| BRCA (tumor) | BRCA (normal) |
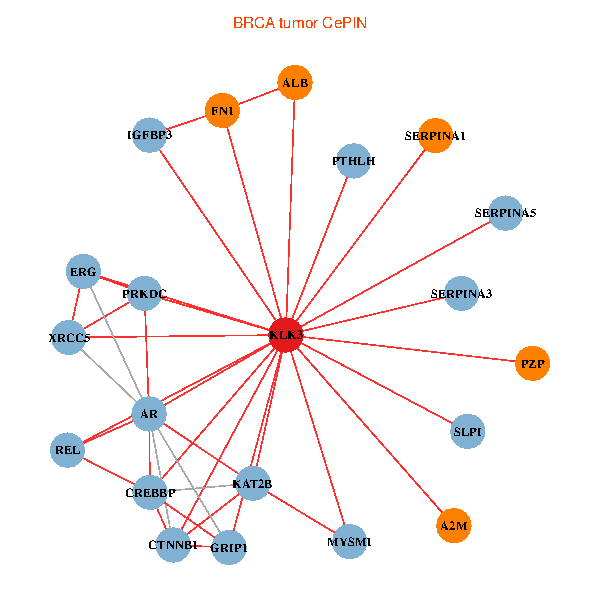
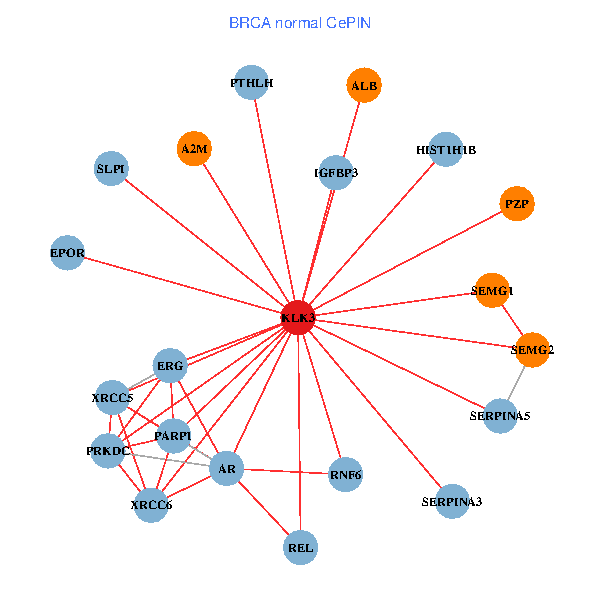
| KLK3, XRCC5, SERPINA1, FN1, CTNNB1, CREBBP, AR, KAT2B, REL, SERPINA3, ALB, A2M, PTHLH, SERPINA5, GRIP1, PRKDC, MYSM1, IGFBP3, ERG, SLPI, PZP (tumor) | KLK3, XRCC5, AR, REL, SERPINA3, ALB, HIST1H1B, A2M, PTHLH, PARP1, SERPINA5, PRKDC, IGFBP3, XRCC6, SEMG1, ERG, EPOR, RNF6, SLPI, PZP, SEMG2 (normal) |
 |  |
|
| COAD (tumor) | COAD (normal) |
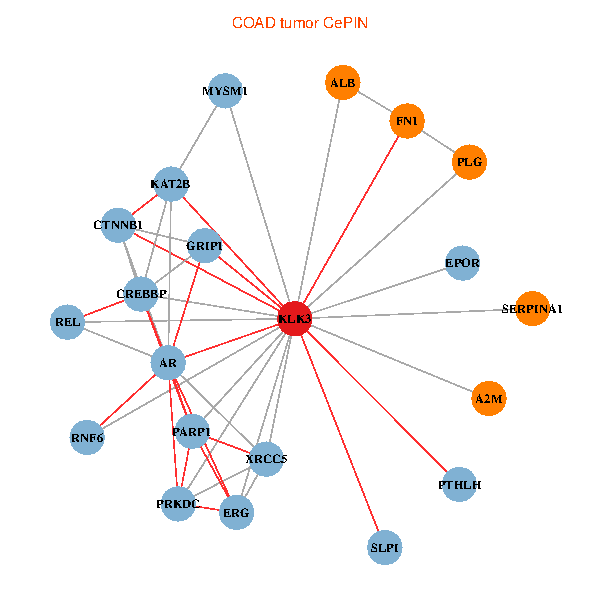
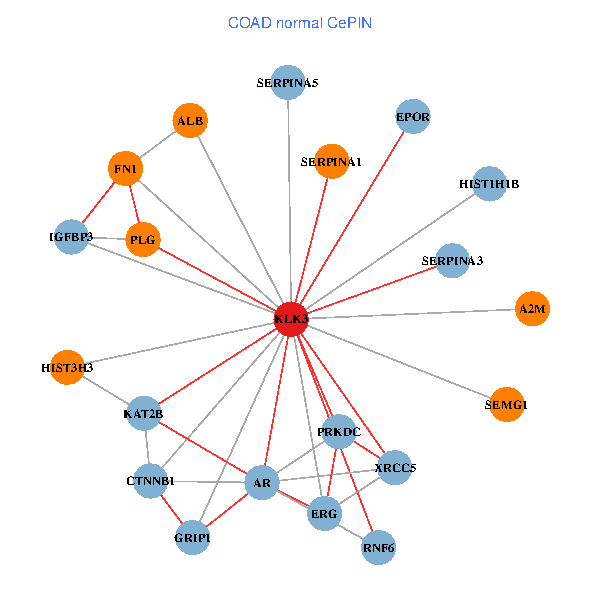
| KLK3, XRCC5, SERPINA1, FN1, CTNNB1, CREBBP, AR, KAT2B, REL, PLG, ALB, A2M, PTHLH, PARP1, GRIP1, PRKDC, MYSM1, ERG, EPOR, RNF6, SLPI (tumor) | KLK3, XRCC5, HIST3H3, SERPINA1, FN1, CTNNB1, AR, KAT2B, SERPINA3, PLG, ALB, HIST1H1B, A2M, SERPINA5, GRIP1, PRKDC, IGFBP3, SEMG1, ERG, EPOR, RNF6 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| KLK3, HIST3H3, SERPINA1, FN1, CTNNB1, CREBBP, AR, KAT2B, SERPINA3, PLG, HIST1H1B, A2M, PTHLH, PARP1, SERPINA5, PRKDC, IGFBP3, XRCC6, EPOR, RNF6, PZP (tumor) | KLK3, XRCC5, HIST3H3, FN1, CREBBP, AR, REL, SERPINA3, PLG, ALB, A2M, PARP1, SERPINA5, GRIP1, PRKDC, IGFBP3, SEMG1, ERG, EPOR, SLPI, SEMG2 (normal) |
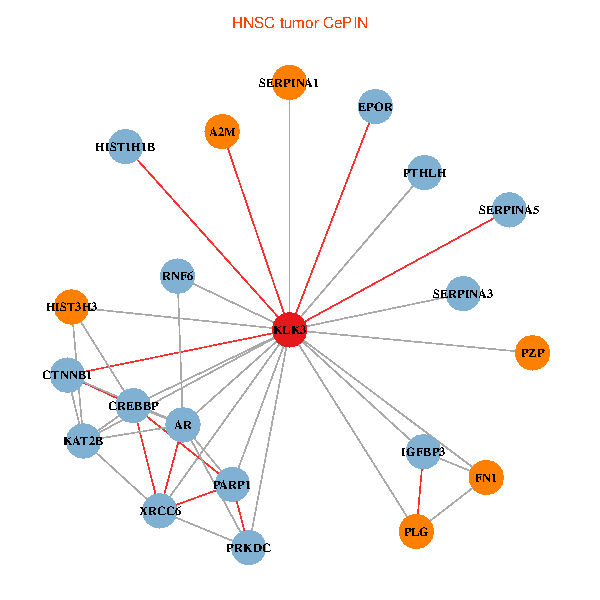 | 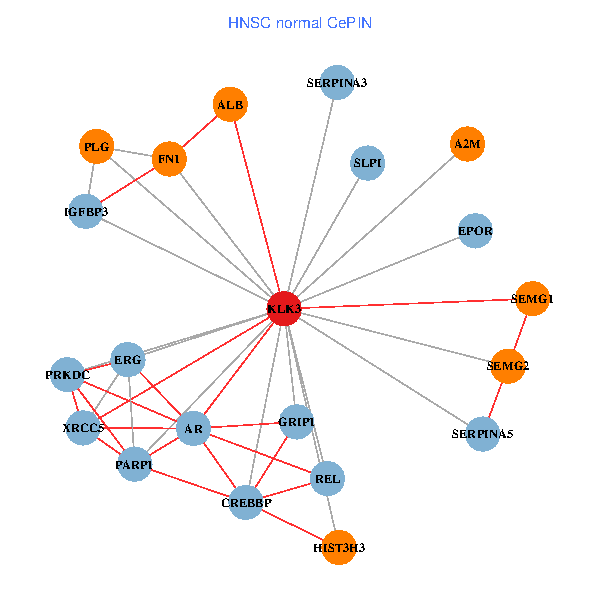 |
|
| KICH (tumor) | KICH (normal) |
| KLK3, XRCC5, HIST3H3, SERPINA1, FN1, AR, KAT2B, REL, PLG, ALB, HIST1H1B, PTHLH, GRIP1, PRKDC, MYSM1, IGFBP3, SEMG1, EPOR, RNF6, SLPI, PZP (tumor) | KLK3, XRCC5, SERPINA1, FN1, AR, KAT2B, REL, SERPINA3, HIST1H1B, A2M, PARP1, GRIP1, PRKDC, MYSM1, IGFBP3, XRCC6, ERG, EPOR, RNF6, SLPI, SEMG2 (normal) |
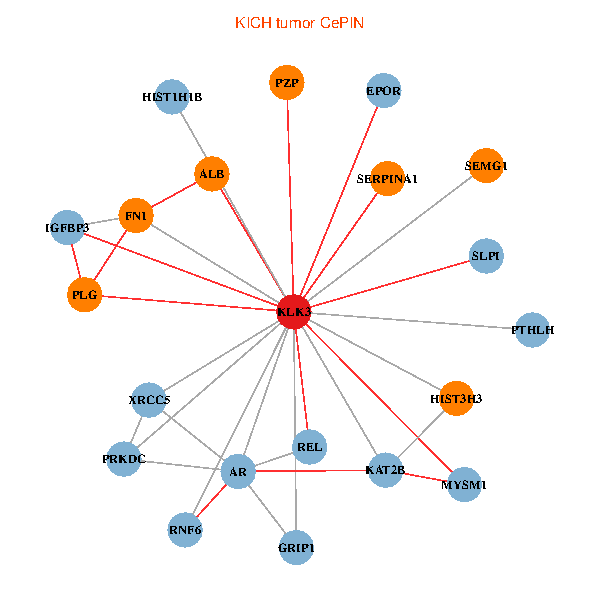 | 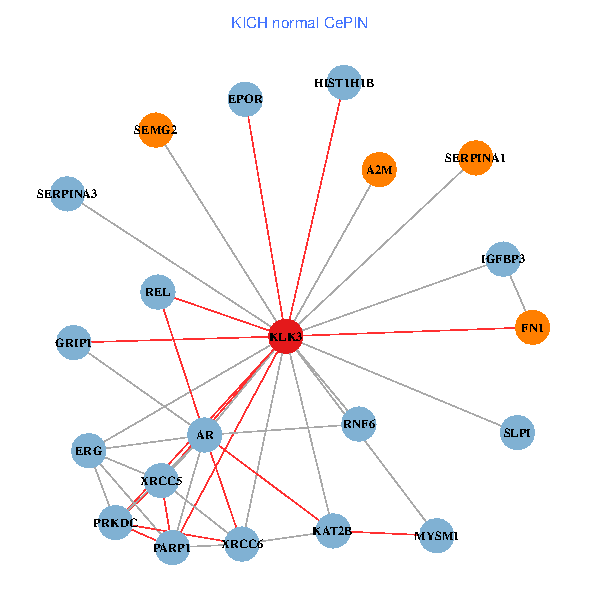 |
|
| KIRC (tumor) | KIRC (normal) |
| KLK3, XRCC5, SERPINA1, FN1, CREBBP, AR, KAT2B, PLG, ALB, A2M, PARP1, SERPINA5, GRIP1, PRKDC, IGFBP3, XRCC6, SEMG1, EPOR, RNF6, SLPI, SEMG2 (tumor) | KLK3, XRCC5, SERPINA1, FN1, CTNNB1, AR, REL, PLG, A2M, PTHLH, PARP1, SERPINA5, GRIP1, MYSM1, IGFBP3, XRCC6, SEMG1, ERG, EPOR, PZP, SEMG2 (normal) |
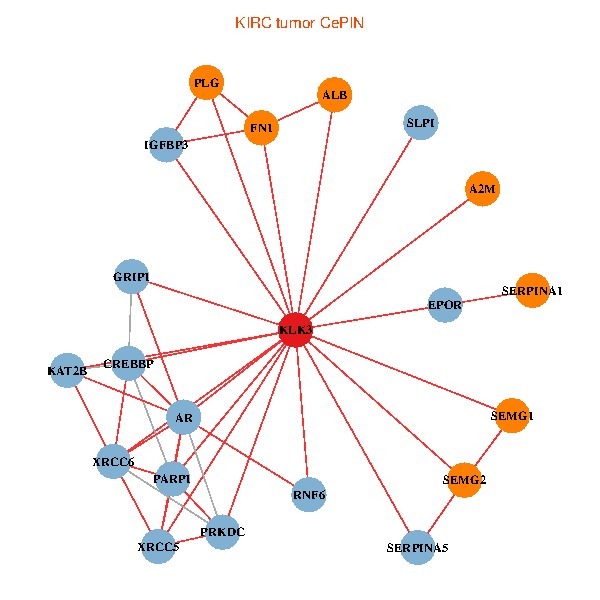 | 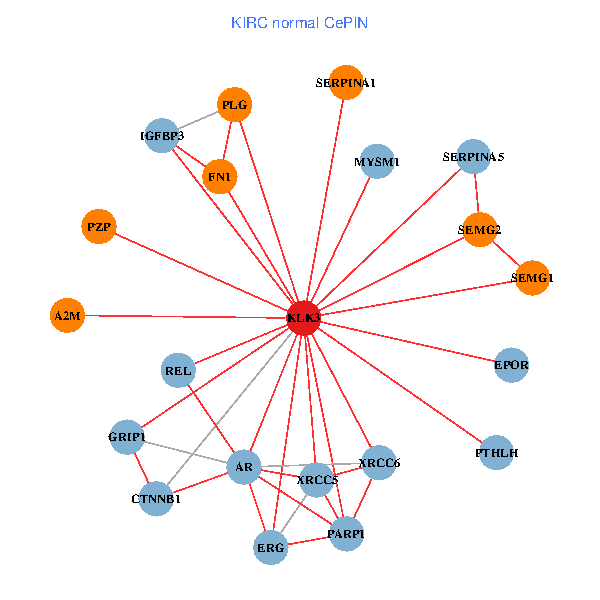 |
|
| KIRP (tumor) | KIRP (normal) |
| KLK3, SERPINA1, CTNNB1, CREBBP, AR, REL, PLG, ALB, HIST1H1B, A2M, PARP1, GRIP1, PRKDC, IGFBP3, XRCC6, ERG, EPOR, RNF6, SLPI, PZP, SEMG2 (tumor) | KLK3, XRCC5, HIST3H3, SERPINA1, CTNNB1, AR, REL, SERPINA3, PLG, ALB, HIST1H1B, A2M, PARP1, SERPINA5, GRIP1, IGFBP3, XRCC6, SEMG1, EPOR, SLPI, SEMG2 (normal) |
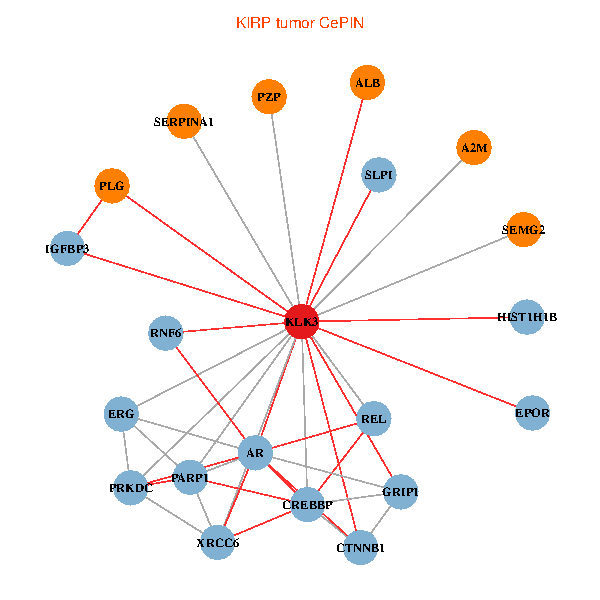 | 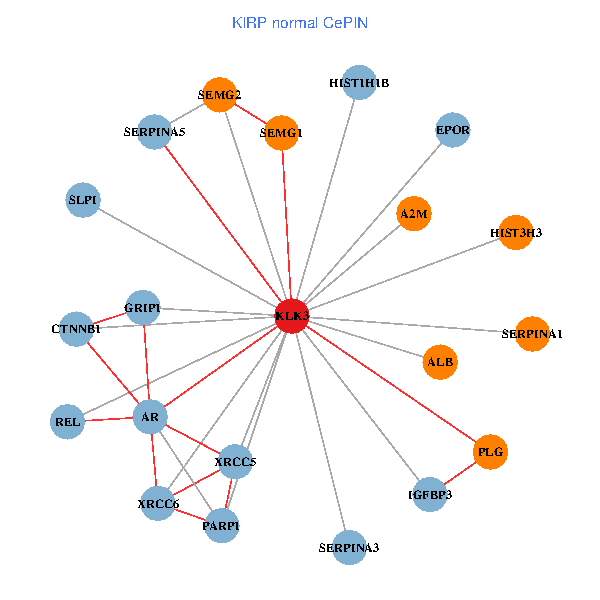 |
|
| LIHC (tumor) | LIHC (normal) |
| KLK3, XRCC5, SERPINA1, CTNNB1, CREBBP, AR, SERPINA3, ALB, A2M, PTHLH, PARP1, SERPINA5, GRIP1, PRKDC, MYSM1, IGFBP3, XRCC6, SEMG1, EPOR, SLPI, PZP (tumor) | KLK3, XRCC5, SERPINA1, FN1, CTNNB1, CREBBP, KAT2B, REL, SERPINA3, ALB, HIST1H1B, A2M, SERPINA5, GRIP1, PRKDC, IGFBP3, SEMG1, ERG, RNF6, PZP, SEMG2 (normal) |
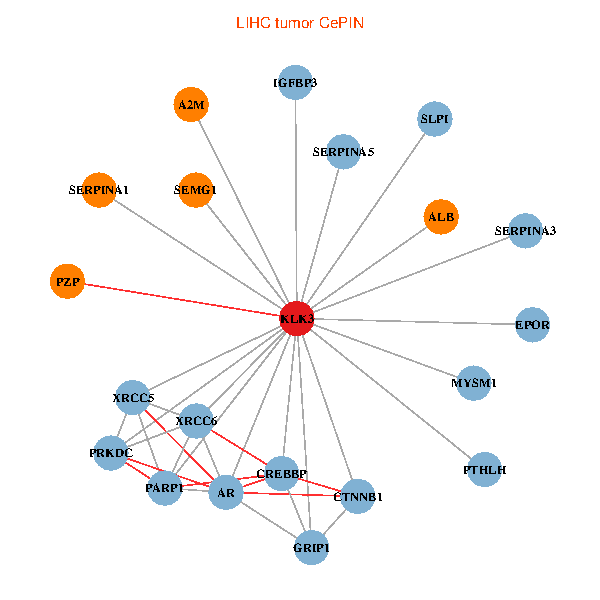 | 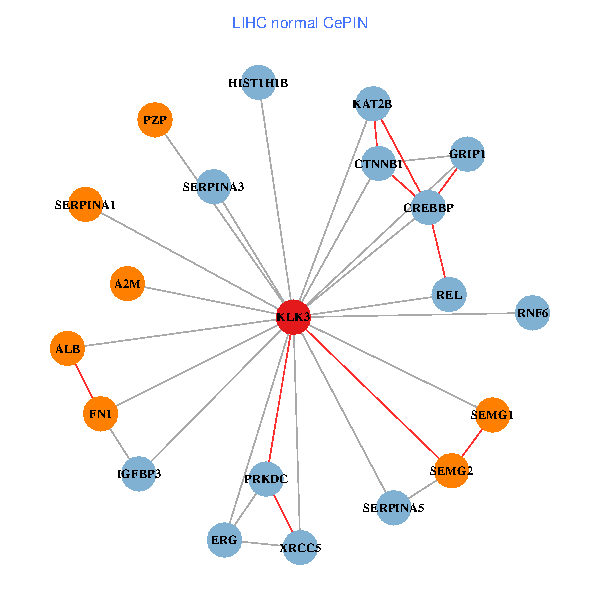 |
|
| LUAD (tumor) | LUAD (normal) |
| KLK3, XRCC5, SERPINA1, CTNNB1, CREBBP, AR, KAT2B, SERPINA3, ALB, HIST1H1B, PTHLH, PARP1, SERPINA5, GRIP1, PRKDC, MYSM1, IGFBP3, XRCC6, EPOR, RNF6, PZP (tumor) | KLK3, XRCC5, HIST3H3, SERPINA1, FN1, CTNNB1, KAT2B, ALB, PTHLH, PARP1, SERPINA5, GRIP1, MYSM1, IGFBP3, XRCC6, SEMG1, EPOR, RNF6, SLPI, PZP, SEMG2 (normal) |
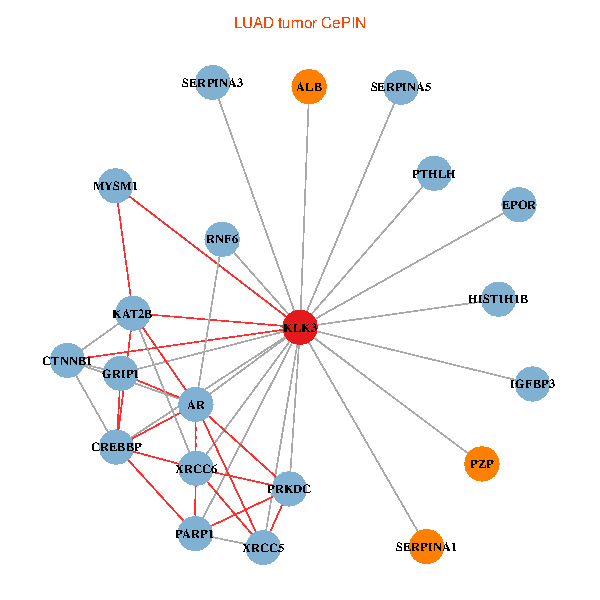 | 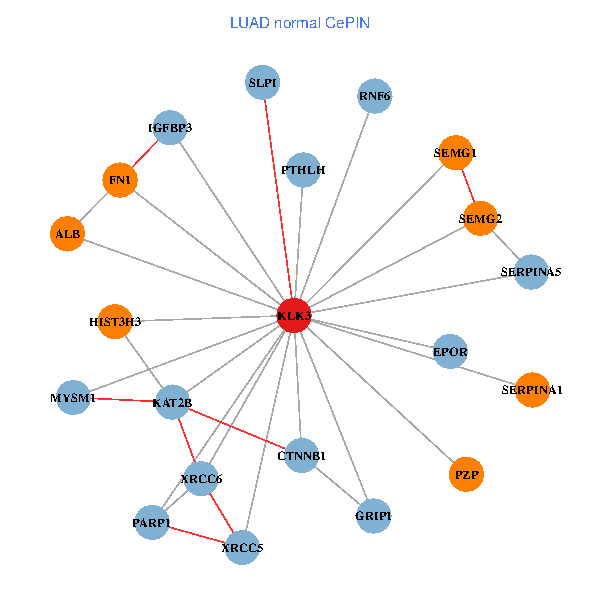 |
|
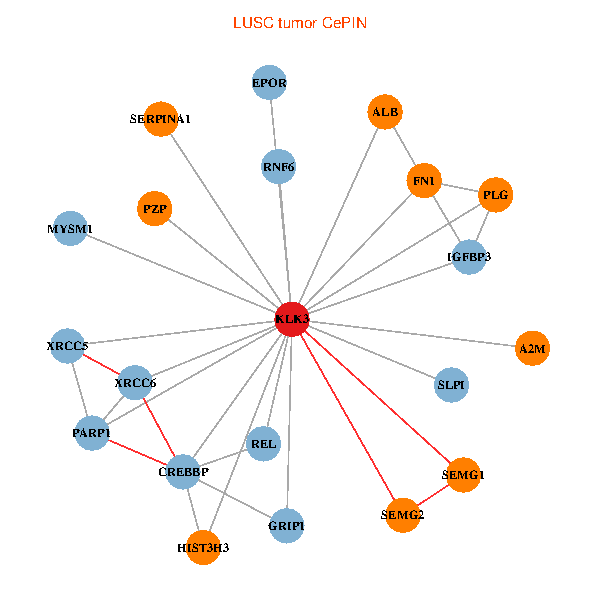
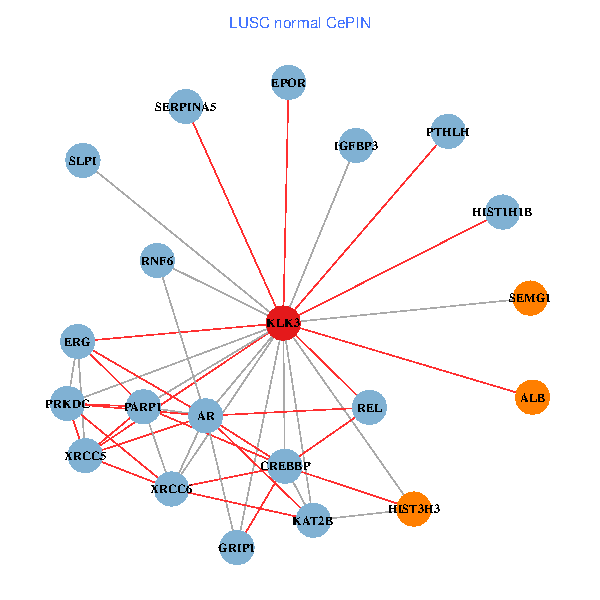
| LUSC (tumor) | LUSC (normal) |
| KLK3, XRCC5, HIST3H3, SERPINA1, FN1, CREBBP, REL, PLG, ALB, A2M, PARP1, GRIP1, MYSM1, IGFBP3, XRCC6, SEMG1, EPOR, RNF6, SLPI, PZP, SEMG2 (tumor) | KLK3, XRCC5, HIST3H3, CREBBP, AR, KAT2B, REL, ALB, HIST1H1B, PTHLH, PARP1, SERPINA5, GRIP1, PRKDC, IGFBP3, XRCC6, SEMG1, ERG, EPOR, RNF6, SLPI (normal) |
 |  |
|
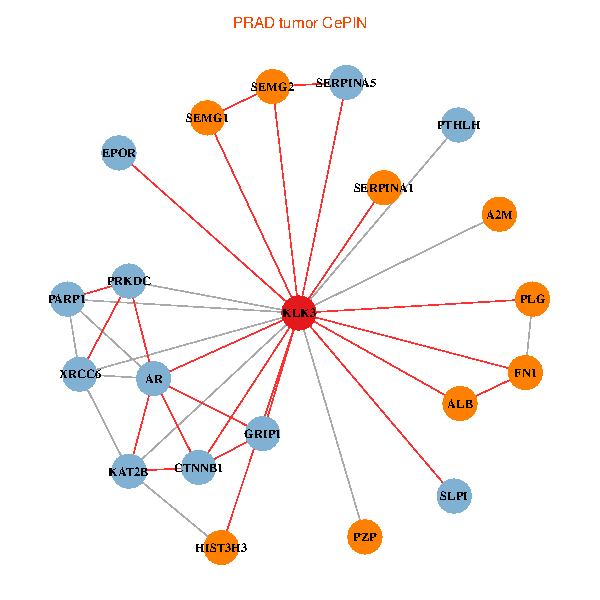
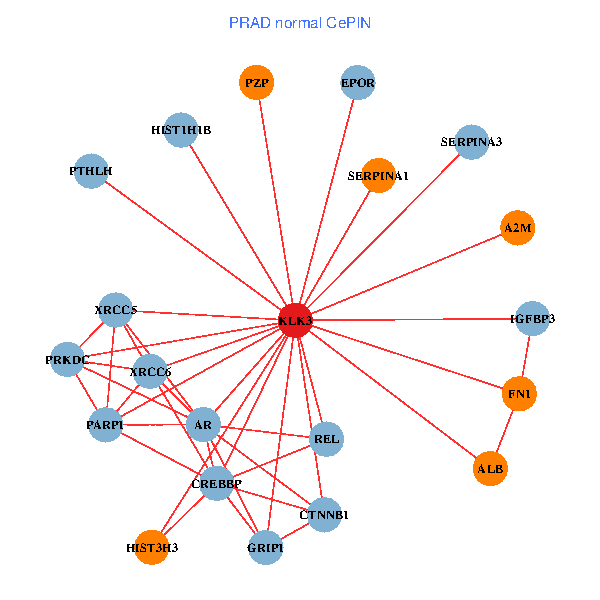
| PRAD (tumor) | PRAD (normal) |
| KLK3, HIST3H3, SERPINA1, FN1, CTNNB1, AR, KAT2B, PLG, ALB, A2M, PTHLH, PARP1, SERPINA5, GRIP1, PRKDC, XRCC6, SEMG1, EPOR, SLPI, PZP, SEMG2 (tumor) | KLK3, XRCC5, HIST3H3, SERPINA1, FN1, CTNNB1, CREBBP, AR, REL, SERPINA3, ALB, HIST1H1B, A2M, PTHLH, PARP1, GRIP1, PRKDC, IGFBP3, XRCC6, EPOR, PZP (normal) |
 |  |
|
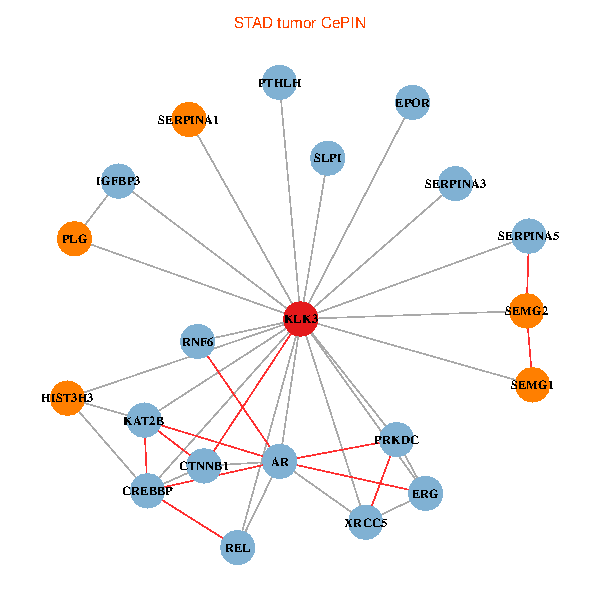
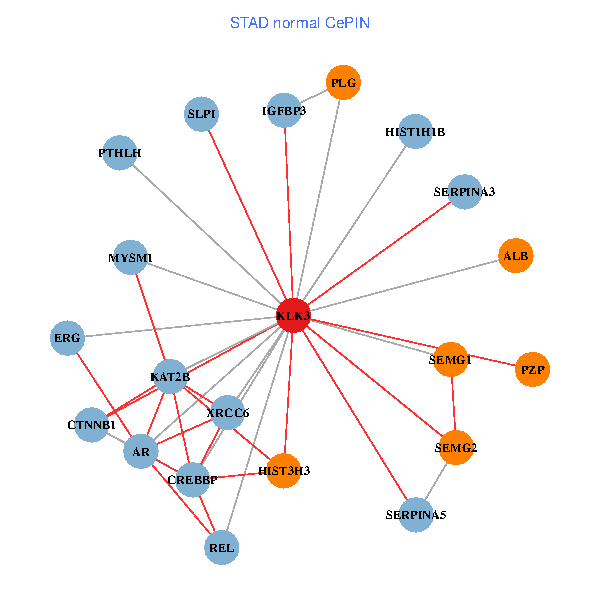
| STAD (tumor) | STAD (normal) |
| KLK3, XRCC5, HIST3H3, SERPINA1, CTNNB1, CREBBP, AR, KAT2B, REL, SERPINA3, PLG, PTHLH, SERPINA5, PRKDC, IGFBP3, SEMG1, ERG, EPOR, RNF6, SLPI, SEMG2 (tumor) | KLK3, HIST3H3, CTNNB1, CREBBP, AR, KAT2B, REL, SERPINA3, PLG, ALB, HIST1H1B, PTHLH, SERPINA5, MYSM1, IGFBP3, XRCC6, SEMG1, ERG, SLPI, PZP, SEMG2 (normal) |
 |  |
|
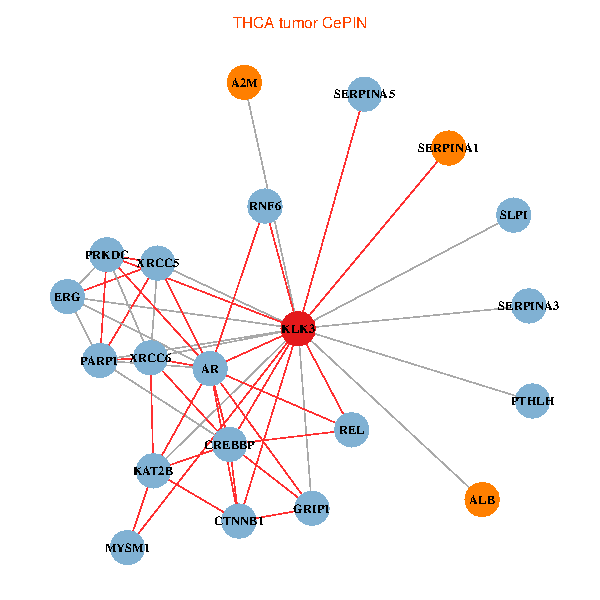
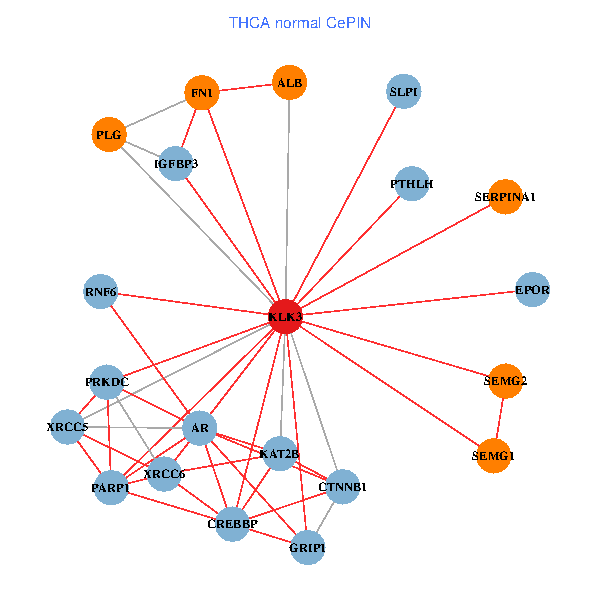
| THCA (tumor) | THCA (normal) |
| KLK3, XRCC5, SERPINA1, CTNNB1, CREBBP, AR, KAT2B, REL, SERPINA3, ALB, A2M, PTHLH, PARP1, SERPINA5, GRIP1, PRKDC, MYSM1, XRCC6, ERG, RNF6, SLPI (tumor) | KLK3, XRCC5, SERPINA1, FN1, CTNNB1, CREBBP, AR, KAT2B, PLG, ALB, PTHLH, PARP1, GRIP1, PRKDC, IGFBP3, XRCC6, SEMG1, EPOR, RNF6, SLPI, SEMG2 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0376358 | Malignant neoplasm of prostate | 781 | BeFree,GAD,GWASCAT |
| umls:C0600139 | Prostate carcinoma | 776 | BeFree |
| umls:C0033578 | Prostatic Neoplasms | 75 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0027627 | Neoplasm Metastasis | 71 | BeFree,GAD |
| umls:C1704272 | Benign Prostatic Hyperplasia | 53 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 31 | BeFree,GAD |
| umls:C0678222 | Breast Carcinoma | 31 | BeFree |
| umls:C0007112 | Adenocarcinoma of prostate | 19 | BeFree |
| umls:C1458155 | Mammary Neoplasms | 17 | BeFree,LHGDN |
| umls:C0178874 | Tumor Progression | 14 | BeFree |
| umls:C0001418 | Adenocarcinoma | 11 | BeFree |
| umls:C0033575 | Prostatic Diseases | 11 | BeFree,LHGDN |
| umls:C0686619 | Secondary malignant neoplasm of lymph node | 10 | BeFree |
| umls:C1328504 | Hormone refractory prostate cancer | 10 | BeFree |
| umls:C0153690 | Secondary malignant neoplasm of bone | 9 | BeFree |
| umls:C0936223 | Metastatic Prostate Carcinoma | 9 | BeFree |
| umls:C1654637 | androgen independent prostate cancer | 9 | BeFree |
| umls:C1519176 | Salivary Gland Pleomorphic Adenoma | 8 | BeFree |
| umls:C0034341 | Pyruvate Carboxylase Deficiency Disease | 7 | BeFree |
| umls:C0278838 | Prostate cancer recurrent | 7 | BeFree |
| umls:C0029925 | Ovarian Carcinoma | 6 | BeFree |
| umls:C2931456 | Prostate cancer, familial | 6 | BeFree |
| umls:C0521158 | Recurrent tumor | 5 | BeFree |
| umls:C0596263 | Carcinogenesis | 5 | BeFree |
| umls:C1140680 | Malignant neoplasm of ovary | 5 | BeFree |
| umls:C2937421 | Prostatic Hyperplasia | 5 | CTD_human,GAD,LHGDN |
| umls:C0019348 | Herpes Simplex Infections | 4 | BeFree |
| umls:C0024141 | Lupus Erythematosus, Systemic | 4 | BeFree |
| umls:C0280280 | stage, prostate cancer | 4 | BeFree |
| umls:C0574785 | Lower Urinary Tract Symptoms | 4 | BeFree |
| umls:C0003469 | Anxiety Disorders | 3 | BeFree |
| umls:C0009402 | Colorectal Carcinoma | 3 | BeFree |
| umls:C0038587 | Substance Withdrawal Syndrome | 3 | BeFree |
| umls:C0085110 | Severe Combined Immunodeficiency | 3 | BeFree |
| umls:C0085278 | Antiphospholipid Syndrome | 3 | BeFree |
| umls:C1527249 | Colorectal Cancer | 3 | BeFree |
| umls:C1739363 | Prostatic Hypertrophy | 3 | BeFree |
| umls:C0004364 | Autoimmune Diseases | 2 | BeFree |
| umls:C0007104 | Female Breast Carcinoma | 2 | BeFree |
| umls:C0007134 | Renal Cell Carcinoma | 2 | BeFree |
| umls:C0016629 | Fowlpox | 2 | BeFree |
| umls:C0022661 | Kidney Failure, Chronic | 2 | BeFree |
| umls:C0022735 | Klinefelter Syndrome | 2 | BeFree |
| umls:C0024115 | Lung diseases | 2 | BeFree |
| umls:C0024121 | Lung Neoplasms | 2 | BeFree |
| umls:C0024143 | Lupus Nephritis | 2 | BeFree |
| umls:C0032002 | Pituitary Diseases | 2 | BeFree |
| umls:C0032460 | Polycystic Ovary Syndrome | 2 | LHGDN |
| umls:C0033581 | prostatitis | 2 | BeFree |
| umls:C0035078 | Kidney Failure | 2 | BeFree |
| umls:C0042029 | Urinary tract infection | 2 | BeFree |
| umls:C0085860 | Autoimmune Syndrome Type II, Polyglandular | 2 | BeFree |
| umls:C0151514 | Atrophic condition of skin | 2 | BeFree |
| umls:C0235653 | Malignant neoplasm of female breast | 2 | BeFree |
| umls:C0238339 | Hereditary pancreatitis | 2 | BeFree |
| umls:C0272138 | Erythroblastosis | 2 | BeFree |
| umls:C0282612 | Prostatic Intraepithelial Neoplasias | 2 | BeFree |
| umls:C0398623 | Thrombophilia | 2 | BeFree |
| umls:C0432474 | Klinefelter's syndrome - male with more than two X chromosomes | 2 | BeFree |
| umls:C0677932 | Progressive Neoplastic Disease | 2 | BeFree |
| umls:C1168327 | High-Grade Prostatic Intraepithelial Neoplasia | 2 | BeFree |
| umls:C1516170 | Cancer Cell Growth | 2 | BeFree |
| umls:C1519680 | Tumor Immunity | 2 | BeFree |
| umls:C1520166 | Xenograft Model | 2 | BeFree |
| umls:C1558141 | Urinary Retention Adverse Event | 2 | BeFree |
| umls:C3539781 | Progressive cGVHD | 2 | BeFree |
| umls:C0002395 | Alzheimer's Disease | 1 | BeFree |
| umls:C0002793 | Anaplasia | 1 | BeFree |
| umls:C0004277 | Tooth Attrition | 1 | BeFree |
| umls:C0005779 | Blood Coagulation Disorders | 1 | BeFree |
| umls:C0006118 | Brain Neoplasms | 1 | BeFree |
| umls:C0007138 | Carcinoma, Transitional Cell | 1 | BeFree |
| umls:C0008354 | Cholera | 1 | BeFree |
| umls:C0009319 | Colitis | 1 | BeFree |
| umls:C0010046 | Corn of toe | 1 | BeFree |
| umls:C0012546 | Diphtheria | 1 | BeFree |
| umls:C0014836 | Escherichia coli Infections | 1 | BeFree |
| umls:C0015230 | Exanthema | 1 | BeFree |
| umls:C0018204 | Granulomatous prostatitis | 1 | BeFree |
| umls:C0020507 | Hyperplasia | 1 | LHGDN |
| umls:C0021051 | Immunologic Deficiency Syndromes | 1 | BeFree |
| umls:C0022650 | Kidney Calculi | 1 | BeFree |
| umls:C0024232 | Lymphatic Metastasis | 1 | GAD |
| umls:C0024305 | Lymphoma, Non-Hodgkin | 1 | LHGDN |
| umls:C0024623 | Malignant neoplasm of stomach | 1 | BeFree |
| umls:C0024897 | Mastocytoma | 1 | BeFree |
| umls:C0025202 | melanoma | 1 | BeFree |
| umls:C0027643 | Neoplasm Recurrence, Local | 1 | CTD_human |
| umls:C0027708 | Nephroblastoma | 1 | BeFree |
| umls:C0027819 | Neuroblastoma | 1 | BeFree |
| umls:C0030201 | Pain, Postoperative | 1 | BeFree |
| umls:C0032580 | Adenomatous Polyposis Coli | 1 | BeFree |
| umls:C0033579 | Prostate nodule | 1 | BeFree |
| umls:C0033860 | Psoriasis | 1 | BeFree |
| umls:C0035222 | Respiratory Distress Syndrome, Adult | 1 | BeFree |
| umls:C0036875 | Disorders of Sex Development | 1 | BeFree |
| umls:C0037286 | Skin Neoplasms | 1 | BeFree |
| umls:C0085215 | Ovarian Failure, Premature | 1 | BeFree |
| umls:C0152013 | Adenocarcinoma of lung (disorder) | 1 | BeFree |
| umls:C0153633 | Malignant neoplasm of brain | 1 | BeFree |
| umls:C0162311 | Androgenetic Alopecia | 1 | BeFree |
| umls:C0221406 | Pituitary-dependent Cushing's disease | 1 | LHGDN |
| umls:C0242594 | Residual Cancer | 1 | BeFree |
| umls:C0242656 | Disease Progression | 1 | CTD_human |
| umls:C0242966 | Systemic Inflammatory Response Syndrome | 1 | BeFree |
| umls:C0262584 | Carcinoma, Small Cell | 1 | BeFree |
| umls:C0263361 | Psoriasis vulgaris | 1 | BeFree |
| umls:C0263978 | Disorder of soft tissue | 1 | BeFree |
| umls:C0278996 | Cancer of Head and Neck | 1 | BeFree |
| umls:C0279672 | Cervical Adenocarcinoma | 1 | BeFree |
| umls:C0334664 | Mast Cell Neoplasm | 1 | BeFree |
| umls:C0346255 | Oncocytoma, renal | 1 | BeFree |
| umls:C0549379 | Recurrent Carcinoma | 1 | BeFree |
| umls:C0600502 | Vascular Hemostatic Disorders | 1 | BeFree |
| umls:C0677886 | Epithelial ovarian cancer | 1 | BeFree |
| umls:C0677949 | Stage III Colorectal Cancer | 1 | BeFree |
| umls:C0677984 | Locally Advanced Malignant Neoplasm | 1 | BeFree |
| umls:C0699791 | Stomach Carcinoma | 1 | BeFree |
| umls:C0700095 | Central neuroblastoma | 1 | BeFree |
| umls:C0796563 | Localized Malignant Neoplasm | 1 | BeFree |
| umls:C0848332 | Spots on skin | 1 | BeFree |
| umls:C0877373 | Advanced cancer | 1 | BeFree |
| umls:C0919267 | ovarian neoplasm | 1 | BeFree |
| umls:C0920420 | cancer recurrence | 1 | BeFree |
| umls:C1261473 | Sarcoma | 1 | BeFree |
| umls:C1266042 | Chromophobe Renal Cell Carcinoma | 1 | BeFree |
| umls:C1334407 | Localized Carcinoma | 1 | BeFree |
| umls:C1334615 | Malignant Phyllodes Tumor of Prostate | 1 | CTD_human |
| umls:C1335729 | Refractory Neoplasm | 1 | BeFree |
| umls:C1658953 | tumor vasculature | 1 | BeFree |
| umls:C1853195 | Prostate Cancer, Hereditary, 7 | 1 | BeFree |
| umls:C1861355 | Syndactyly, Type IV | 1 | BeFree |
| umls:C2242987 | Benign Mastocytoma | 1 | BeFree |
| umls:C2316810 | Chronic kidney disease stage 5 | 1 | BeFree |
| umls:C2945759 | aggressive cancer | 1 | BeFree |
| umls:C3146250 | Stage III Colorectal Cancer AJCC v7 | 1 | BeFree |
| umls:C3146265 | Prostate cancer stage D | 1 | BeFree |
| umls:C3273239 | Proliferative Inflammatory Atrophy | 1 | BeFree |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))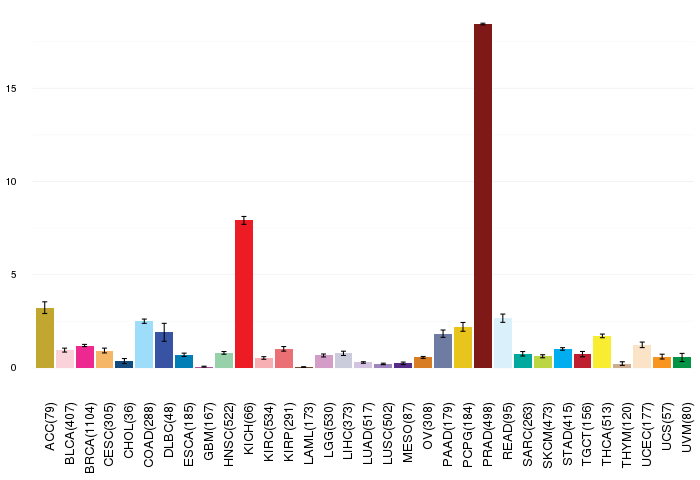
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))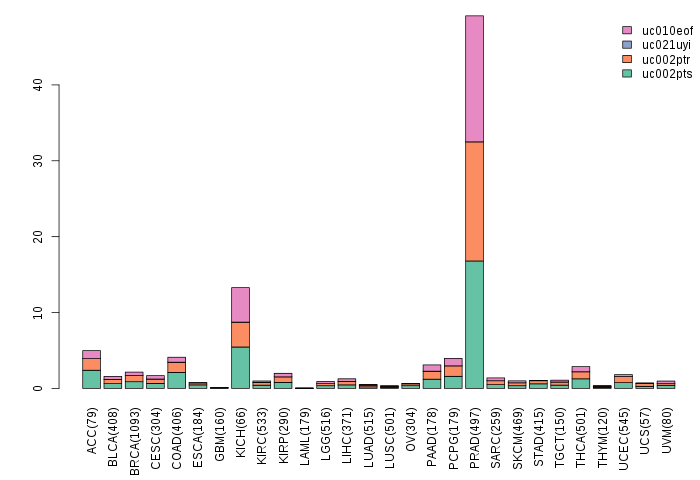
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data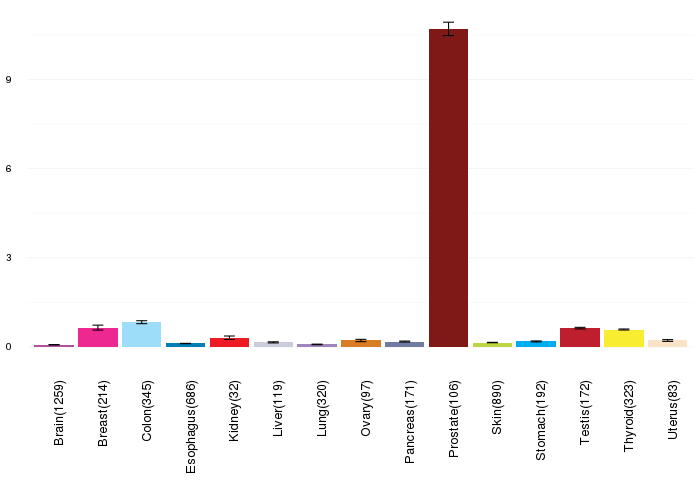
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))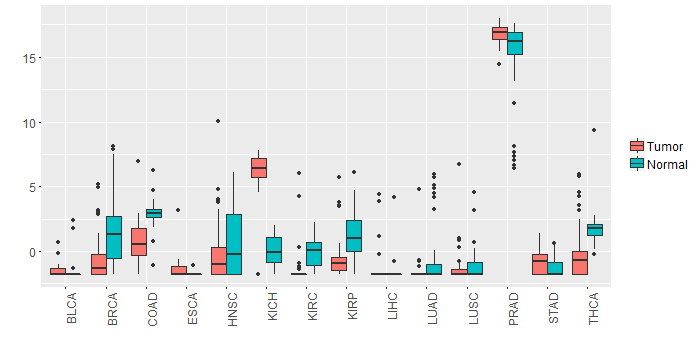
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.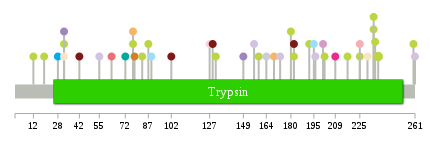

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 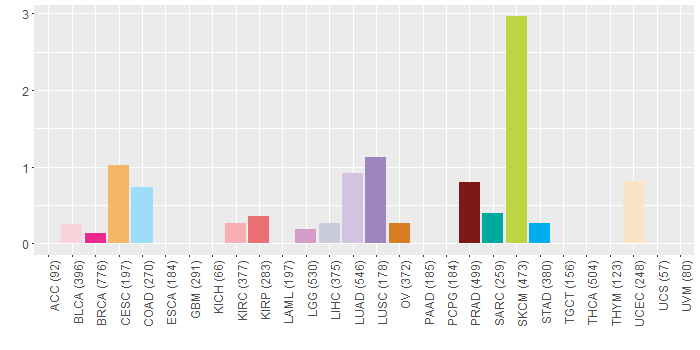
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)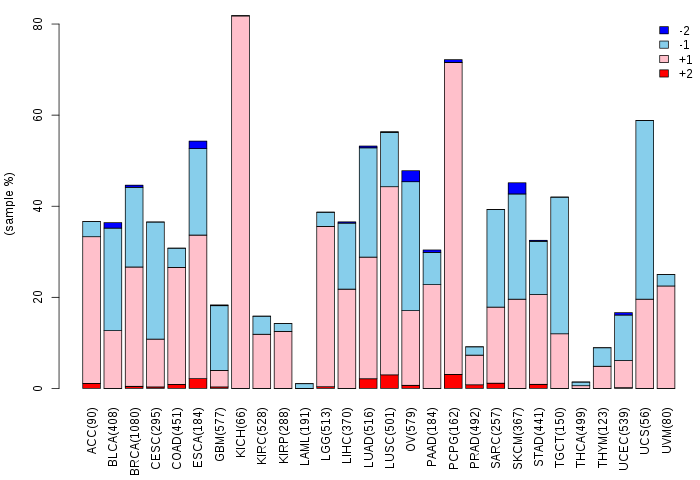
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 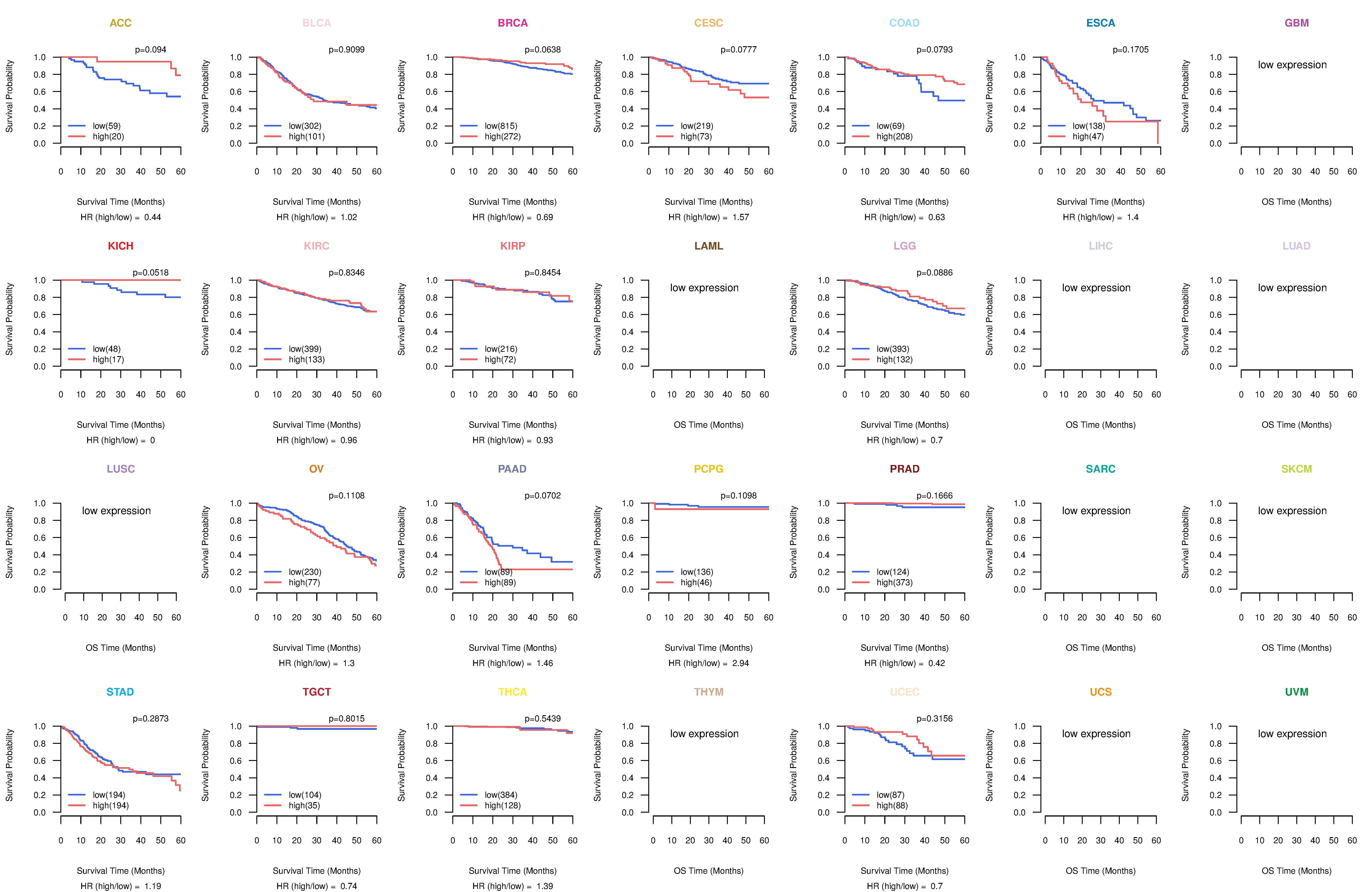
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 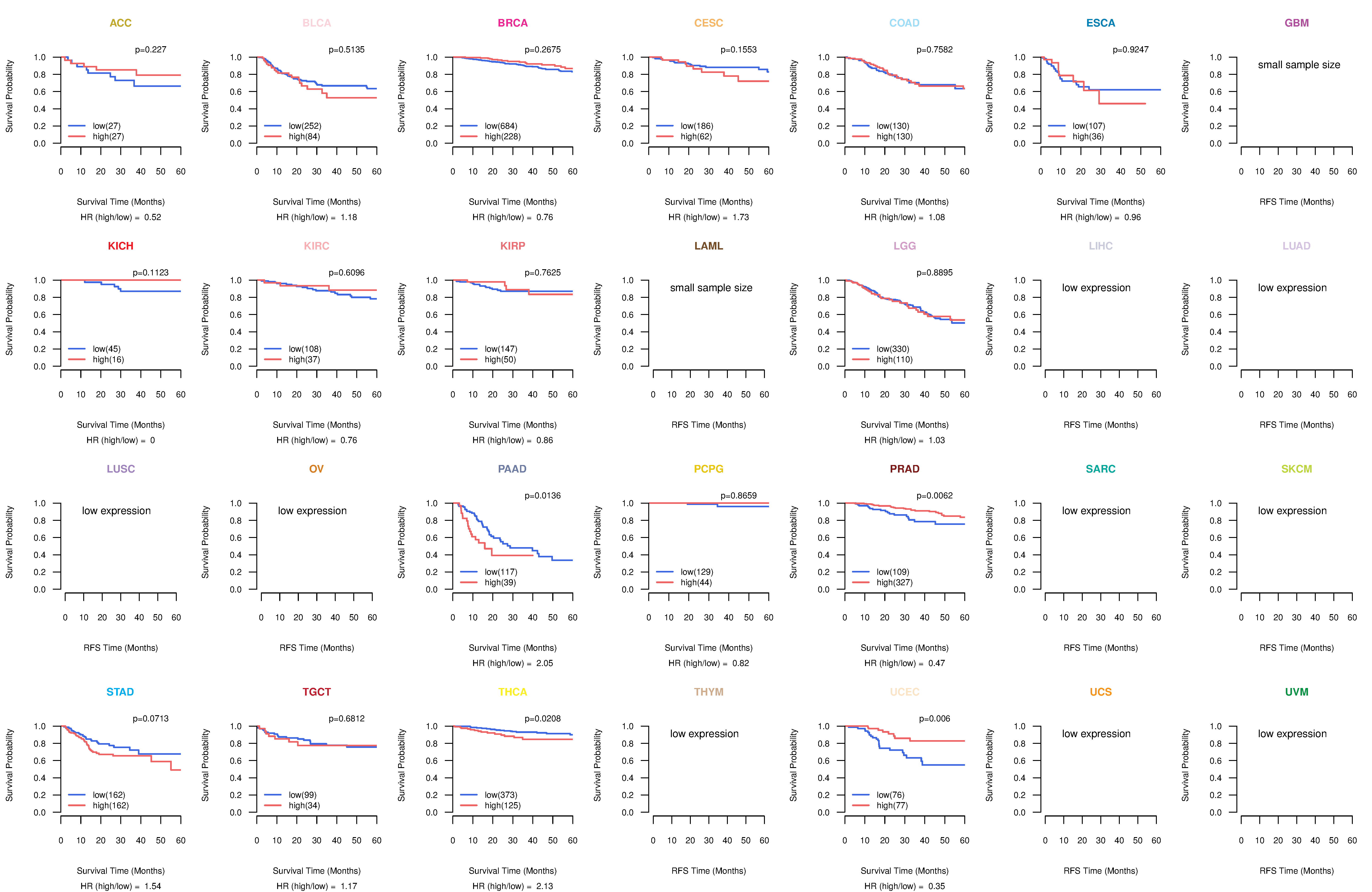
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 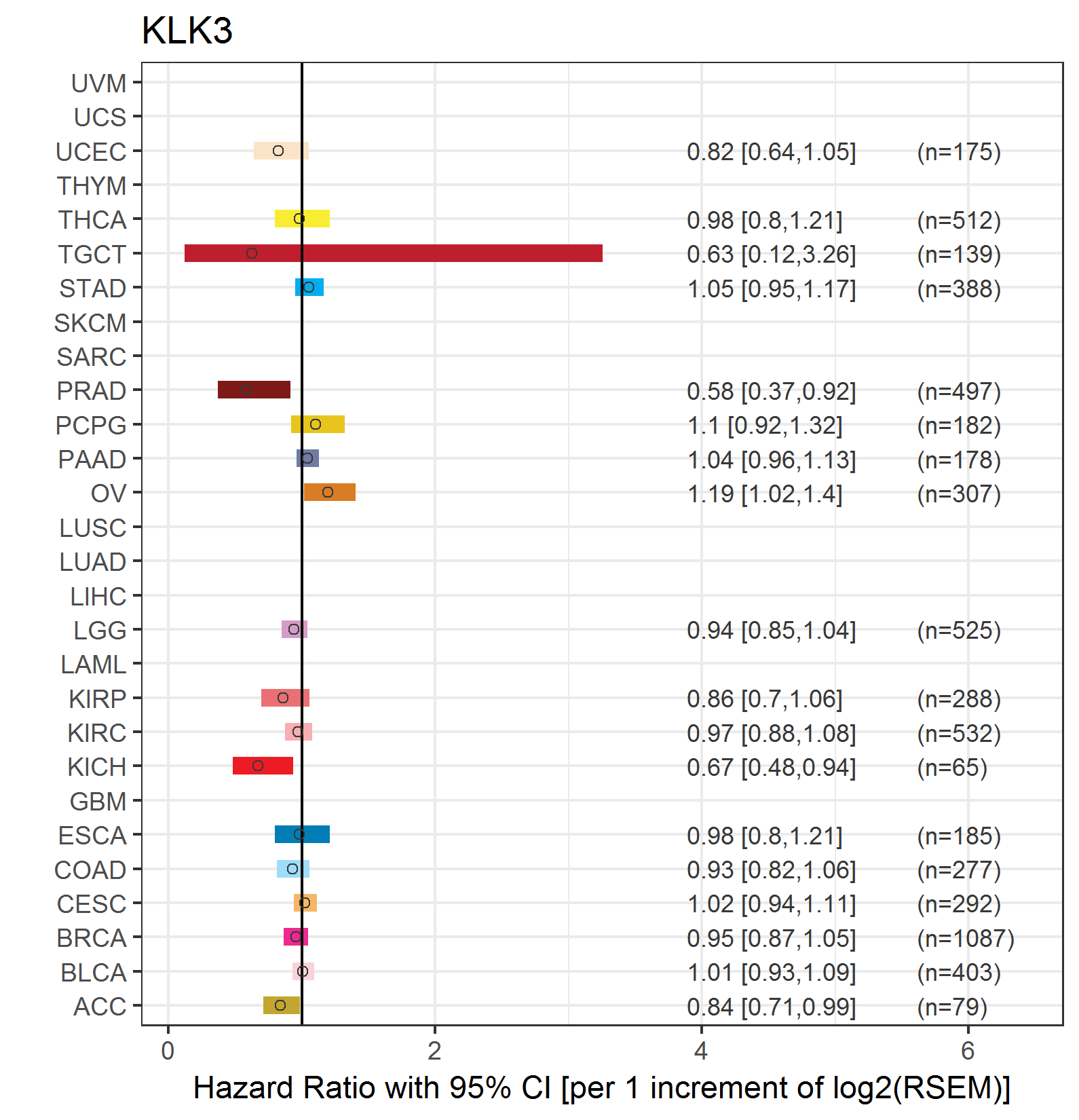
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 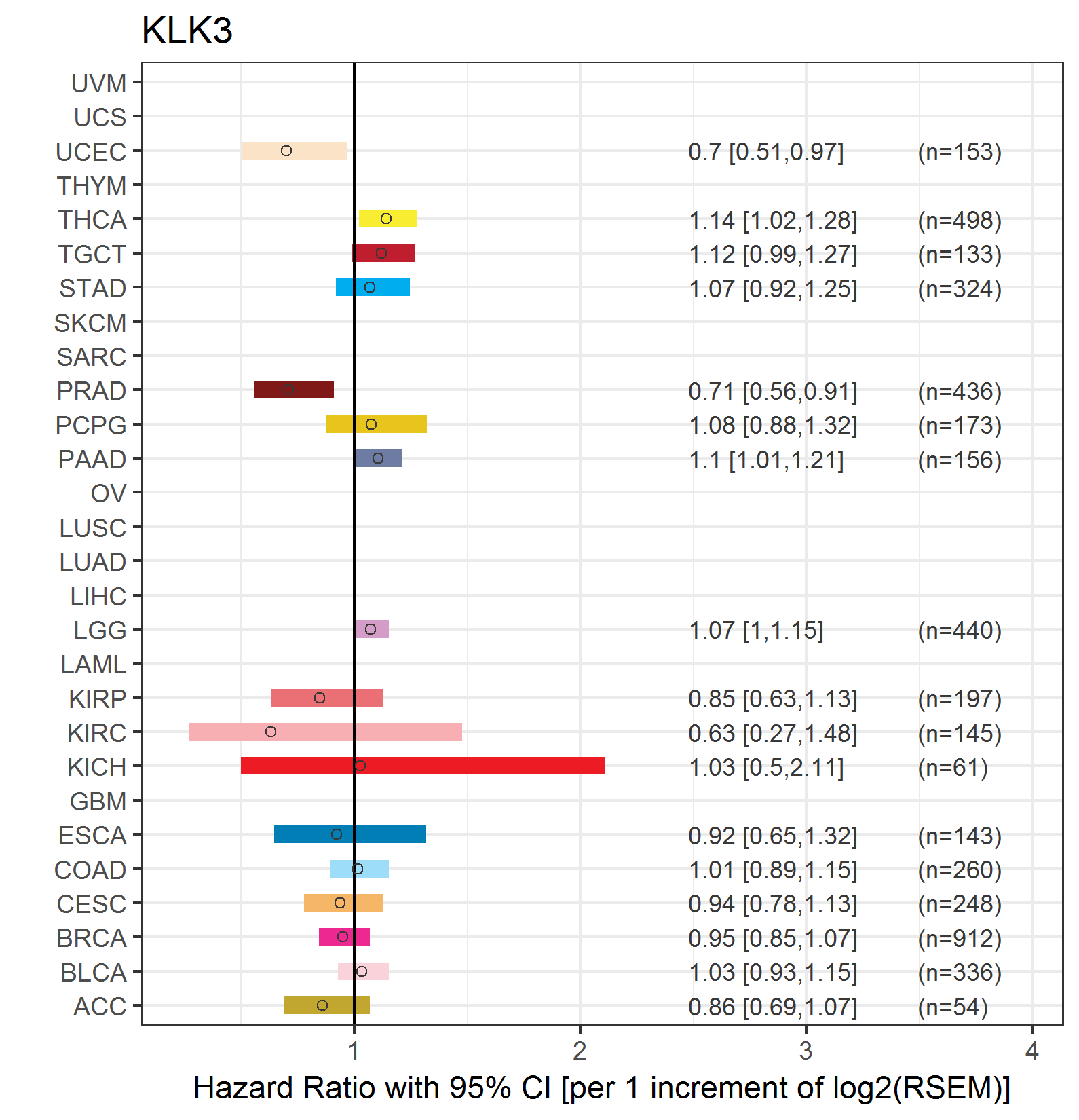
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























