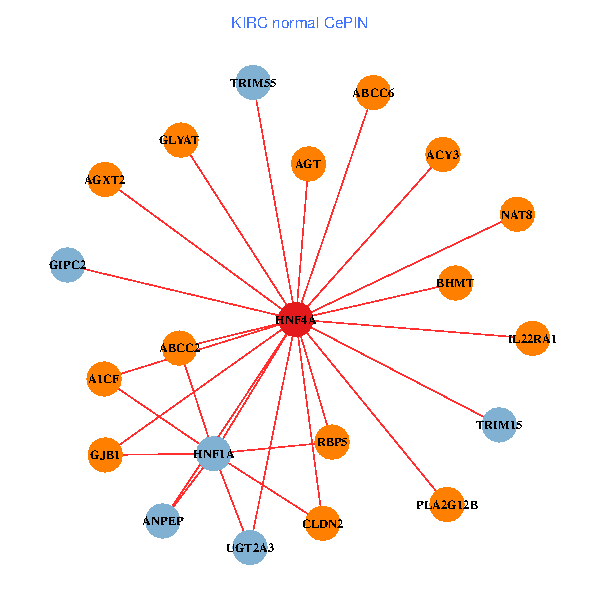
| KIRC (tumor) | KIRC (normal) |
| HNF4A, GIPC2, ADH6, HNF1A, ACY3, ANPEP, PCK1, ABCC2, TRIM15, GLYAT, CYP27A1, AGXT2, BHMT, A1CF, ALDH1L1, G6PC, RBP5, NAT8, APOM, SLC22A7, PLA2G12B (tumor) | HNF4A, TRIM55, GIPC2, HNF1A, GJB1, CLDN2, ACY3, ANPEP, ABCC2, AGT, TRIM15, UGT2A3, GLYAT, AGXT2, ABCC6, IL22RA1, BHMT, A1CF, RBP5, NAT8, PLA2G12B (normal) |
 |  |
|
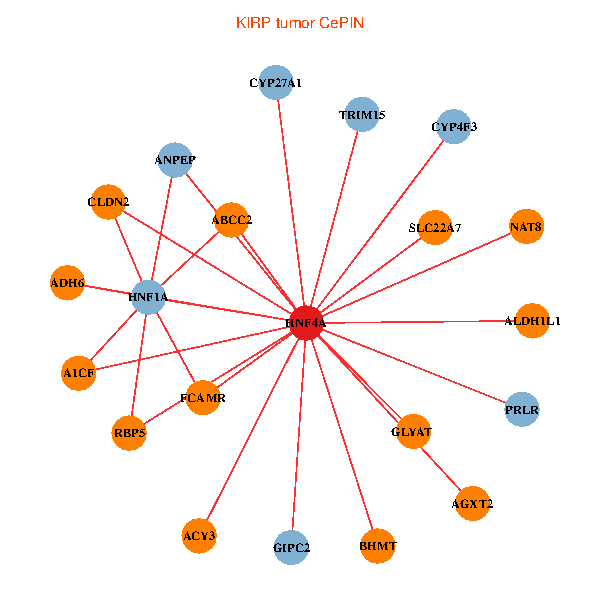
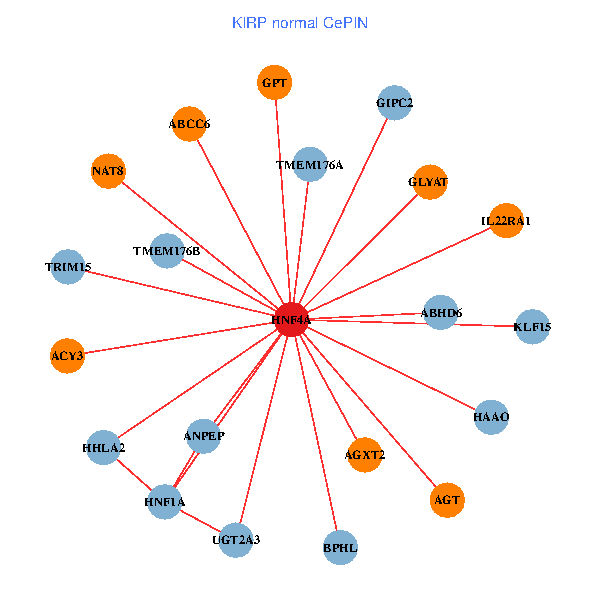
| KIRP (tumor) | KIRP (normal) |
| HNF4A, GIPC2, ADH6, HNF1A, CLDN2, ACY3, ANPEP, ABCC2, PRLR, TRIM15, GLYAT, CYP27A1, AGXT2, BHMT, A1CF, ALDH1L1, FCAMR, RBP5, NAT8, CYP4F3, SLC22A7 (tumor) | HNF4A, GIPC2, HNF1A, ACY3, HHLA2, ANPEP, AGT, TRIM15, UGT2A3, GLYAT, AGXT2, ABCC6, IL22RA1, GPT, TMEM176A, HAAO, ABHD6, NAT8, TMEM176B, KLF15, BPHL (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| HNF4A, ARPC5, SUB1, UBE2I, NR1I2, WTAP, RASSF1, BACE1, PGM1, FTSJ1, MED10, SRP19, CYTH2, ABCC6, A1CF, DCAF13, DCAF11, L2HGDH, UTP11L, SLC26A1, CYP4F3 (tumor) | HNF4A, ADH6, HNF1A, APOB, GJB1, CPB2, F2, KNG1, TF, ABCC2, F11, SERPINA10, SERPINC1, ABCC6, A1CF, HGD, F12, CYP2J2, HAO1, CYP4F3, KLF15 (normal) |
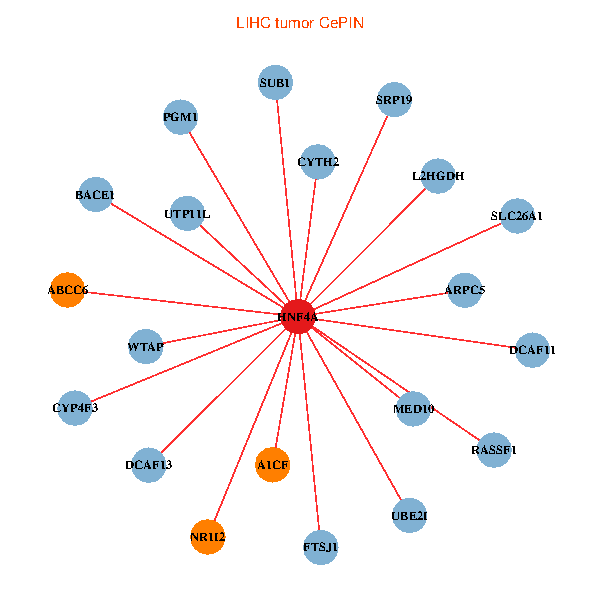 | 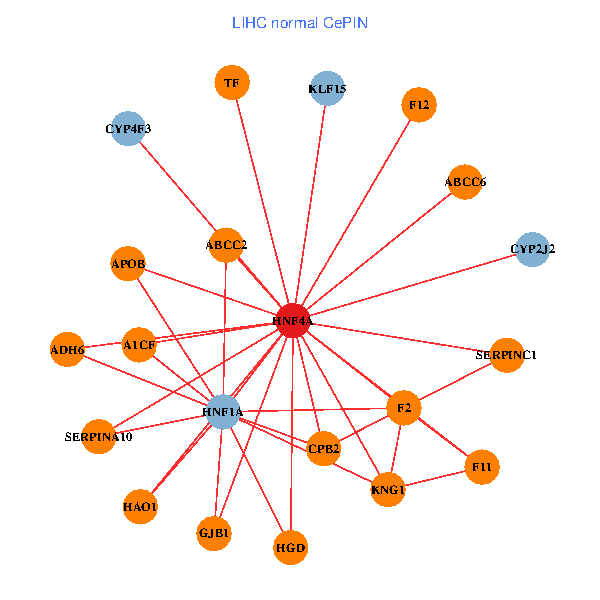 |
|
| LUAD (tumor) | LUAD (normal) |
| HNF4A, ADH6, HNF1A, HHLA2, HSPA4L, UGT2B15, NME7, PZP, BAI2, RDH11, SPATS2L, SLC39A6, CHST9, TTC26, TSNAXIP1, C22orf23, CYP2J2, TMTC4, EFCAB1, LRRC6, CLCNKA (tumor) | HNF4A, NR1I2, ADH6, HNF1A, F2, GPX2, CYP2C9, TRIM15, UGT2B15, UGT2A3, MYO1A, SERPINA4, A1CF, HGD, GUCY2C, SGK2, TM4SF4, CYP3A5, AKR1C4, CYP4F3, PDE11A (normal) |
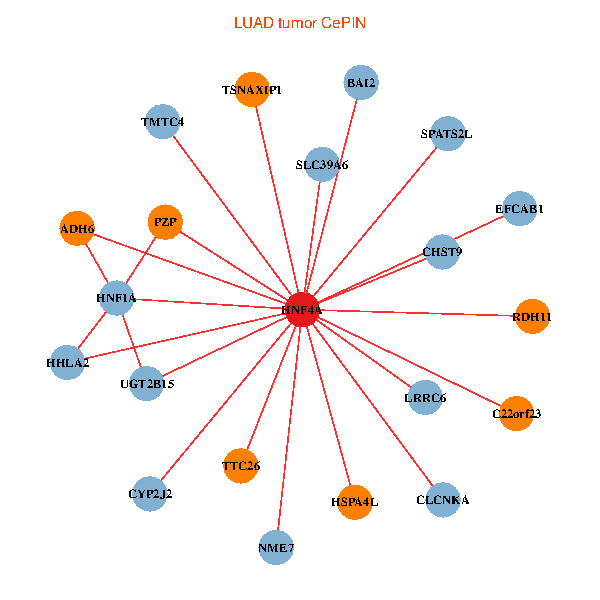 | 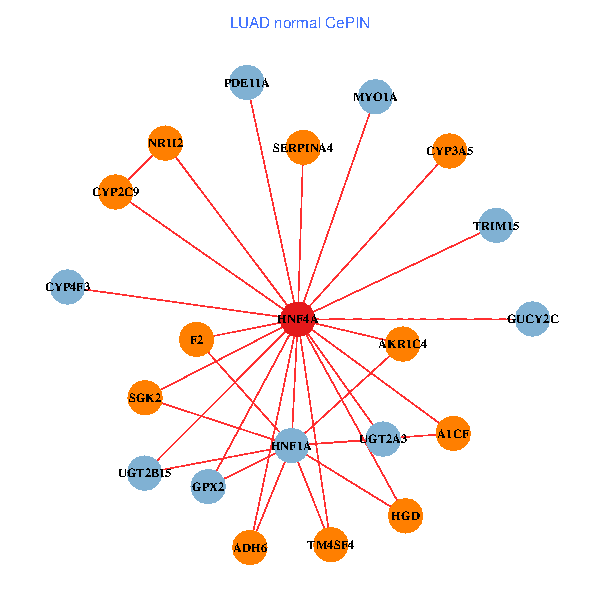 |
|
| LUSC (tumor) | LUSC (normal) |
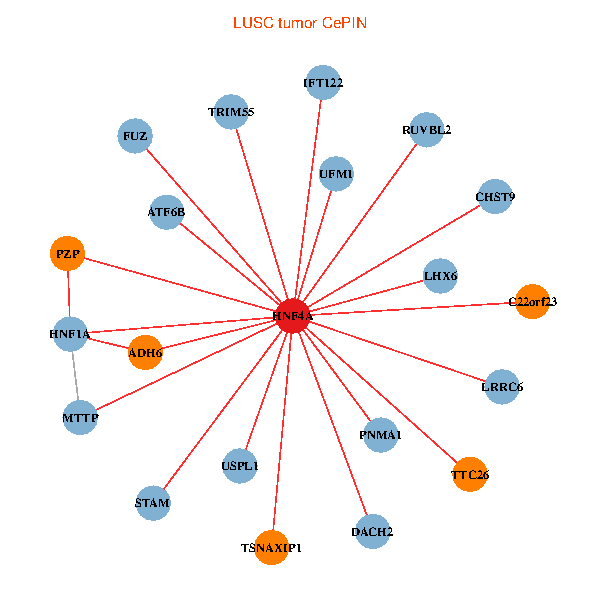
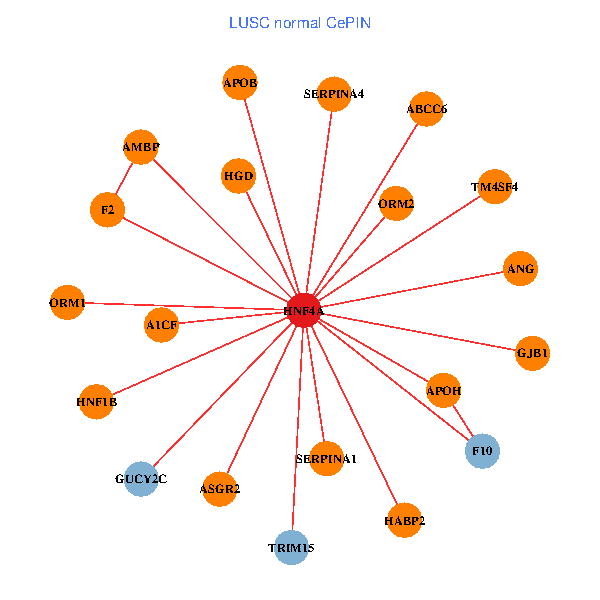
| HNF4A, TRIM55, RUVBL2, ADH6, STAM, HNF1A, UFM1, DACH2, MTTP, PNMA1, PZP, USPL1, CHST9, TTC26, ATF6B, TSNAXIP1, C22orf23, LRRC6, LHX6, IFT122, FUZ (tumor) | HNF4A, SERPINA1, APOB, ASGR2, GJB1, F2, F10, HABP2, TRIM15, AMBP, ABCC6, APOH, SERPINA4, A1CF, ORM1, ANG, HGD, GUCY2C, HNF1B, ORM2, TM4SF4 (normal) |
 |  |
|
| PRAD (tumor) | PRAD (normal) |
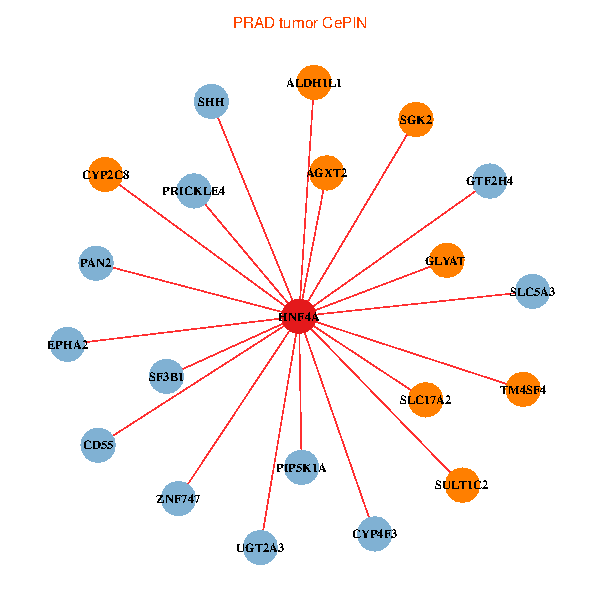
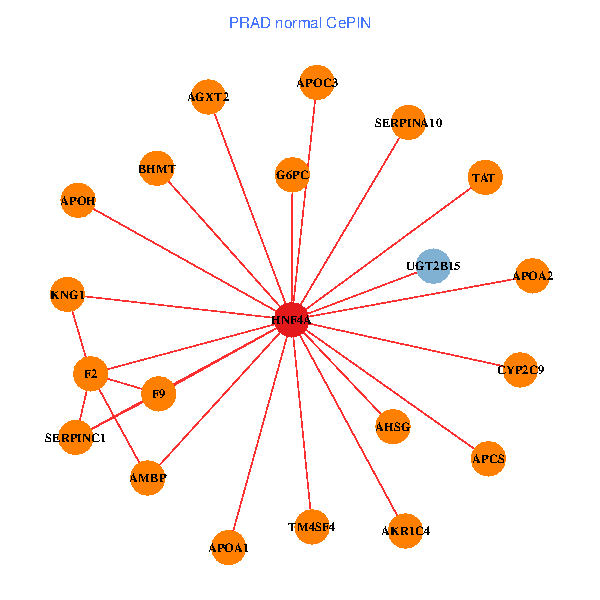
| HNF4A, EPHA2, CD55, GTF2H4, PIP5K1A, SF3B1, SLC5A3, UGT2A3, GLYAT, AGXT2, PAN2, SHH, SULT1C2, ALDH1L1, ZNF747, SGK2, CYP2C8, SLC17A2, TM4SF4, CYP4F3, PRICKLE4 (tumor) | HNF4A, APCS, F2, APOA1, KNG1, APOA2, CYP2C9, AHSG, TAT, SERPINA10, AMBP, UGT2B15, SERPINC1, AGXT2, APOH, F9, BHMT, APOC3, G6PC, TM4SF4, AKR1C4 (normal) |
 |  |
|
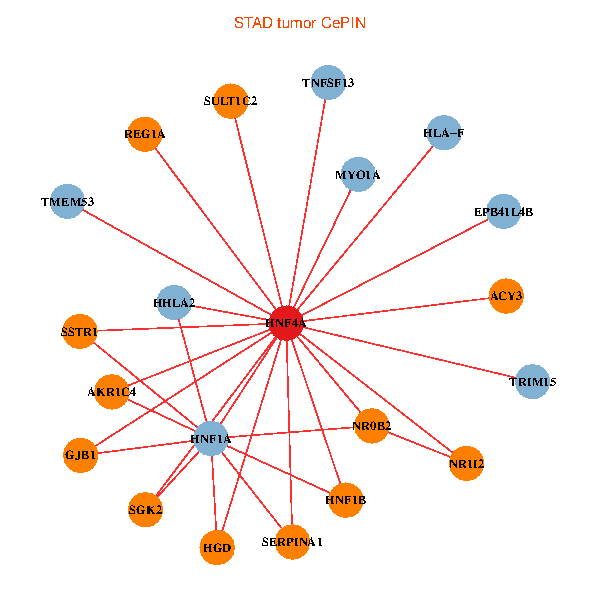
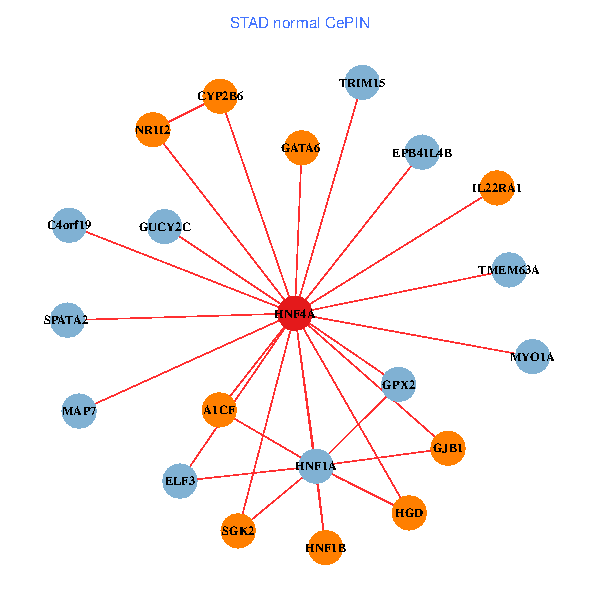
| STAD (tumor) | STAD (normal) |
| HNF4A, NR0B2, SERPINA1, NR1I2, EPB41L4B, HNF1A, GJB1, ACY3, HHLA2, TRIM15, MYO1A, TNFSF13, SULT1C2, SSTR1, HGD, HNF1B, REG1A, SGK2, HLA-F, TMEM53, AKR1C4 (tumor) | HNF4A, SPATA2, NR1I2, MAP7, EPB41L4B, HNF1A, GJB1, GPX2, ELF3, TRIM15, MYO1A, IL22RA1, A1CF, HGD, GATA6, GUCY2C, HNF1B, SGK2, C4orf19, CYP2B6, TMEM63A (normal) |
 |  |
|
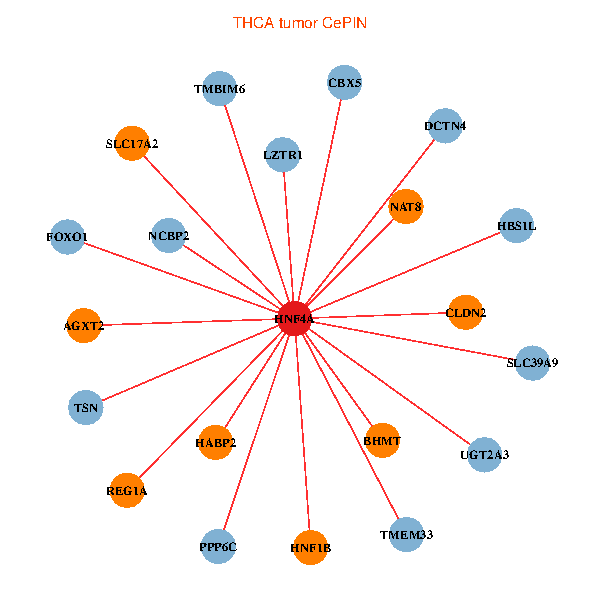
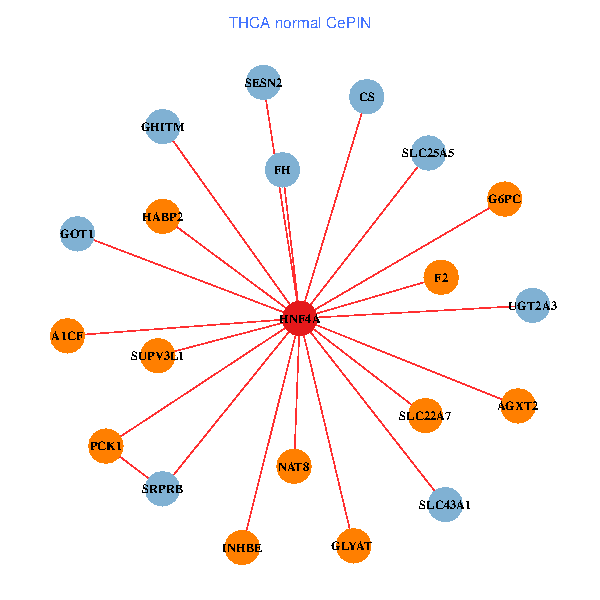
| THCA (tumor) | THCA (normal) |
| HNF4A, FOXO1, TMEM33, CBX5, CLDN2, HABP2, TSN, TMBIM6, PPP6C, LZTR1, UGT2A3, DCTN4, AGXT2, BHMT, HNF1B, NCBP2, REG1A, NAT8, SLC17A2, HBS1L, SLC39A9 (tumor) | HNF4A, SLC25A5, SESN2, SRPRB, F2, GHITM, HABP2, FH, PCK1, CS, SUPV3L1, UGT2A3, GLYAT, AGXT2, GOT1, A1CF, G6PC, INHBE, NAT8, SLC43A1, SLC22A7 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 153 | BeFree,CLINVAR,CTD_human,GAD,GWASCAT,LHGDN,UNIPROT |
| umls:C0011849 | Diabetes Mellitus | 108 | BeFree,GAD |
| umls:C0011847 | Diabetes | 100 | BeFree |
| umls:C0342276 | Maturity onset diabetes mellitus in young | 85 | BeFree,CTD_human,GAD |
| umls:C2239176 | Liver carcinoma | 31 | BeFree,LHGDN |
| umls:C0009402 | Colorectal Carcinoma | 17 | BeFree |
| umls:C0023903 | Liver neoplasms | 17 | BeFree |
| umls:C0011854 | Diabetes Mellitus, Insulin-Dependent | 14 | BeFree,GAD |
| umls:C1527249 | Colorectal Cancer | 14 | BeFree |
| umls:C0596263 | Carcinogenesis | 13 | BeFree |
| umls:C0020456 | Hyperglycemia | 11 | BeFree |
| umls:C0020615 | Hypoglycemia | 11 | BeFree,GAD,LHGDN |
| umls:C0699790 | Colon Carcinoma | 11 | BeFree |
| umls:C0007102 | Malignant tumor of colon | 10 | BeFree |
| umls:C0085207 | Gestational Diabetes | 10 | BeFree,GAD |
| umls:C1852093 | Maturity-Onset Diabetes of the Young, Type 1 | 10 | BeFree,CLINVAR,CTD_human,MGD,UNIPROT |
| umls:C0027627 | Neoplasm Metastasis | 8 | BeFree,LHGDN |
| umls:C0009324 | Ulcerative Colitis | 7 | BeFree,CTD_human,GAD |
| umls:C0019163 | Hepatitis B | 7 | BeFree,LHGDN |
| umls:C0020459 | Hyperinsulinism | 7 | BeFree,CLINVAR,GAD |
| umls:C0032580 | Adenomatous Polyposis Coli | 7 | BeFree |
| umls:C0024623 | Malignant neoplasm of stomach | 6 | BeFree |
| umls:C0158981 | Neonatal diabetes mellitus | 6 | BeFree |
| umls:C0699791 | Stomach Carcinoma | 6 | BeFree |
| umls:C0025202 | melanoma | 5 | BeFree |
| umls:C0271650 | Impaired glucose tolerance | 5 | BeFree |
| umls:C0524620 | Metabolic Syndrome X | 5 | BeFree |
| umls:C0010346 | Crohn Disease | 4 | BeFree |
| umls:C0020474 | Hyperlipidemia, Familial Combined | 4 | BeFree |
| umls:C0028754 | Obesity | 4 | BeFree |
| umls:C0178874 | Tumor Progression | 4 | BeFree |
| umls:C0001418 | Adenocarcinoma | 3 | BeFree,LHGDN |
| umls:C0002793 | Anaplasia | 3 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 3 | BeFree |
| umls:C0007134 | Renal Cell Carcinoma | 3 | BeFree |
| umls:C0011881 | Diabetic Nephropathy | 3 | BeFree,LHGDN,RGD |
| umls:C0015503 | Factor VII Deficiency | 3 | BeFree |
| umls:C0021390 | Inflammatory Bowel Diseases | 3 | BeFree |
| umls:C0022658 | Kidney Diseases | 3 | BeFree |
| umls:C0025517 | Metabolic Diseases | 3 | BeFree |
| umls:C0026764 | Multiple Myeloma | 3 | BeFree |
| umls:C0279000 | Liver and Intrahepatic Biliary Tract Carcinoma | 3 | BeFree |
| umls:C0279626 | Squamous cell carcinoma of esophagus | 3 | BeFree |
| umls:C0342277 | Diabetes mellitus autosomal dominant type II (disorder) | 3 | BeFree |
| umls:C0345904 | Malignant neoplasm of liver | 3 | BeFree |
| umls:C0376358 | Malignant neoplasm of prostate | 3 | BeFree |
| umls:C0600139 | Prostate carcinoma | 3 | BeFree |
| umls:C0678222 | Breast Carcinoma | 3 | BeFree |
| umls:C0004153 | Atherosclerosis | 2 | BeFree,GAD |
| umls:C0005684 | Malignant neoplasm of urinary bladder | 2 | BeFree |
| umls:C0007103 | Malignant neoplasm of endometrium | 2 | BeFree |
| umls:C0008312 | Primary biliary cirrhosis | 2 | BeFree |
| umls:C0009319 | Colitis | 2 | BeFree |
| umls:C0015624 | Fanconi Syndrome | 2 | BeFree |
| umls:C0015625 | Fanconi Anemia | 2 | BeFree |
| umls:C0015938 | Fetal Macrosomia | 2 | BeFree |
| umls:C0019693 | HIV Infections | 2 | BeFree |
| umls:C0022661 | Kidney Failure, Chronic | 2 | BeFree,GAD |
| umls:C0027819 | Neuroblastoma | 2 | BeFree |
| umls:C0029925 | Ovarian Carcinoma | 2 | BeFree |
| umls:C0205734 | Diabetes, Autoimmune | 2 | BeFree |
| umls:C0442874 | Neuropathy | 2 | BeFree |
| umls:C0476089 | Endometrial Carcinoma | 2 | BeFree |
| umls:C0699885 | Carcinoma of bladder | 2 | BeFree |
| umls:C0700095 | Central neuroblastoma | 2 | BeFree |
| umls:C0740457 | Malignant neoplasm of kidney | 2 | BeFree |
| umls:C1140680 | Malignant neoplasm of ovary | 2 | BeFree |
| umls:C1368683 | Epithelioma | 2 | BeFree |
| umls:C1378703 | Renal carcinoma | 2 | BeFree |
| umls:C1857395 | De Toni-Debre-Fanconi Syndrome | 2 | BeFree |
| umls:C1883486 | Uterine Corpus Cancer | 2 | BeFree |
| umls:C0000768 | Congenital Abnormality | 1 | BeFree |
| umls:C0001430 | Adenoma | 1 | BeFree |
| umls:C0002395 | Alzheimer's Disease | 1 | GAD |
| umls:C0003850 | Arteriosclerosis | 1 | BeFree |
| umls:C0004114 | Astrocytoma | 1 | BeFree |
| umls:C0005779 | Blood Coagulation Disorders | 1 | BeFree |
| umls:C0005967 | Bone neoplasms | 1 | BeFree |
| umls:C0006625 | Cachexia | 1 | BeFree |
| umls:C0007131 | Non-Small Cell Lung Carcinoma | 1 | BeFree |
| umls:C0007222 | Cardiovascular Diseases | 1 | BeFree |
| umls:C0008350 | Cholelithiasis | 1 | BeFree |
| umls:C0008370 | Cholestasis | 1 | BeFree |
| umls:C0011853 | Diabetes Mellitus, Experimental | 1 | RGD |
| umls:C0011859 | Lipoatrophic Diabetes Mellitus | 1 | BeFree |
| umls:C0011880 | Diabetic Ketoacidosis | 1 | BeFree |
| umls:C0014544 | Epilepsy | 1 | BeFree |
| umls:C0014556 | Epilepsy, Temporal Lobe | 1 | BeFree |
| umls:C0015645 | Fasciitis | 1 | BeFree |
| umls:C0017638 | Glioma | 1 | BeFree |
| umls:C0018799 | Heart Diseases | 1 | BeFree |
| umls:C0019069 | Hemophilia A | 1 | BeFree |
| umls:C0020538 | Hypertensive disease | 1 | BeFree |
| umls:C0021670 | insulinoma | 1 | BeFree |
| umls:C0023467 | Leukemia, Myelocytic, Acute | 1 | BeFree |
| umls:C0023470 | Myeloid Leukemia | 1 | BeFree |
| umls:C0023530 | Leukopenia | 1 | BeFree |
| umls:C0023890 | Liver Cirrhosis | 1 | BeFree |
| umls:C0024121 | Lung Neoplasms | 1 | BeFree |
| umls:C0025149 | Medulloblastoma | 1 | BeFree |
| umls:C0027709 | Nephrocalcinosis | 1 | BeFree |
| umls:C0027773 | Nesidioblastosis | 1 | BeFree |
| umls:C0027947 | Neutropenia | 1 | BeFree,GAD |
| umls:C0029002 | Onchocerciasis, Ocular | 1 | BeFree |
| umls:C0029408 | Degenerative polyarthritis | 1 | BeFree |
| umls:C0029463 | Osteosarcoma | 1 | BeFree |
| umls:C0030567 | Parkinson Disease | 1 | BeFree |
| umls:C0032584 | polyps | 1 | BeFree |
| umls:C0036572 | Seizures | 1 | BeFree |
| umls:C0040336 | Tobacco Use Disorder | 1 | GAD |
| umls:C0151747 | Renal tubular disorder | 1 | BeFree |
| umls:C0153676 | Secondary malignant neoplasm of lung | 1 | BeFree |
| umls:C0162429 | Malnutrition | 1 | BeFree |
| umls:C0205851 | Germ cell tumor | 1 | BeFree |
| umls:C0206624 | Hepatoblastoma | 1 | BeFree |
| umls:C0221032 | Familial generalized lipodystrophy | 1 | BeFree |
| umls:C0221715 | Intestinal carcinoma | 1 | BeFree |
| umls:C0233794 | Memory impairment | 1 | BeFree |
| umls:C0238052 | Xanthomatosis, Cerebrotendinous | 1 | BeFree |
| umls:C0239946 | Fibrosis, Liver | 1 | BeFree |
| umls:C0242339 | Dyslipidemias | 1 | GAD |
| umls:C0262401 | Carcinoma of ampulla of Vater | 1 | BeFree |
| umls:C0268800 | Simple renal cyst | 1 | BeFree |
| umls:C0271694 | Familial partial lipodystrophy | 1 | BeFree |
| umls:C0280100 | Solid tumour | 1 | BeFree |
| umls:C0346429 | Multiple malignancy | 1 | BeFree |
| umls:C0362046 | Prediabetes syndrome | 1 | BeFree |
| umls:C0376544 | Hematopoietic Neoplasms | 1 | BeFree |
| umls:C0400966 | Non-alcoholic Fatty Liver Disease | 1 | BeFree |
| umls:C0497406 | Overweight | 1 | BeFree |
| umls:C0524909 | Hepatitis B, Chronic | 1 | BeFree |
| umls:C0585442 | Osteosarcoma of bone | 1 | BeFree |
| umls:C0684275 | Hemophilia, NOS | 1 | BeFree |
| umls:C0687120 | Nephronophthisis | 1 | BeFree |
| umls:C0750952 | Biliary Tract Cancer | 1 | BeFree |
| umls:C0947622 | Cholecystolithiasis | 1 | BeFree |
| umls:C1458155 | Mammary Neoplasms | 1 | GAD |
| umls:C1512409 | Hepatocarcinogenesis | 1 | BeFree |
| umls:C1569637 | Adenocarcinoma, Endometrioid | 1 | BeFree |
| umls:C1623038 | Cirrhosis | 1 | BeFree |
| umls:C1720860 | Familial Partial Lipodystrophy, Type 2 | 1 | BeFree |
| umls:C1838100 | MATURITY-ONSET DIABETES OF THE YOUNG, TYPE 3 (disorder) | 1 | BeFree |
| umls:C2316810 | Chronic kidney disease stage 5 | 1 | BeFree |
| umls:C2718001 | Protein Misfolding Disorders | 1 | BeFree |
| umls:C2931833 | Hyperinsulinemic hypoglycemia, familial, 2 | 1 | BeFree |
| umls:C3825462 | Diabetes in youth | 1 | BeFree |
| umls:C3826457 | Diabetes in children | 1 | BeFree |
| umls:C3888018 | Congenital Hyperinsulinism | 1 | GAD |
| umls:C4014962 | FANCONI RENOTUBULAR SYNDROME 4 WITH MATURITY-ONSET DIABETES OF THE YOUNG | 1 | UNIPROT |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))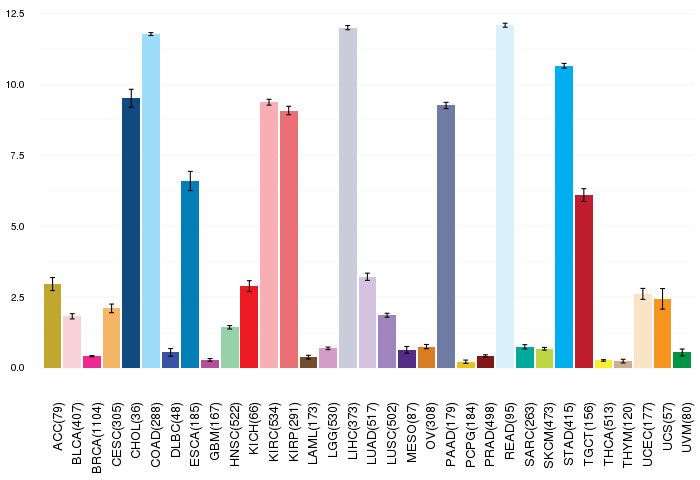
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))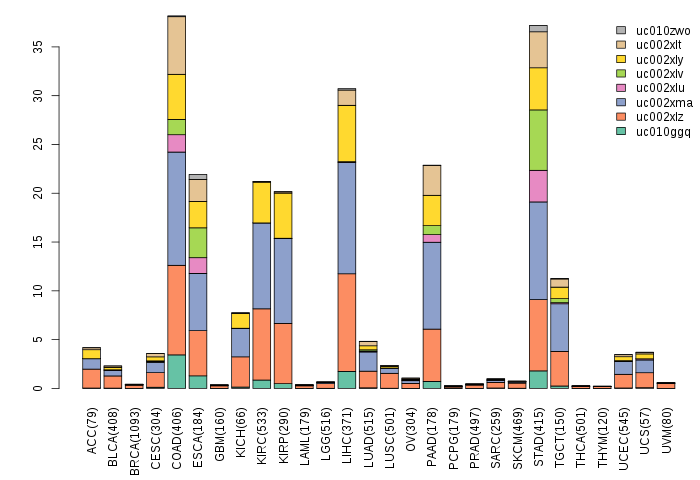
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data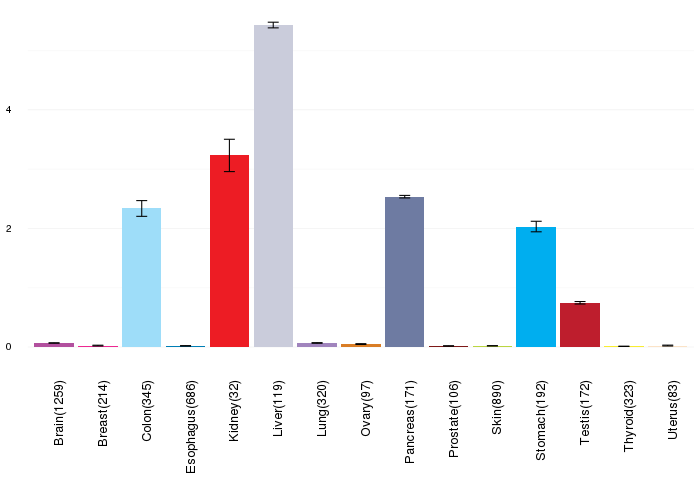
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))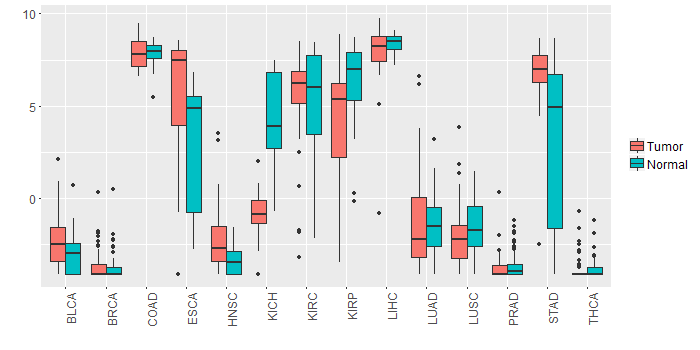
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.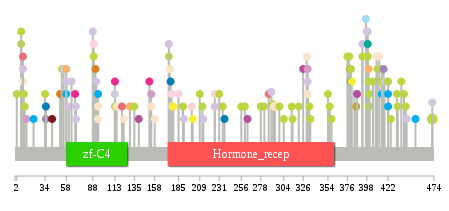

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 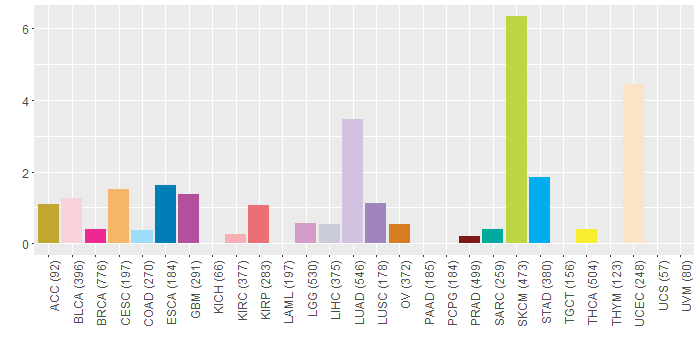
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)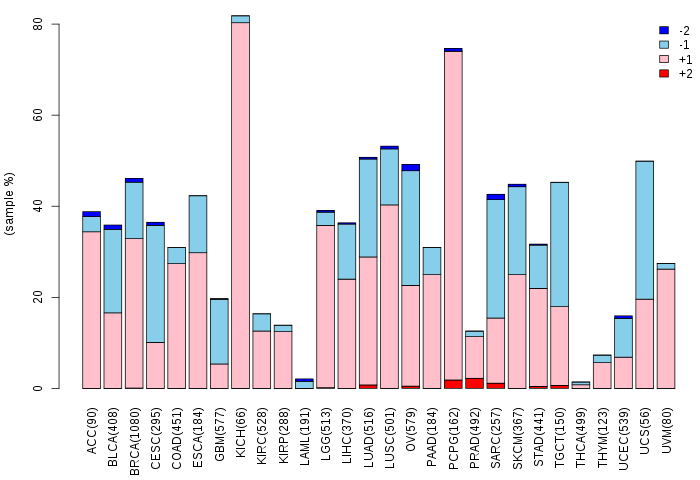
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 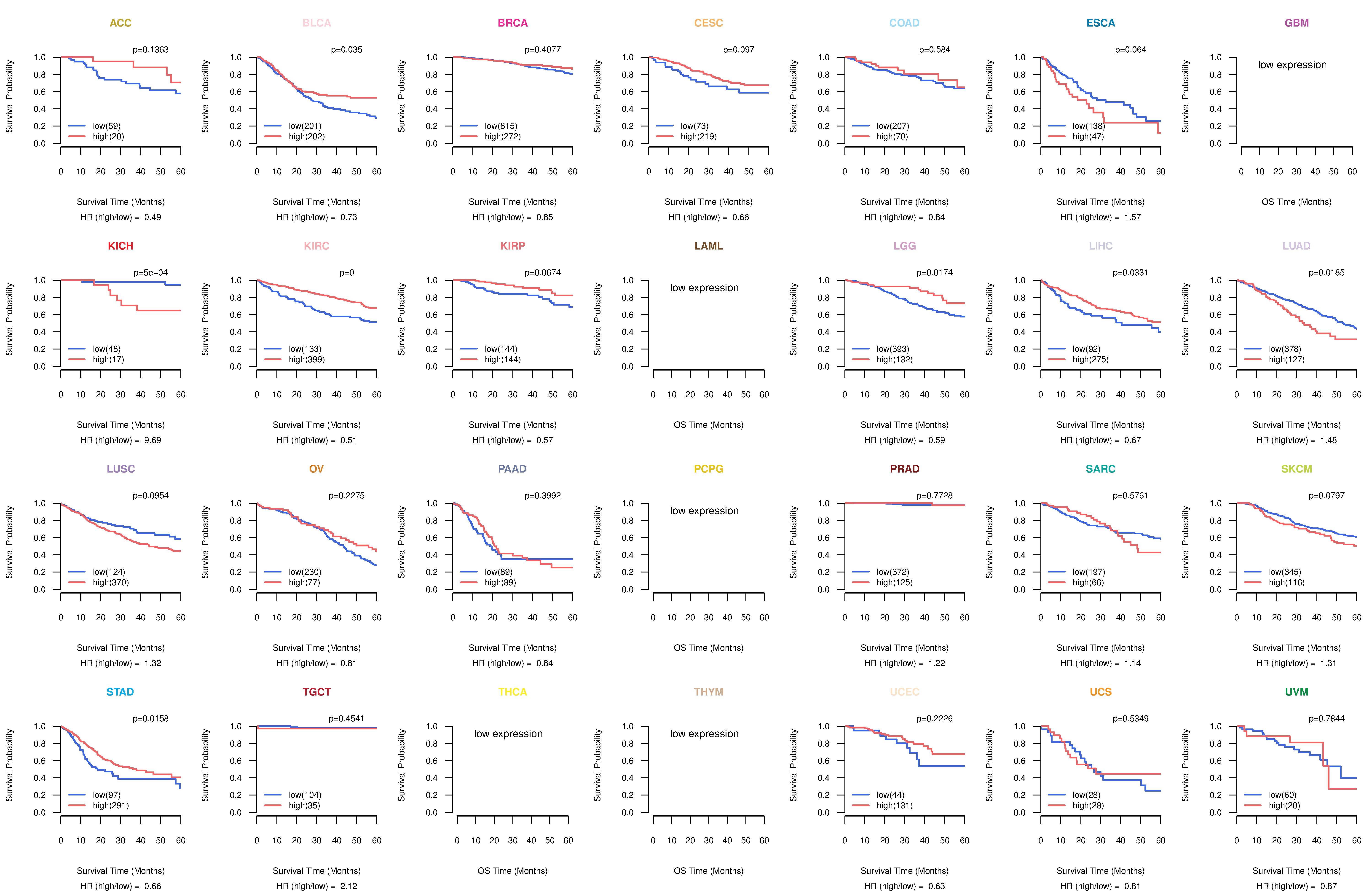
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 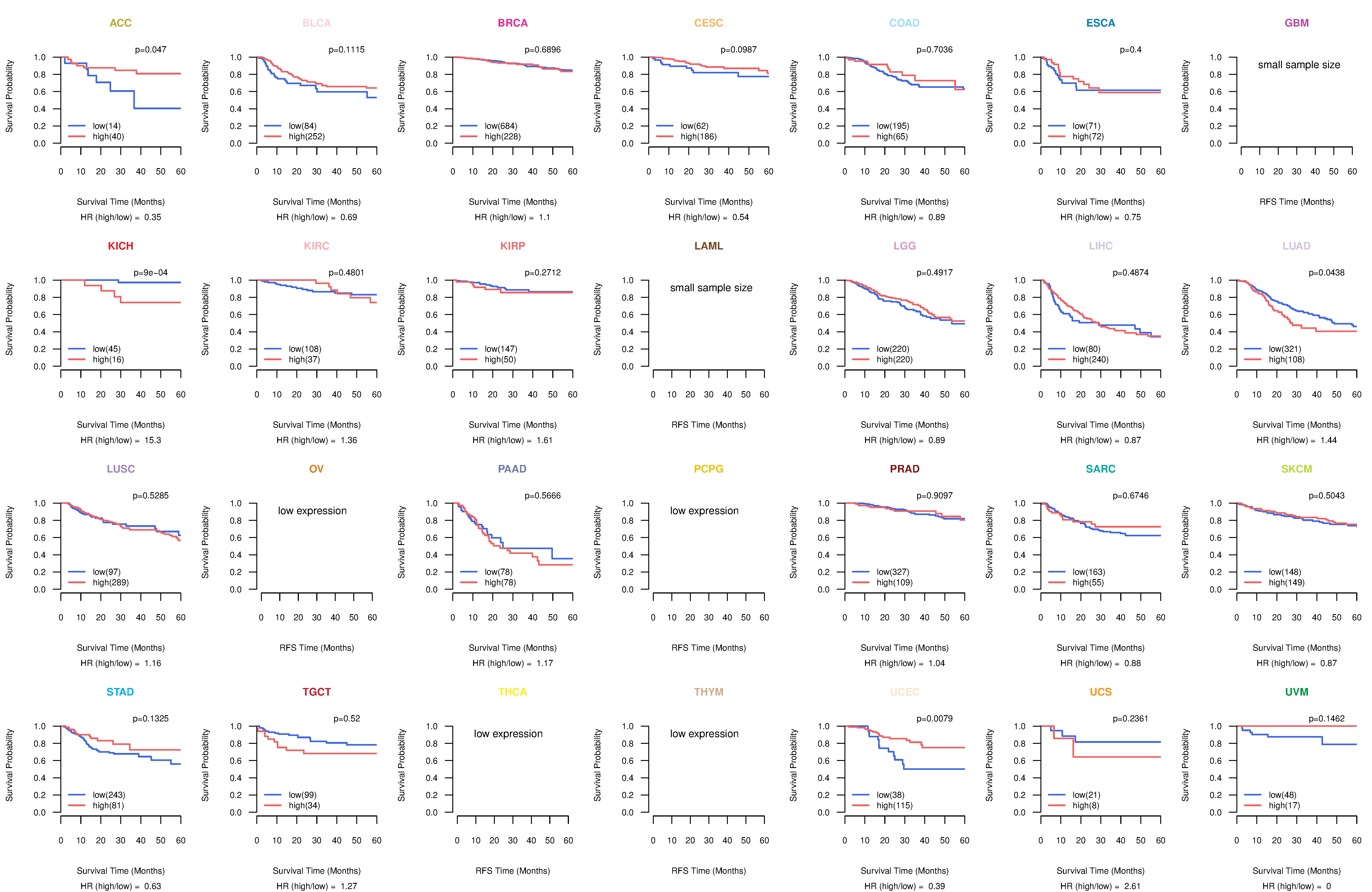
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 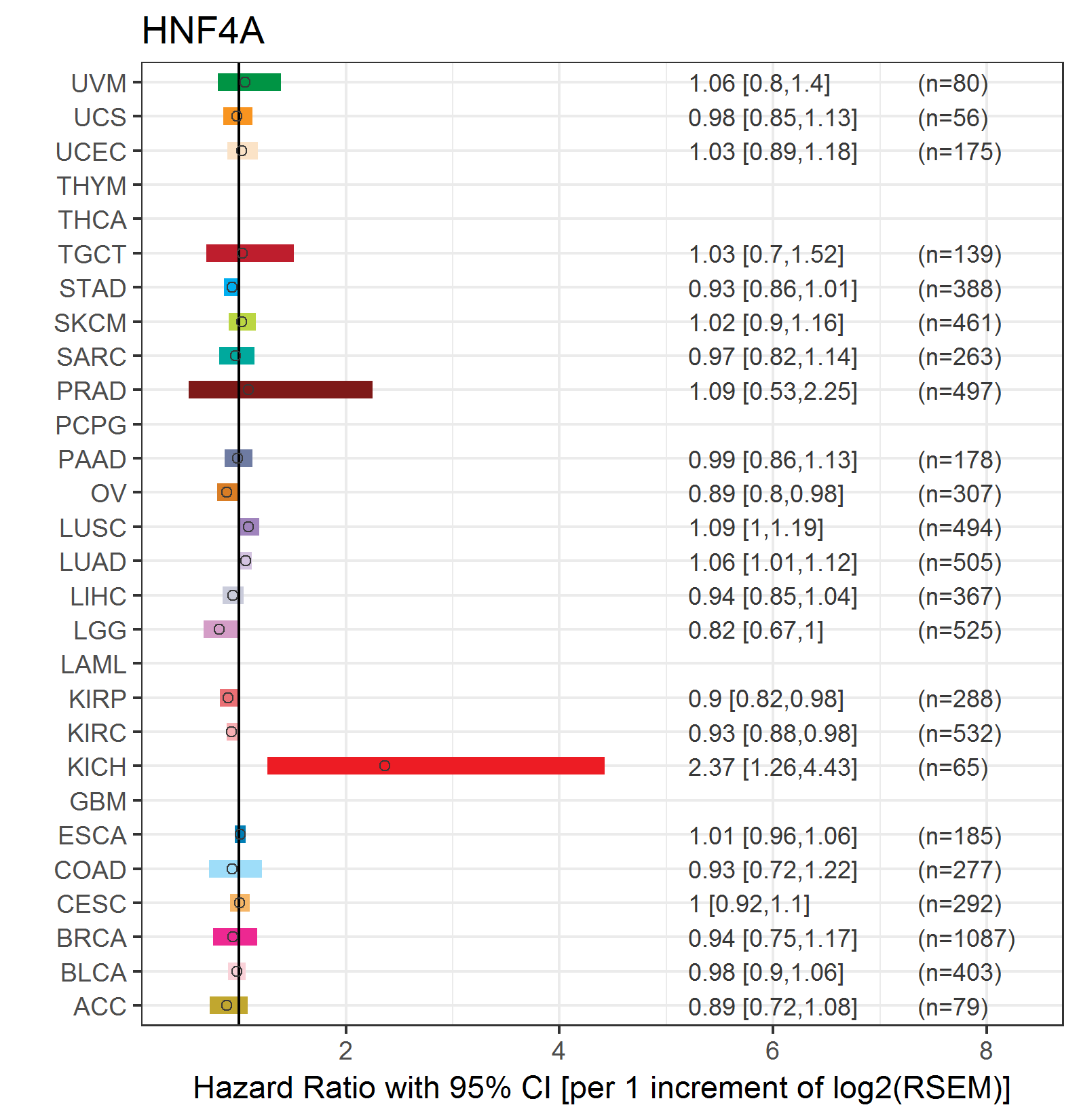
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 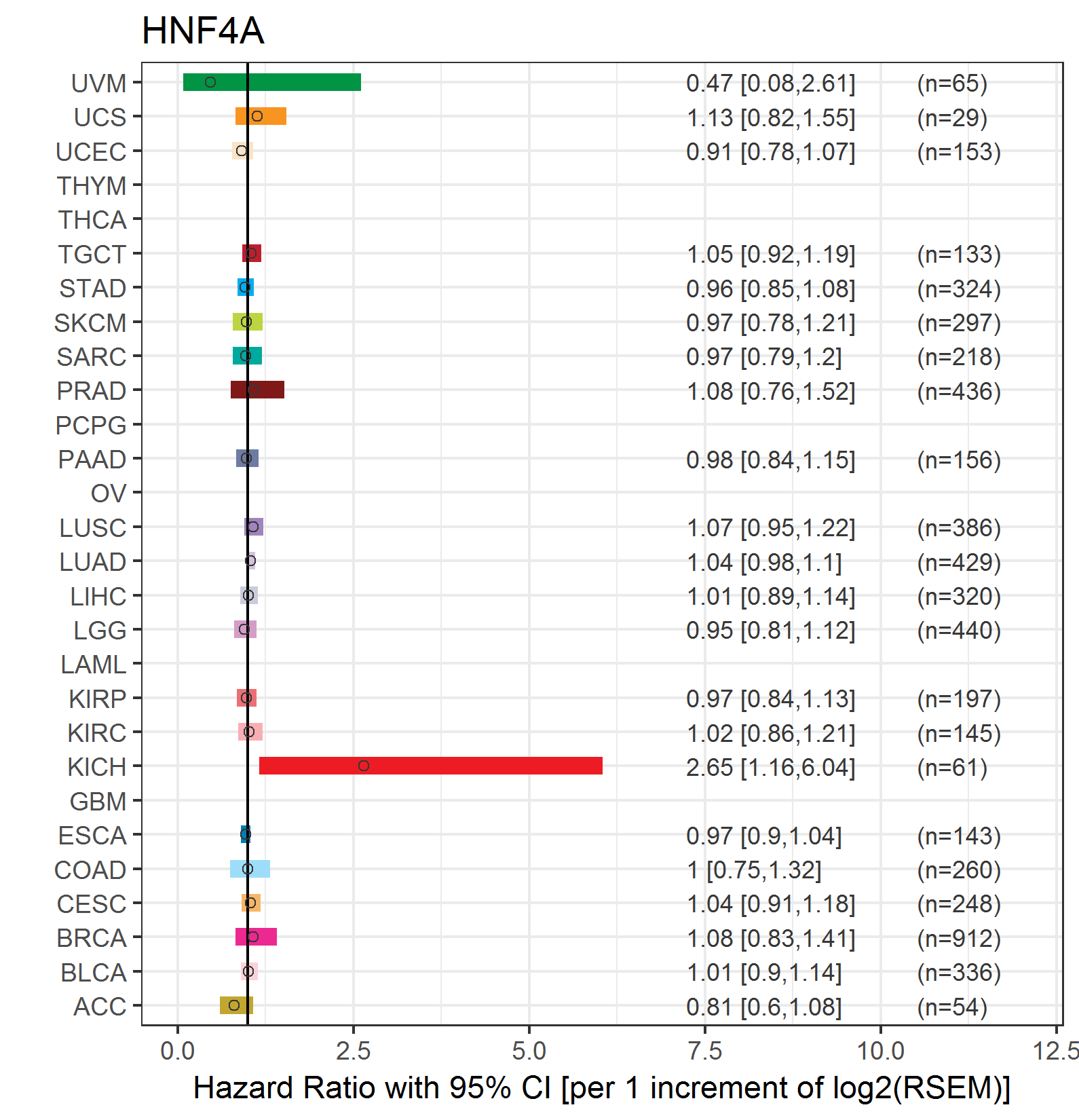
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























