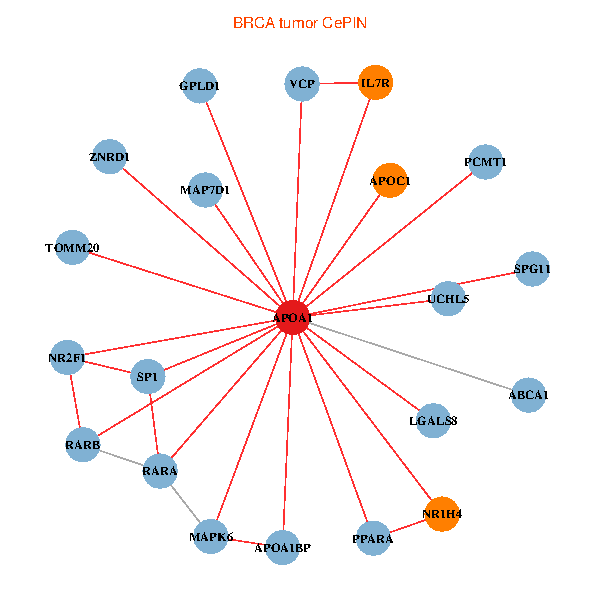
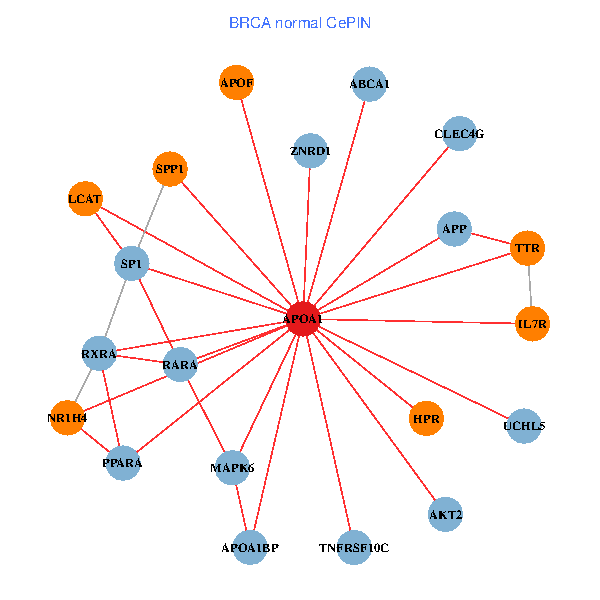
| BRCA (tumor) | BRCA (normal) |
| APOA1, SP1, RARA, VCP, RARB, ABCA1, UCHL5, IL7R, LGALS8, NR2F1, PPARA, MAPK6, TOMM20, ZNRD1, NR1H4, PCMT1, MAP7D1, GPLD1, APOC1, APOA1BP, SPG11 (tumor) | APOA1, SP1, SPP1, RARA, RXRA, APP, ABCA1, UCHL5, AKT2, IL7R, TNFRSF10C, TTR, PPARA, MAPK6, CLEC4G, ZNRD1, NR1H4, LCAT, HPR, APOF, APOA1BP (normal) |
 |  |
|
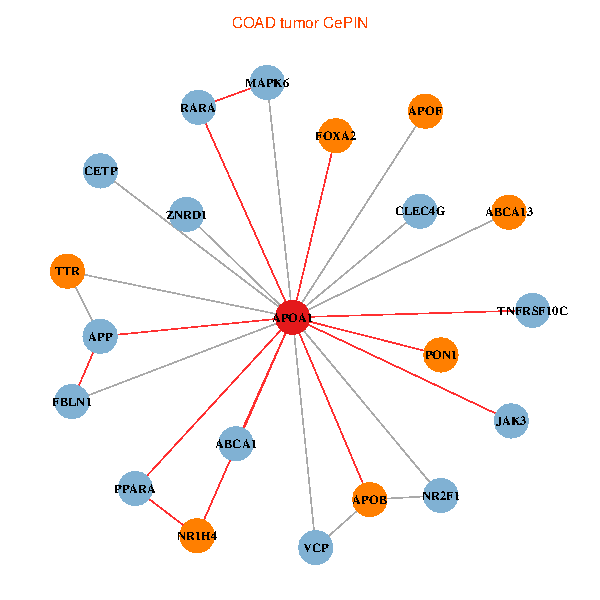
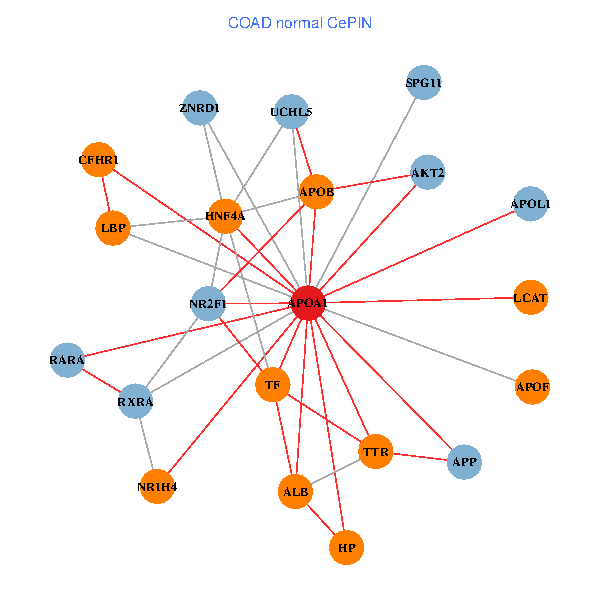
| COAD (tumor) | COAD (normal) |
| APOA1, RARA, VCP, APP, ABCA1, APOB, JAK3, TNFRSF10C, TTR, NR2F1, PPARA, CETP, MAPK6, FBLN1, CLEC4G, ZNRD1, ABCA13, NR1H4, FOXA2, APOF, PON1 (tumor) | APOA1, HNF4A, RARA, RXRA, APP, ALB, UCHL5, AKT2, APOB, TTR, NR2F1, TF, HP, ZNRD1, NR1H4, LCAT, LBP, APOF, APOL1, CFHR1, SPG11 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| APOA1, SP1, VCP, RARB, ALB, UCHL5, APOB, JAK3, TTR, PPARA, CETP, CLEC4G, LCAT, HPR, SCARB1, GPLD1, PON1, APOC1, APOA1BP, CFHR1, SPG11 (tumor) | APOA1, SP1, APP, ALB, ABCA1, UCHL5, APOB, IL7R, TTR, PPARA, CETP, MAPK6, HP, ZNRD1, NR1H4, LCAT, HPR, APOF, PON1, APOA1BP, SPG11 (normal) |
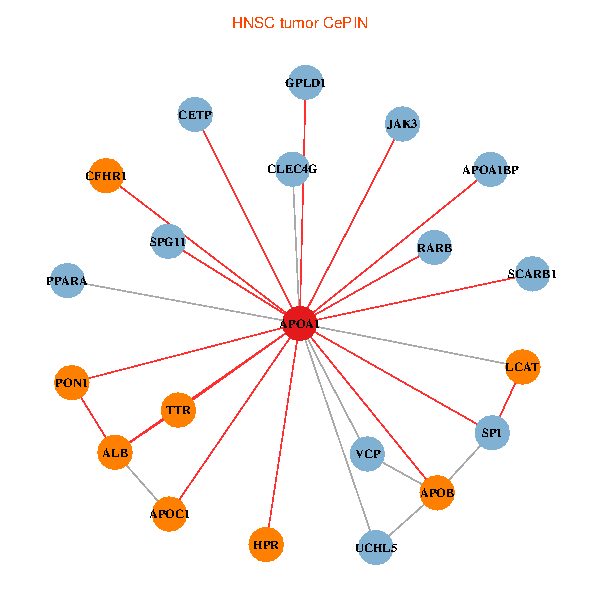 | 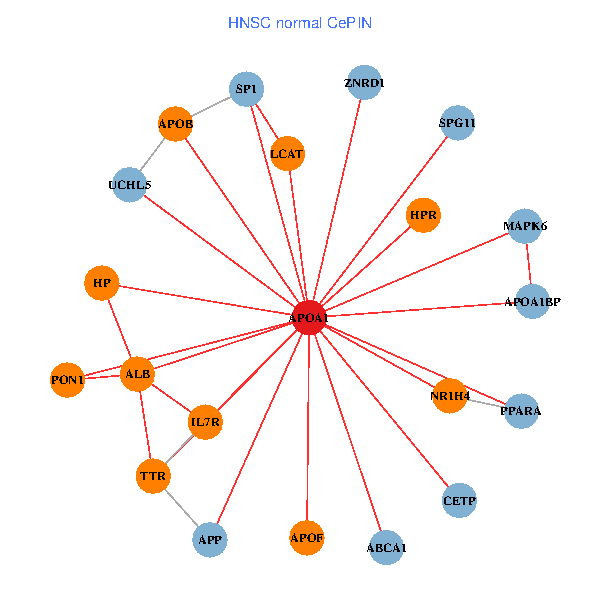 |
|
| KICH (tumor) | KICH (normal) |
| APOA1, HNF4A, SP1, SPP1, APP, ALB, AKT2, APOB, IL7R, TTR, CETP, MAPK6, ABCA13, NR1H4, FOXA2, HPR, LBP, PLTP, APOF, APOC1, CFHR1 (tumor) | APOA1, SP1, RARA, RARB, UCHL5, JAK3, TNFRSF10C, LGALS8, NR2F1, PPARA, ATP5B, ZNRD1, PCMT1, MAP7D1, FOXA2, LCAT, LBP, PLTP, APOF, PDE1A, SPG11 (normal) |
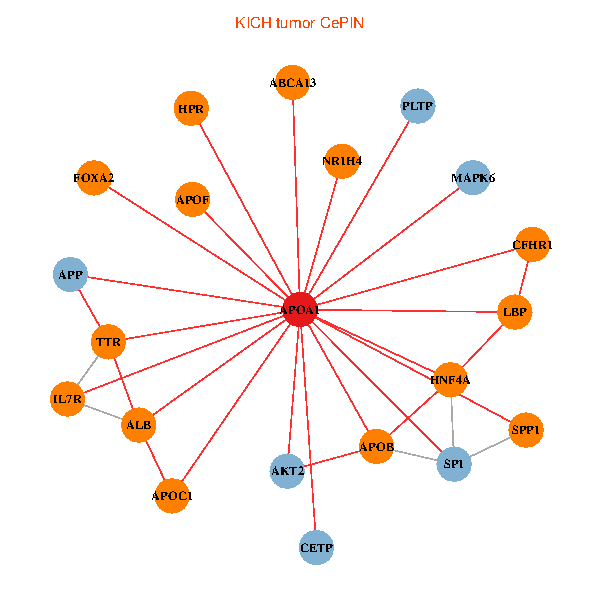 | 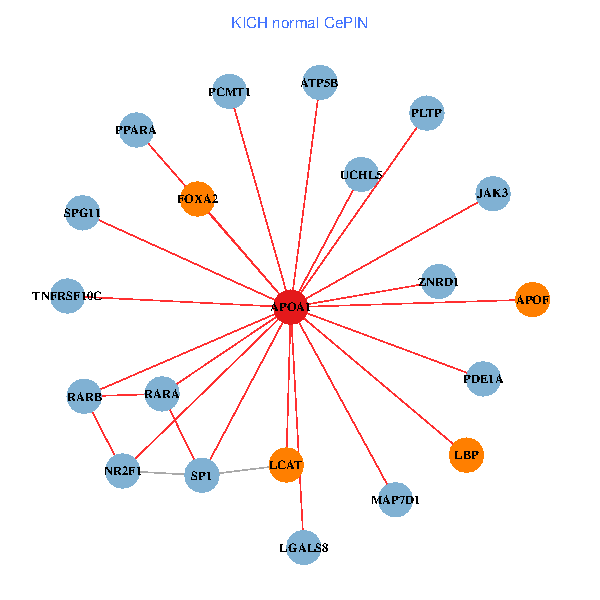 |
|
| KIRC (tumor) | KIRC (normal) |
| APOA1, HNF4A, RXRA, APP, PITX3, ALB, AKT2, APOB, TNFRSF10C, TTR, PPARA, CETP, MAPK6, TF, ABCA13, FOXA2, HPR, LBP, PDE1A, APOC1, SPG11 (tumor) | APOA1, LGALS3, RARB, ALB, UCHL5, APOB, TTR, LGALS8, SAA1, TF, HP, ZNRD1, SAA2, HPR, LBP, APOF, PON1, APOC1, APOA1BP, CFHR1, SPG11 (normal) |
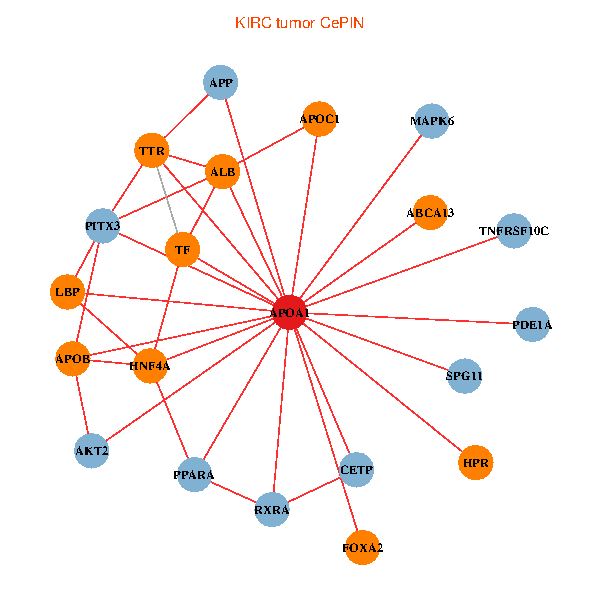 | 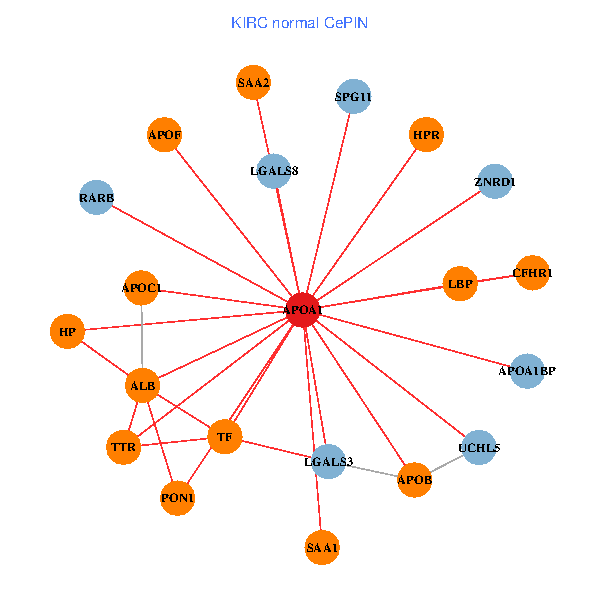 |
|
| KIRP (tumor) | KIRP (normal) |
| APOA1, HNF4A, RXRA, APP, PITX3, ALB, AKT2, APOB, TNFRSF10C, TTR, PPARA, CETP, MAPK6, ABCA13, PCMT1, FOXA2, LCAT, HPR, LBP, SCARB1, PDE1A (tumor) | APOA1, HNF4A, SPP1, LGALS3, APP, ALB, AKT2, APOB, TTR, NR2F2, TF, CLEC4G, HP, PCMT1, HPR, LBP, SCARB1, APOL1, PON1, APOC1, SPG11 (normal) |
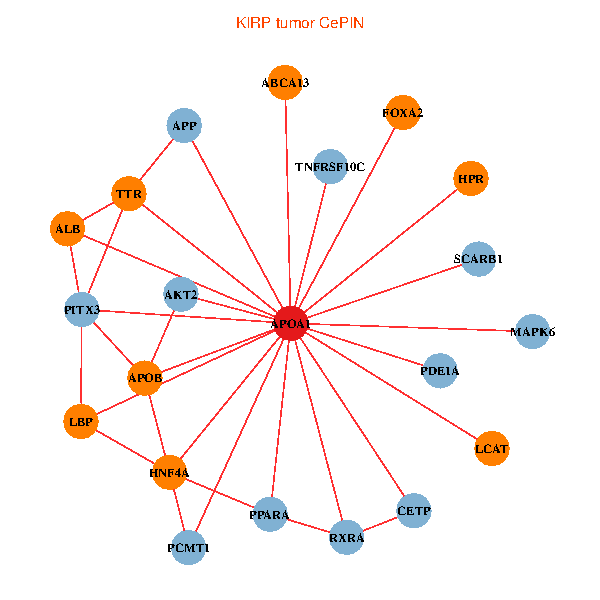 | 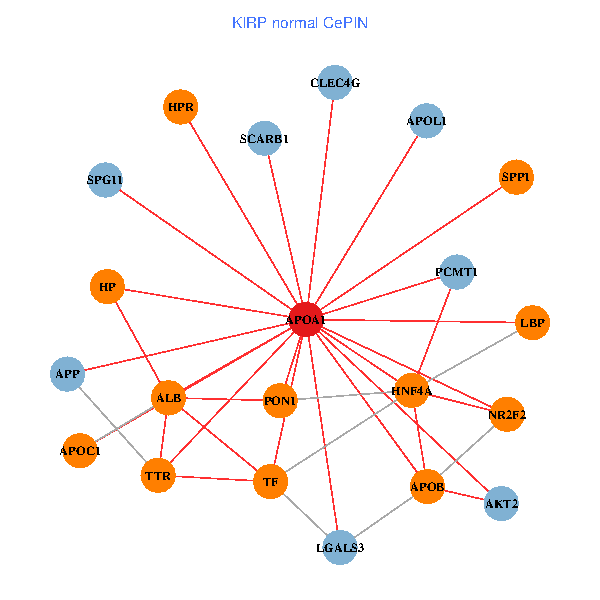 |
|
| LIHC (tumor) | LIHC (normal) |
| APOA1, SP1, ALB, ABCA1, IL7R, JAK3, TTR, LGALS8, NR2F1, CETP, CD40LG, TF, CLEC4G, MIS12, LCAT, PLTP, APOL1, PDE1A, APOC1, APOA1BP, SPG11 (tumor) | APOA1, HNF4A, SP1, RXRA, ALB, APOB, TTR, NR2F2, TF, HP, NR1H4, MAP7D1, FOXA2, LCAT, HPR, SCARB1, GPLD1, APOF, PON1, APOC1, CFHR1 (normal) |
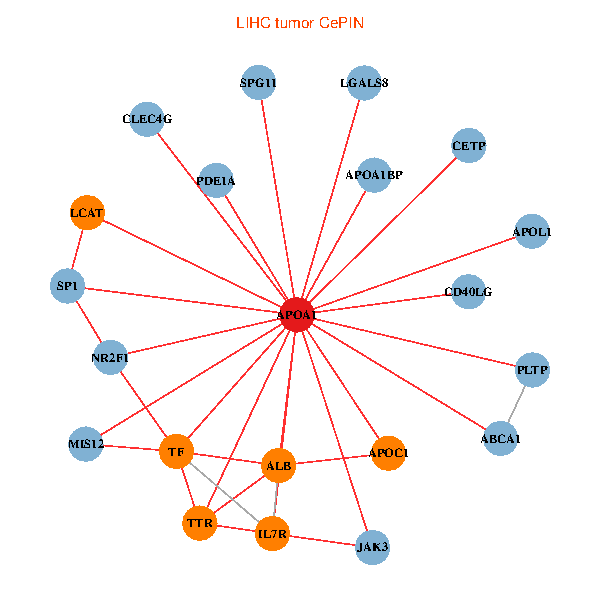 | 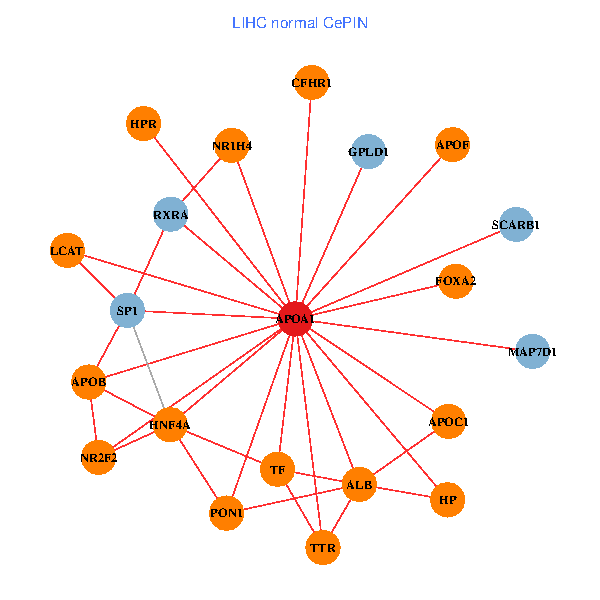 |
|
| LUAD (tumor) | LUAD (normal) |
| APOA1, HNF4A, SP1, LGALS3, UCHL5, IL7R, TNFRSF10C, LGALS8, PPARA, CETP, ATP5B, FBLN1, TF, PCMT1, MAP7D1, FOXA2, GPLD1, PDE1A, APOC1, APOA1BP, SPG11 (tumor) | APOA1, HNF4A, SPP1, APP, ALB, UCHL5, APOB, TTR, CETP, SAA1, TF, NR1H4, SAA2, FOXA2, LCAT, HPR, LBP, SCARB1, APOF, APOC1, CFHR1 (normal) |
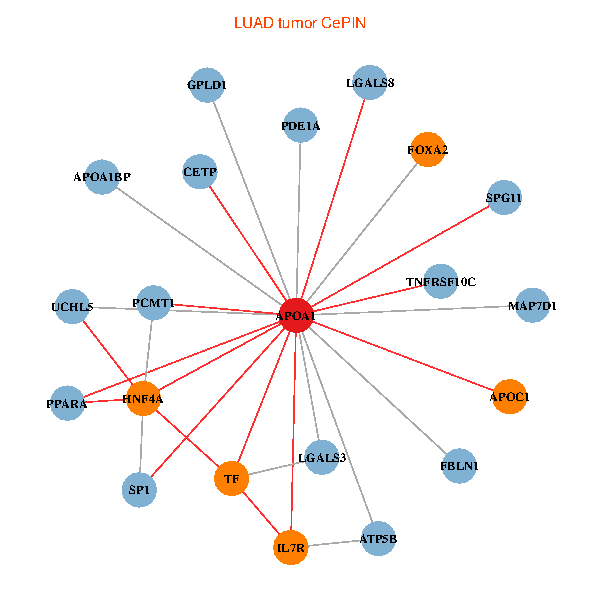 | 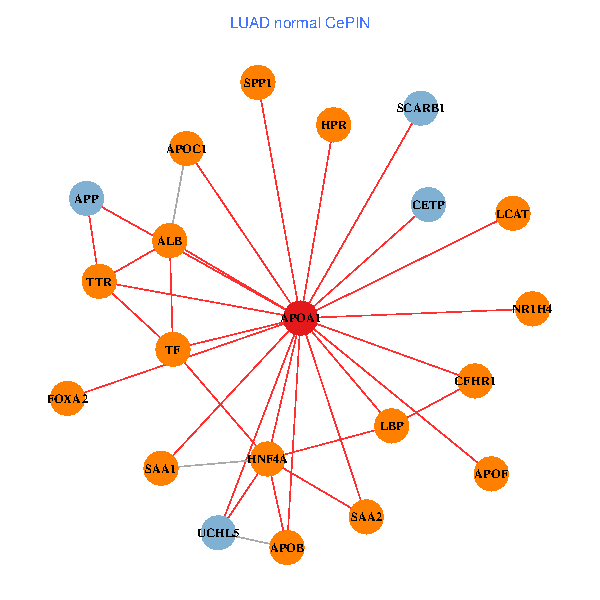 |
|
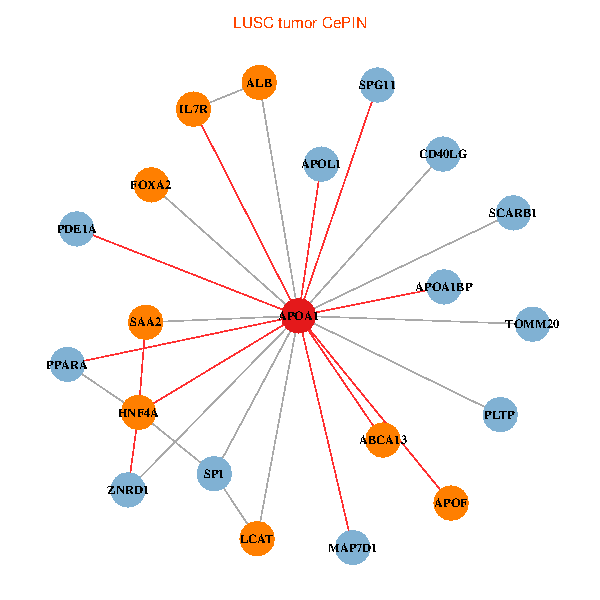
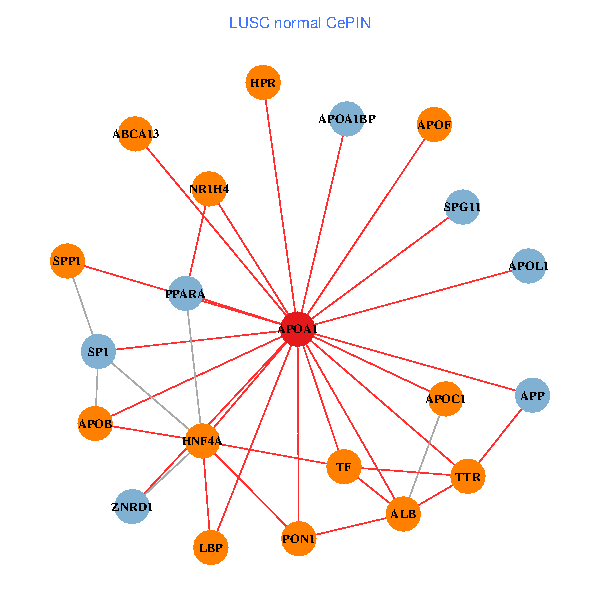
| LUSC (tumor) | LUSC (normal) |
| APOA1, HNF4A, SP1, ALB, IL7R, PPARA, TOMM20, CD40LG, ZNRD1, ABCA13, SAA2, MAP7D1, FOXA2, LCAT, PLTP, SCARB1, APOF, APOL1, PDE1A, APOA1BP, SPG11 (tumor) | APOA1, HNF4A, SP1, SPP1, APP, ALB, APOB, TTR, PPARA, TF, ZNRD1, ABCA13, NR1H4, HPR, LBP, APOF, APOL1, PON1, APOC1, APOA1BP, SPG11 (normal) |
 |  |
|
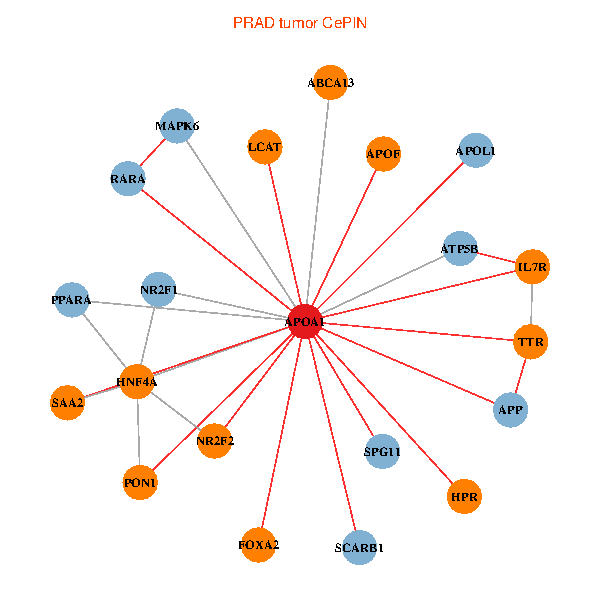
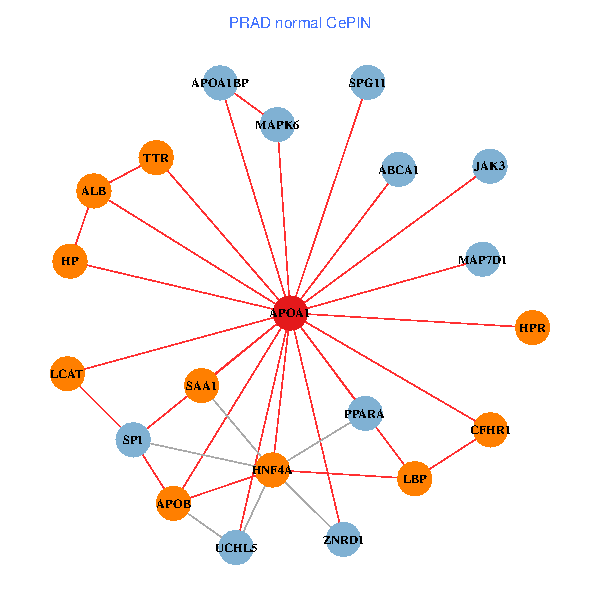
| PRAD (tumor) | PRAD (normal) |
| APOA1, HNF4A, RARA, APP, IL7R, TTR, NR2F1, PPARA, ATP5B, NR2F2, MAPK6, ABCA13, SAA2, FOXA2, LCAT, HPR, SCARB1, APOF, APOL1, PON1, SPG11 (tumor) | APOA1, HNF4A, SP1, ALB, ABCA1, UCHL5, APOB, JAK3, TTR, PPARA, MAPK6, SAA1, HP, ZNRD1, MAP7D1, LCAT, HPR, LBP, APOA1BP, CFHR1, SPG11 (normal) |
 |  |
|
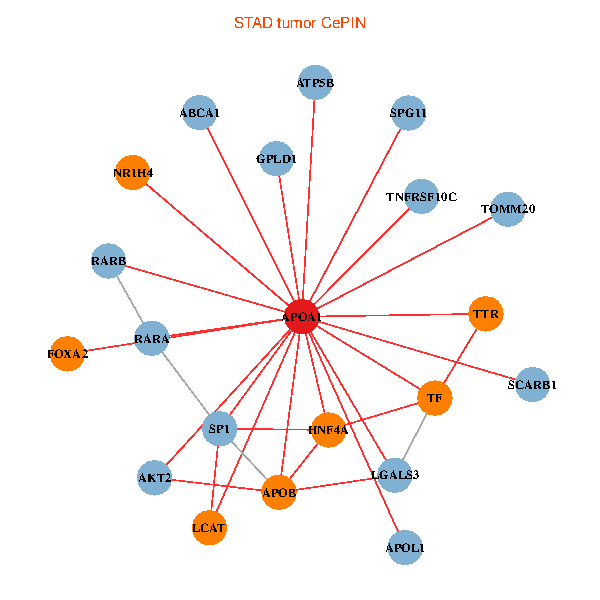
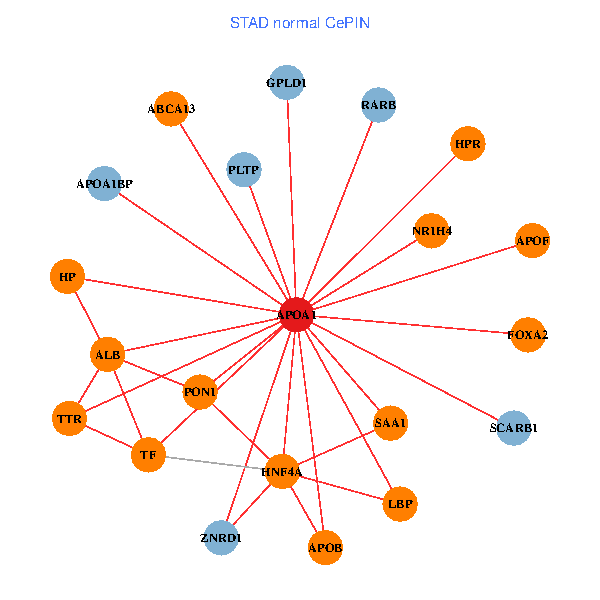
| STAD (tumor) | STAD (normal) |
| APOA1, HNF4A, SP1, RARA, LGALS3, RARB, ABCA1, AKT2, APOB, TNFRSF10C, TTR, ATP5B, TOMM20, TF, NR1H4, FOXA2, LCAT, SCARB1, GPLD1, APOL1, SPG11 (tumor) | APOA1, HNF4A, RARB, ALB, APOB, TTR, SAA1, TF, HP, ZNRD1, ABCA13, NR1H4, FOXA2, HPR, LBP, PLTP, SCARB1, GPLD1, APOF, PON1, APOA1BP (normal) |
 |  |
|
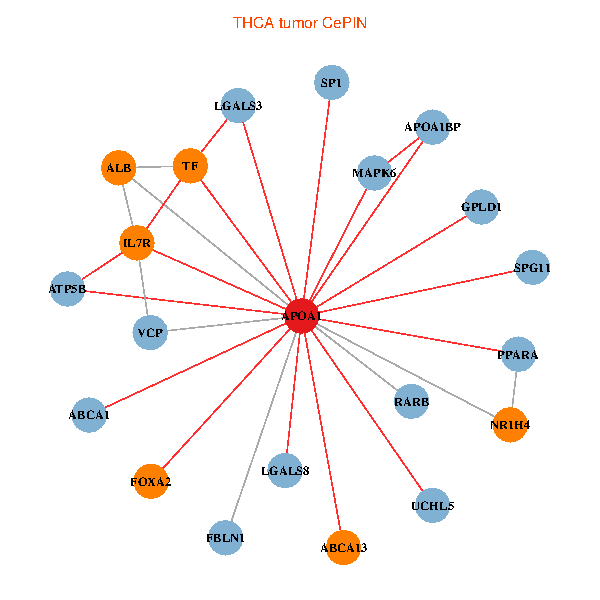
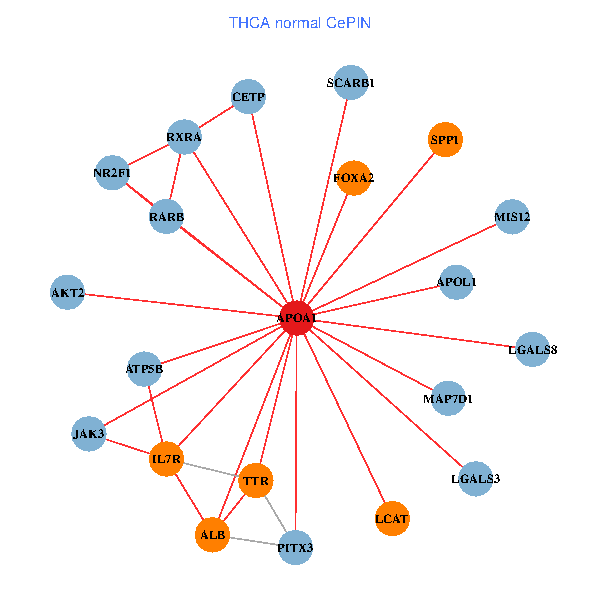
| THCA (tumor) | THCA (normal) |
| APOA1, SP1, VCP, LGALS3, RARB, ALB, ABCA1, UCHL5, IL7R, LGALS8, PPARA, ATP5B, MAPK6, FBLN1, TF, ABCA13, NR1H4, FOXA2, GPLD1, APOA1BP, SPG11 (tumor) | APOA1, SPP1, RXRA, LGALS3, RARB, PITX3, ALB, AKT2, IL7R, JAK3, TTR, LGALS8, NR2F1, CETP, ATP5B, MIS12, MAP7D1, FOXA2, LCAT, SCARB1, APOL1 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0004153 | Atherosclerosis | 65 | BeFree,GAD,LHGDN |
| umls:C0003850 | Arteriosclerosis | 62 | BeFree,LHGDN,RGD |
| umls:C0010054 | Coronary Arteriosclerosis | 50 | BeFree,GAD |
| umls:C0010068 | Coronary heart disease | 50 | BeFree,GAD,LHGDN |
| umls:C1956346 | Coronary Artery Disease | 43 | BeFree,GAD,LHGDN |
| umls:C0002726 | Amyloidosis | 23 | BeFree,LHGDN |
| umls:C0007222 | Cardiovascular Diseases | 23 | BeFree,GAD,LHGDN |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 22 | BeFree,GAD |
| umls:C0020474 | Hyperlipidemia, Familial Combined | 20 | BeFree,GAD,LHGDN |
| umls:C0027051 | Myocardial Infarction | 20 | BeFree,GAD,LHGDN |
| umls:C2239176 | Liver carcinoma | 16 | BeFree,CTD_human |
| umls:C0039292 | Tangier Disease | 15 | BeFree,LHGDN |
| umls:C0524620 | Metabolic Syndrome X | 15 | BeFree,GAD,RGD |
| umls:C0028754 | Obesity | 14 | BeFree,GAD |
| umls:C0020473 | Hyperlipidemia | 12 | BeFree,GAD |
| umls:C0023903 | Liver neoplasms | 10 | BeFree |
| umls:C0242339 | Dyslipidemias | 9 | BeFree,GAD |
| umls:C0020538 | Hypertensive disease | 8 | BeFree,CTD_human,GAD,LHGDN,RGD |
| umls:C0268380 | Systemic amyloidosis | 8 | BeFree |
| umls:C0281479 | Primary Systemic Amyloidosis | 8 | BeFree |
| umls:C0342883 | Cholesteryl Ester Transfer Protein Deficiency | 8 | BeFree,GAD |
| umls:C1704429 | Hypoalphalipoproteinemia, Familial | 8 | BeFree,CLINVAR,ORPHANET |
| umls:C2936349 | Plaque, Amyloid | 8 | BeFree |
| umls:C0002395 | Alzheimer's Disease | 7 | BeFree,GAD |
| umls:C0011849 | Diabetes Mellitus | 7 | BeFree |
| umls:C0020445 | Hypercholesterolemia, Familial | 7 | BeFree,LHGDN |
| umls:C0948089 | Acute Coronary Syndrome | 7 | BeFree,GAD,LHGDN |
| umls:C2931838 | Familial HDL deficiency | 6 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 5 | BeFree |
| umls:C0011847 | Diabetes | 5 | BeFree |
| umls:C0020443 | Hypercholesterolemia | 5 | BeFree,GAD |
| umls:C0038454 | Cerebrovascular accident | 5 | CTD_human,GAD,LHGDN |
| umls:C0678222 | Breast Carcinoma | 5 | BeFree |
| umls:C3165209 | High density lipoprotein deficiency | 5 | BeFree |
| umls:C0019693 | HIV Infections | 4 | BeFree,CTD_human |
| umls:C0023195 | Lecithin Acyltransferase Deficiency | 4 | BeFree |
| umls:C0023895 | Liver diseases | 4 | BeFree,GAD |
| umls:C0042373 | Vascular Diseases | 4 | BeFree,LHGDN |
| umls:C0151744 | Myocardial Ischemia | 4 | BeFree,GAD,LHGDN |
| umls:C0497327 | Dementia | 4 | BeFree,GAD,LHGDN |
| umls:C0011265 | Presenile dementia | 3 | BeFree |
| umls:C0020557 | Hypertriglyceridemia | 3 | GAD,LHGDN |
| umls:C0021368 | Inflammation | 3 | CTD_human,GAD,LHGDN |
| umls:C0027720 | Nephrosis | 3 | BeFree |
| umls:C0027726 | Nephrotic Syndrome | 3 | BeFree,RGD |
| umls:C0029925 | Ovarian Carcinoma | 3 | BeFree |
| umls:C0033860 | Psoriasis | 3 | BeFree |
| umls:C0035309 | Retinal Diseases | 3 | BeFree,GAD |
| umls:C0038356 | Stomach Neoplasms | 3 | BeFree,CTD_human |
| umls:C0206246 | Amyloidosis, Hereditary | 3 | BeFree |
| umls:C0206624 | Hepatoblastoma | 3 | BeFree |
| umls:C0268389 | Amyloidosis, familial visceral | 3 | BeFree,CLINVAR,CTD_human,UNIPROT |
| umls:C0342895 | Fish-Eye Disease | 3 | BeFree |
| umls:C1522137 | Hypertriglyceridemia result | 3 | GAD |
| umls:C2712907 | Combined hyperlipidemia | 3 | BeFree |
| umls:C3149462 | HYPERALPHALIPOPROTEINEMIA 1 | 3 | BeFree |
| umls:C0001418 | Adenocarcinoma | 2 | CTD_human,LHGDN |
| umls:C0008350 | Cholelithiasis | 2 | BeFree,GAD |
| umls:C0011884 | Diabetic Retinopathy | 2 | BeFree,LHGDN |
| umls:C0013080 | Down Syndrome | 2 | BeFree |
| umls:C0015230 | Exanthema | 2 | BeFree |
| umls:C0015695 | Fatty Liver | 2 | BeFree |
| umls:C0016977 | Gall Bladder Diseases | 2 | BeFree,LHGDN |
| umls:C0019196 | Hepatitis C | 2 | BeFree,GAD |
| umls:C0020476 | Hyperlipoproteinemias | 2 | BeFree |
| umls:C0020480 | Hyperlipoproteinemia Type IV | 2 | BeFree |
| umls:C0022661 | Kidney Failure, Chronic | 2 | BeFree,GAD |
| umls:C0022876 | Premature Obstetric Labor | 2 | GAD |
| umls:C0023890 | Liver Cirrhosis | 2 | BeFree |
| umls:C0024121 | Lung Neoplasms | 2 | CTD_human,LHGDN |
| umls:C0026769 | Multiple Sclerosis | 2 | BeFree,GAD |
| umls:C0027627 | Neoplasm Metastasis | 2 | BeFree,LHGDN |
| umls:C0029408 | Degenerative polyarthritis | 2 | BeFree |
| umls:C0029928 | Ovarian Diseases | 2 | BeFree |
| umls:C0032460 | Polycystic Ovary Syndrome | 2 | BeFree |
| umls:C0032580 | Adenomatous Polyposis Coli | 2 | BeFree |
| umls:C0036341 | Schizophrenia | 2 | LHGDN |
| umls:C0085220 | Cerebral Amyloid Angiopathy | 2 | BeFree |
| umls:C0178874 | Tumor Progression | 2 | BeFree |
| umls:C0206245 | Amyloid Neuropathies, Familial | 2 | BeFree |
| umls:C0233794 | Memory impairment | 2 | BeFree |
| umls:C0243026 | Sepsis | 2 | BeFree,RGD |
| umls:C0302314 | Xanthoma | 2 | BeFree |
| umls:C0311277 | Obesity, Abdominal | 2 | BeFree,GAD |
| umls:C0338656 | Impaired cognition | 2 | BeFree |
| umls:C0473527 | Hypoalphalipoproteinemias | 2 | CTD_human,GAD,LHGDN |
| umls:C0497406 | Overweight | 2 | BeFree |
| umls:C0577631 | Carotid Atherosclerosis | 2 | GAD |
| umls:C0848332 | Spots on skin | 2 | BeFree |
| umls:C0947622 | Cholecystolithiasis | 2 | GAD |
| umls:C0947751 | Vascular inflammations | 2 | BeFree |
| umls:C1140680 | Malignant neoplasm of ovary | 2 | BeFree |
| umls:C1272641 | Systemic arterial pressure | 2 | GAD |
| umls:C2711227 | Steatohepatitis | 2 | BeFree |
| umls:C0002895 | Anemia, Sickle Cell | 1 | GAD |
| umls:C0003130 | Anoxia | 1 | CTD_human |
| umls:C0003742 | Arcus Senilis | 1 | BeFree |
| umls:C0003864 | Arthritis | 1 | BeFree |
| umls:C0003868 | Arthritis, Gouty | 1 | BeFree |
| umls:C0003873 | Rheumatoid Arthritis | 1 | BeFree |
| umls:C0004096 | Asthma | 1 | BeFree |
| umls:C0004364 | Autoimmune Diseases | 1 | LHGDN |
| umls:C0004943 | Behcet Syndrome | 1 | CTD_human |
| umls:C0006111 | Brain Diseases | 1 | BeFree |
| umls:C0007102 | Malignant tumor of colon | 1 | BeFree |
| umls:C0007138 | Carcinoma, Transitional Cell | 1 | RGD |
| umls:C0007282 | Carotid Stenosis | 1 | BeFree |
| umls:C0007785 | Cerebral Infarction | 1 | GAD |
| umls:C0007820 | Cerebrovascular Disorders | 1 | GAD |
| umls:C0008495 | Chorioamnionitis | 1 | GAD |
| umls:C0010346 | Crohn Disease | 1 | BeFree |
| umls:C0011065 | Cessation of life | 1 | CTD_human |
| umls:C0011609 | Drug Eruptions | 1 | CTD_human |
| umls:C0011853 | Diabetes Mellitus, Experimental | 1 | RGD |
| umls:C0013604 | Edema | 1 | CTD_human |
| umls:C0015814 | Femur Head Necrosis | 1 | GAD |
| umls:C0015944 | Fetal Membranes, Premature Rupture | 1 | GAD |
| umls:C0017636 | Glioblastoma | 1 | BeFree |
| umls:C0017665 | Membranous glomerulonephritis | 1 | CTD_human |
| umls:C0018099 | Gout | 1 | GAD |
| umls:C0018801 | Heart failure | 1 | BeFree |
| umls:C0018802 | Congestive heart failure | 1 | BeFree |
| umls:C0019163 | Hepatitis B | 1 | LHGDN |
| umls:C0020479 | Hyperlipoproteinemia Type III | 1 | BeFree |
| umls:C0020542 | Pulmonary Hypertension | 1 | BeFree |
| umls:C0020550 | Hyperthyroidism | 1 | RGD |
| umls:C0020639 | Hypoproteinemia | 1 | RGD |
| umls:C0021390 | Inflammatory Bowel Diseases | 1 | GAD |
| umls:C0021655 | Insulin Resistance | 1 | GAD |
| umls:C0023891 | Liver Cirrhosis, Alcoholic | 1 | BeFree |
| umls:C0023893 | Liver Cirrhosis, Experimental | 1 | RGD |
| umls:C0024141 | Lupus Erythematosus, Systemic | 1 | BeFree |
| umls:C0024623 | Malignant neoplasm of stomach | 1 | BeFree |
| umls:C0025286 | Meningioma | 1 | BeFree |
| umls:C0025517 | Metabolic Diseases | 1 | BeFree |
| umls:C0028259 | Nodule | 1 | BeFree |
| umls:C0029445 | Bone necrosis | 1 | GAD |
| umls:C0032285 | Pneumonia | 1 | BeFree |
| umls:C0032914 | Pre-Eclampsia | 1 | GAD |
| umls:C0034069 | Pulmonary Fibrosis | 1 | BeFree |
| umls:C0036572 | Seizures | 1 | BeFree |
| umls:C0037116 | Silicosis | 1 | BeFree |
| umls:C0040028 | Thrombocythemia, Essential | 1 | BeFree |
| umls:C0041349 | Nephritis, Tubulointerstitial | 1 | BeFree |
| umls:C0041948 | Uremia | 1 | BeFree |
| umls:C0043119 | Werner Syndrome | 1 | BeFree |
| umls:C0149896 | Primary gout | 1 | BeFree |
| umls:C0149940 | Sciatic Neuropathy | 1 | RGD |
| umls:C0151526 | Premature Birth | 1 | GAD |
| umls:C0152013 | Adenocarcinoma of lung (disorder) | 1 | CTD_human |
| umls:C0152025 | Polyneuropathy | 1 | BeFree |
| umls:C0152096 | Complete trisomy 18 syndrome | 1 | BeFree |
| umls:C0155626 | Acute myocardial infarction | 1 | BeFree |
| umls:C0155668 | Old myocardial infarction | 1 | BeFree |
| umls:C0206701 | Cystadenocarcinoma, Serous | 1 | BeFree |
| umls:C0242216 | Biliary calculi | 1 | LHGDN |
| umls:C0242488 | Acute Lung Injury | 1 | CTD_human |
| umls:C0264683 | Coronary artery atheroma | 1 | BeFree |
| umls:C0266258 | Congenital absence of liver | 1 | BeFree |
| umls:C0268381 | Primary amyloidosis | 1 | BeFree |
| umls:C0268392 | Localized amyloidosis | 1 | BeFree |
| umls:C0268407 | Senile cardiac amyloidosis | 1 | BeFree |
| umls:C0271650 | Impaired glucose tolerance | 1 | BeFree |
| umls:C0333463 | Senile Plaques | 1 | BeFree |
| umls:C0340288 | Stable angina | 1 | BeFree |
| umls:C0375023 | Respiratory syncytial virus (RSV) infection in conditions classified elsewhere and of unspecified site | 1 | BeFree |
| umls:C0376358 | Malignant neoplasm of prostate | 1 | BeFree |
| umls:C0376618 | Endotoxemia | 1 | RGD |
| umls:C0518010 | body mass | 1 | GAD |
| umls:C0524909 | Hepatitis B, Chronic | 1 | LHGDN |
| umls:C0553573 | Primary infertility | 1 | BeFree |
| umls:C0553692 | Brain hemorrhage | 1 | GAD |
| umls:C0600139 | Prostate carcinoma | 1 | BeFree |
| umls:C0677886 | Epithelial ovarian cancer | 1 | BeFree |
| umls:C0699790 | Colon Carcinoma | 1 | BeFree |
| umls:C0699791 | Stomach Carcinoma | 1 | BeFree |
| umls:C0740394 | Hyperuricemia | 1 | GAD,LHGDN |
| umls:C0751003 | Brain Aneurysm | 1 | GAD |
| umls:C0751955 | Brain Infarction | 1 | BeFree |
| umls:C0795956 | Chylomicron retention disease | 1 | BeFree |
| umls:C0853897 | Diabetic Cardiomyopathies | 1 | BeFree,CTD_human |
| umls:C0860207 | Drug-Induced Liver Injury | 1 | CTD_human |
| umls:C0948008 | Ischemic stroke | 1 | GAD |
| umls:C0948896 | Primary hypogonadism | 1 | BeFree |
| umls:C1269683 | Major Depressive Disorder | 1 | RGD |
| umls:C1271104 | Blood pressure finding | 1 | GAD |
| umls:C1335177 | Ovarian Serous Adenocarcinoma | 1 | BeFree |
| umls:C1519670 | Tumor Angiogenesis | 1 | BeFree |
| umls:C1520166 | Xenograft Model | 1 | BeFree |
| umls:C1527249 | Colorectal Cancer | 1 | GAD |
| umls:C1623038 | Cirrhosis | 1 | BeFree |
| umls:C1704417 | Hyperlipoproteinemia Type IIb | 1 | BeFree |
| umls:C1719315 | Hereditary cardiac amyloidosis | 1 | BeFree |
| umls:C1800706 | Idiopathic Pulmonary Fibrosis | 1 | BeFree |
| umls:C1848533 | Ataxia with vitamin E deficiency | 1 | CTD_human |
| umls:C1867403 | Pyloric Stenosis, Infantile Hypertrophic 1 | 1 | BeFree |
| umls:C2062908 | Gout acute | 1 | BeFree |
| umls:C2316810 | Chronic kidney disease stage 5 | 1 | BeFree |
| umls:C2711472 | Infection by Trypanosoma brucei brucei | 1 | BeFree |
| umls:C2919032 | Infection of amniotic sac and membranes, unspecified, unspecified trimester, not applicable or unspecified | 1 | GAD |
| umls:C3665444 | Neutrophilia (disorder) | 1 | BeFree |
| umls:C3714514 | Infection | 1 | LHGDN |
| umls:C0342898 | Apolipoprotein A-I deficiency | 0 | CLINVAR,ORPHANET |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))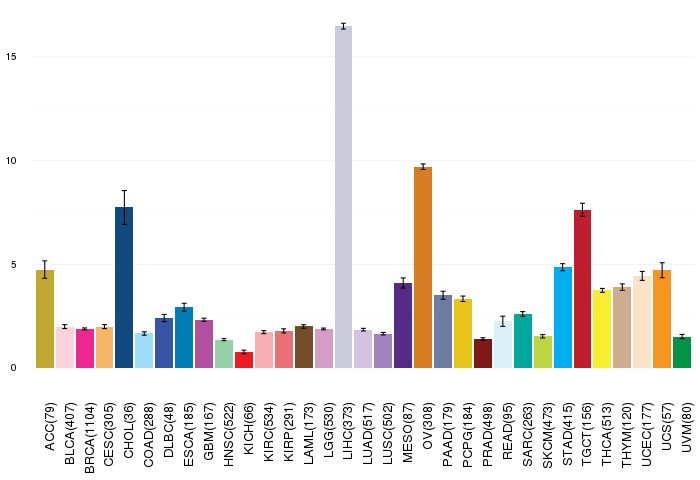
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))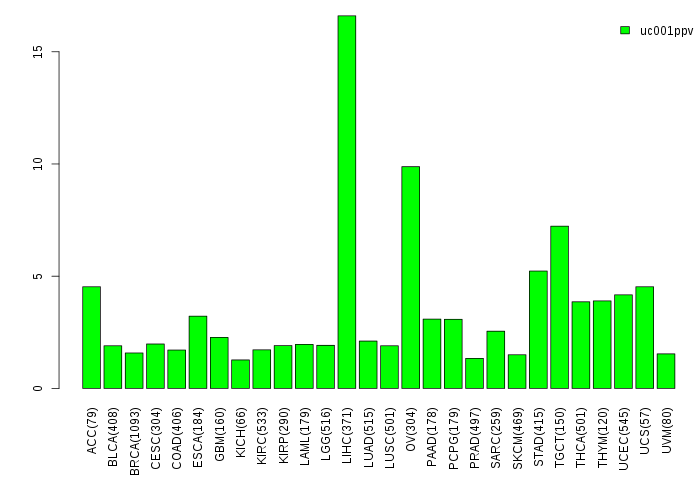
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data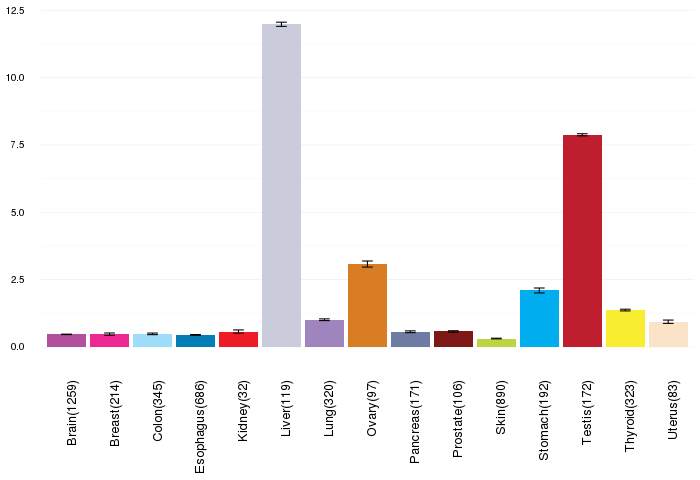
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))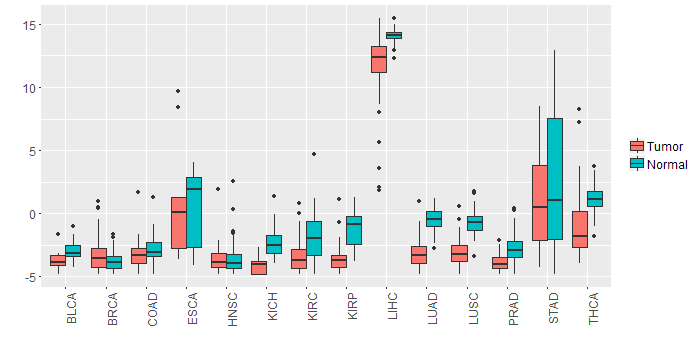
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.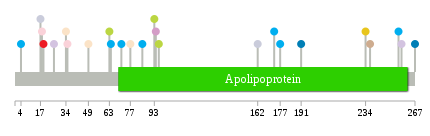

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 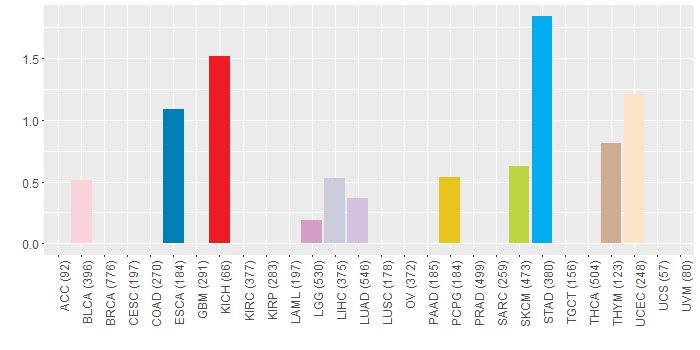
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)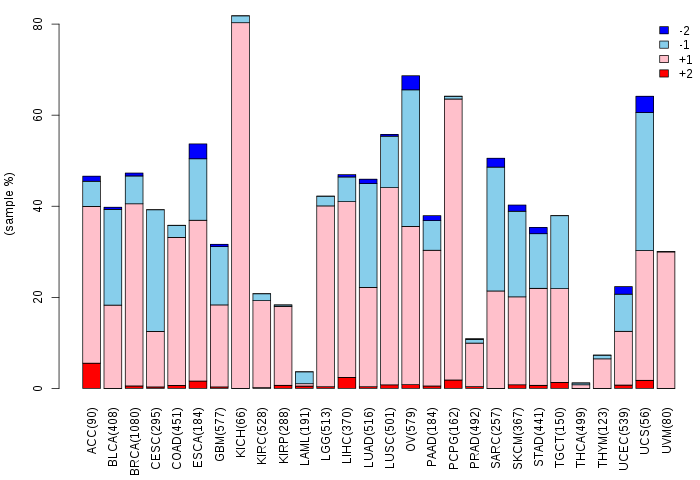
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 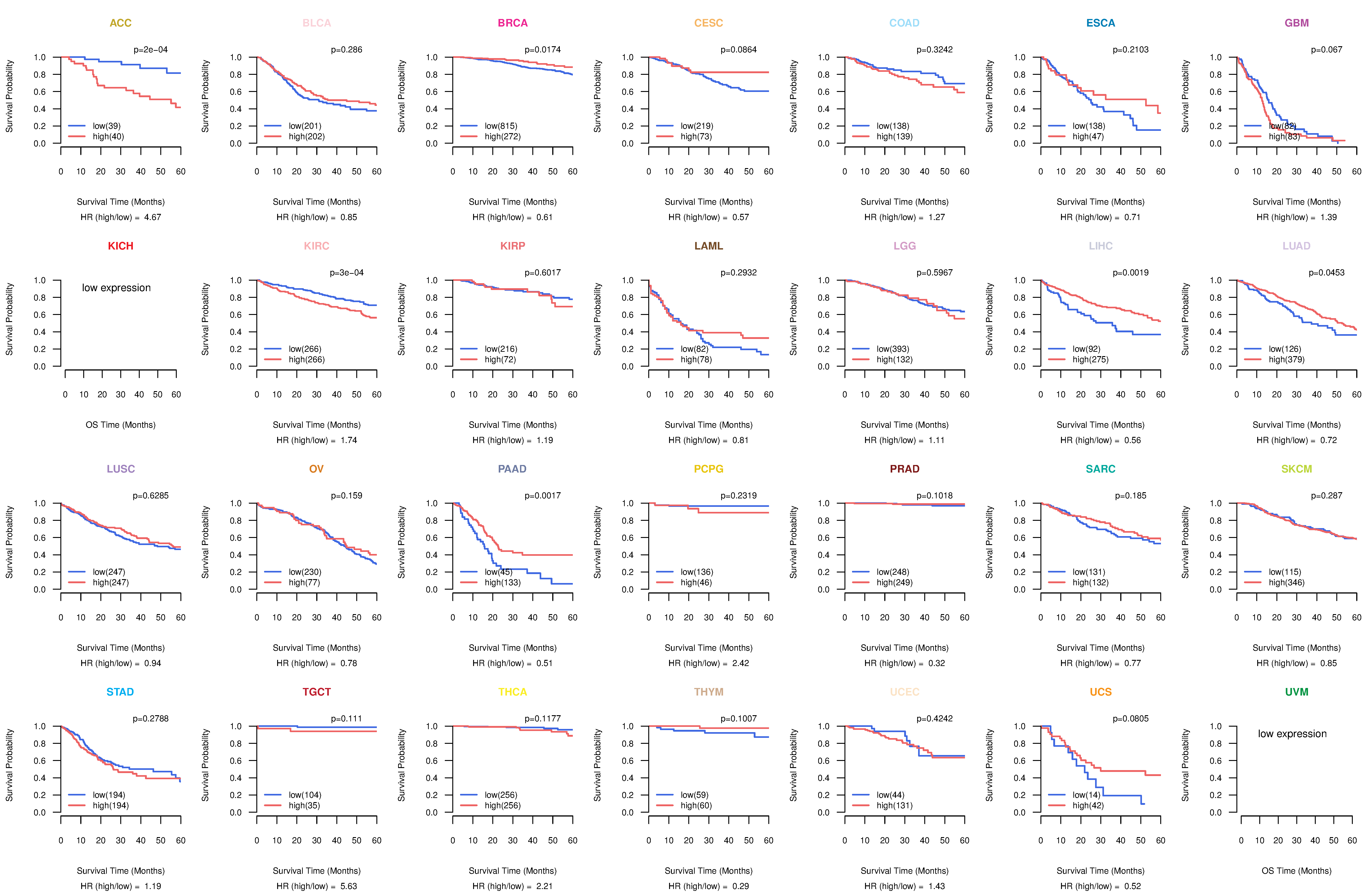
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 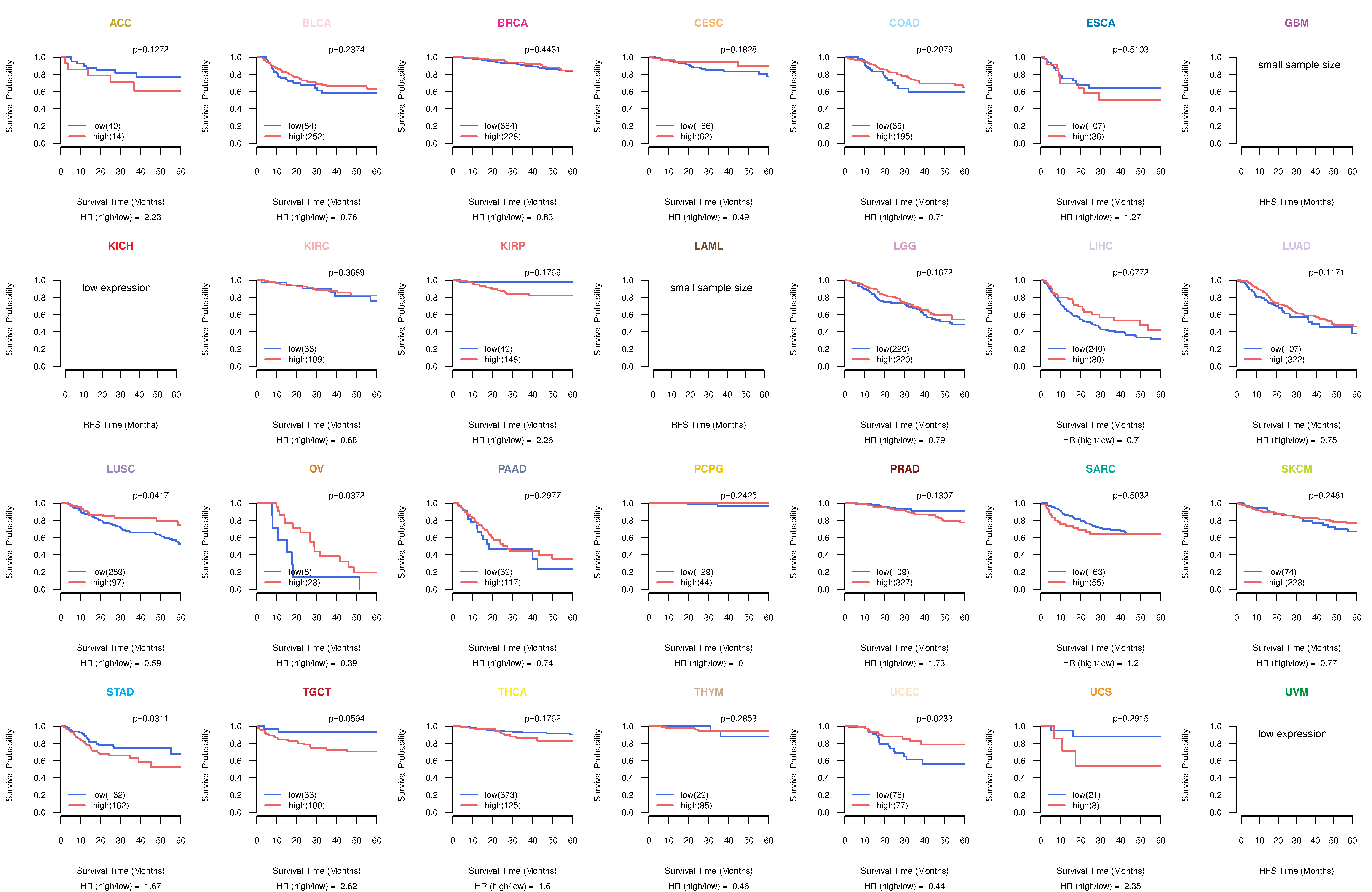
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 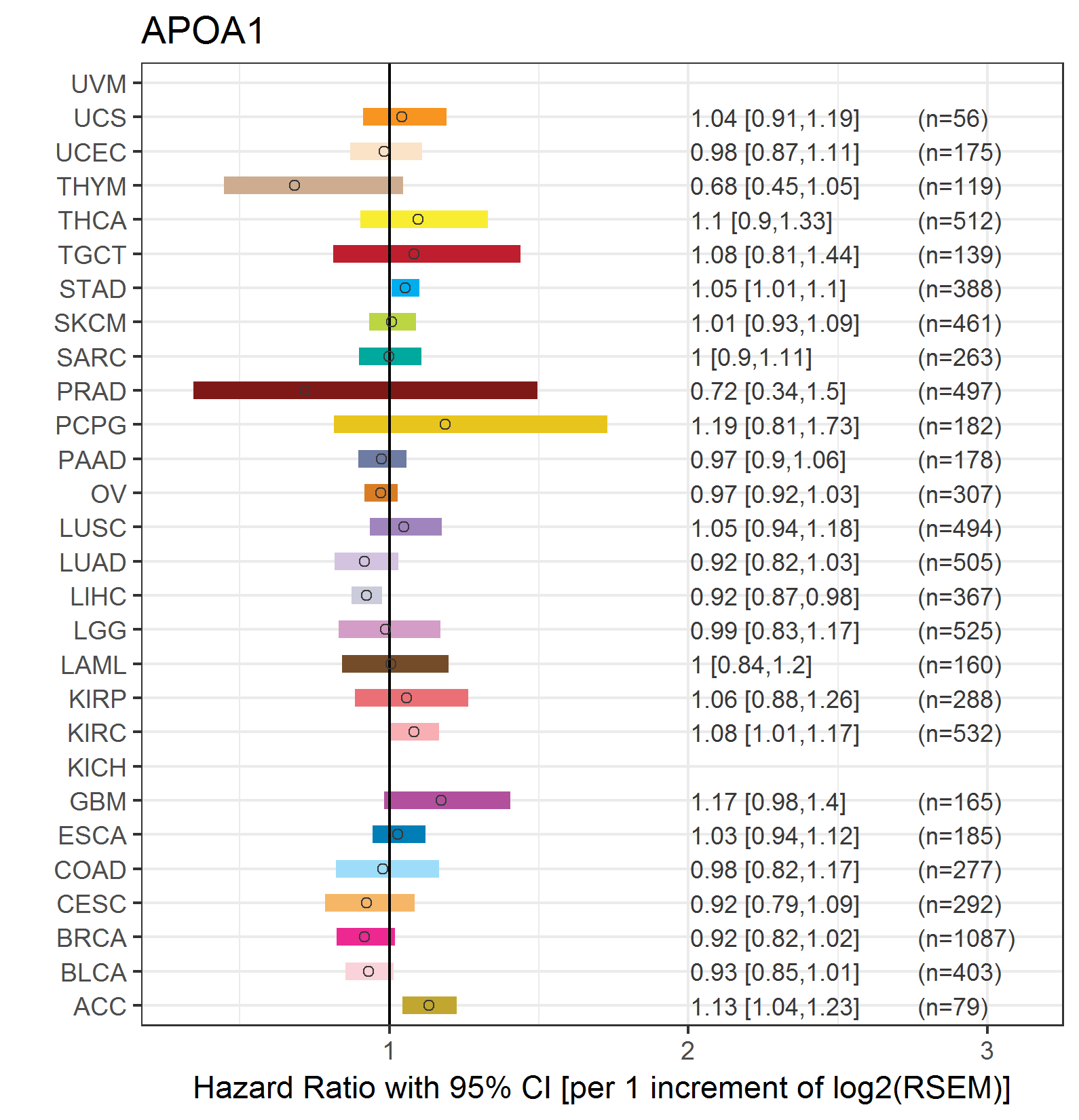
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 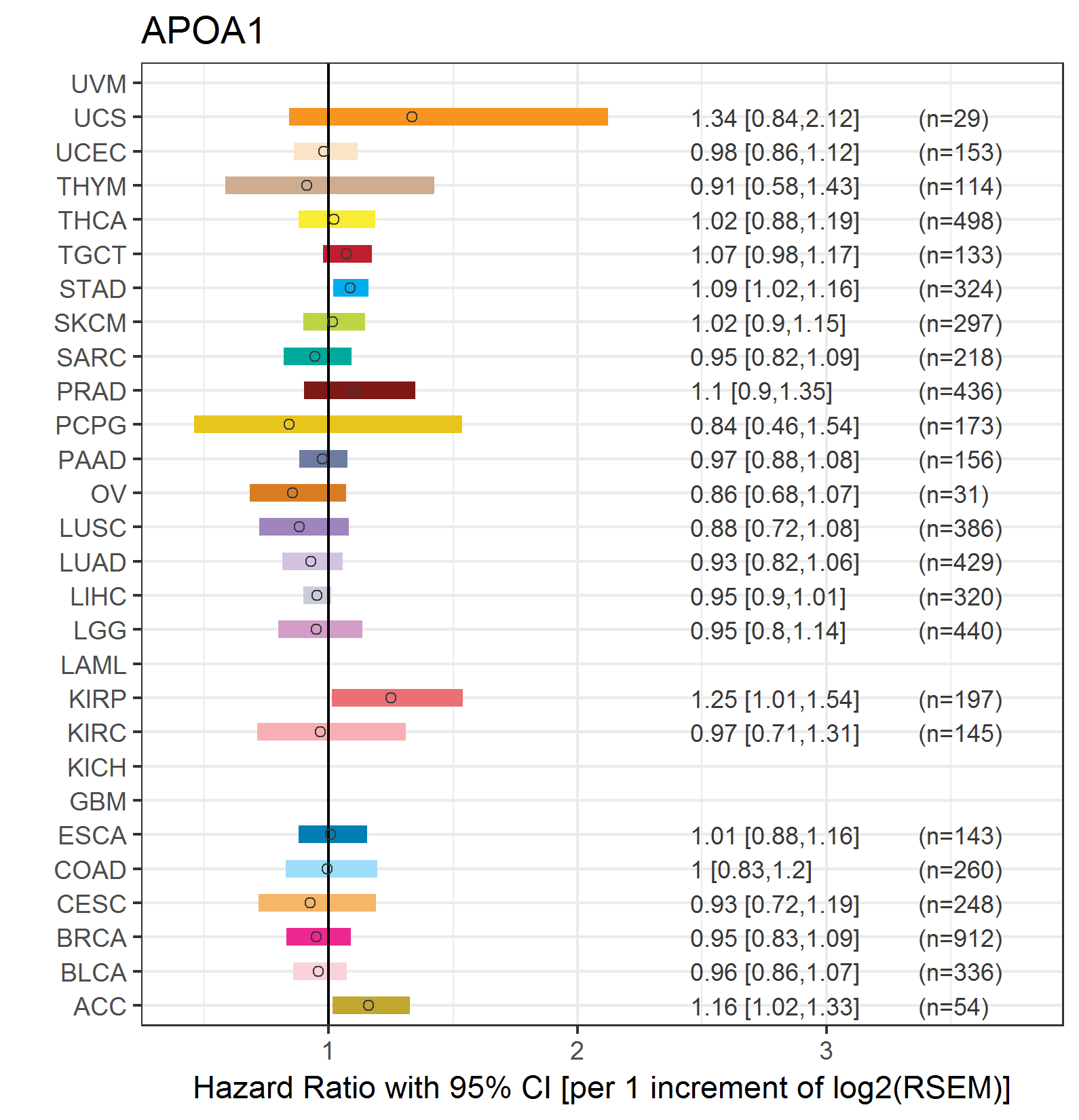
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























