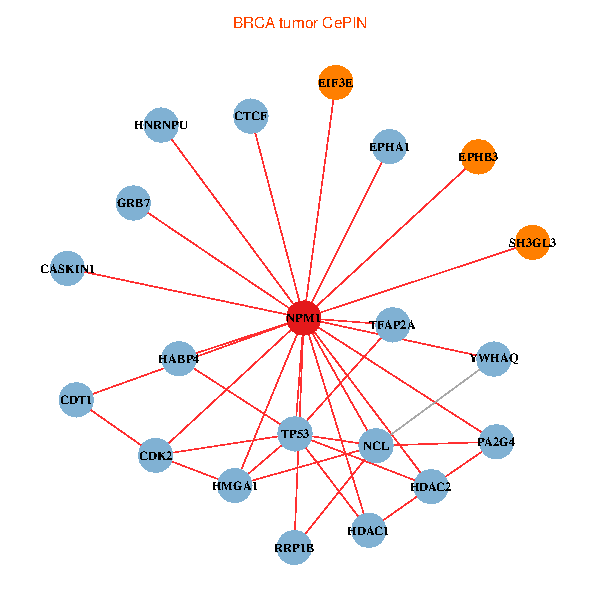
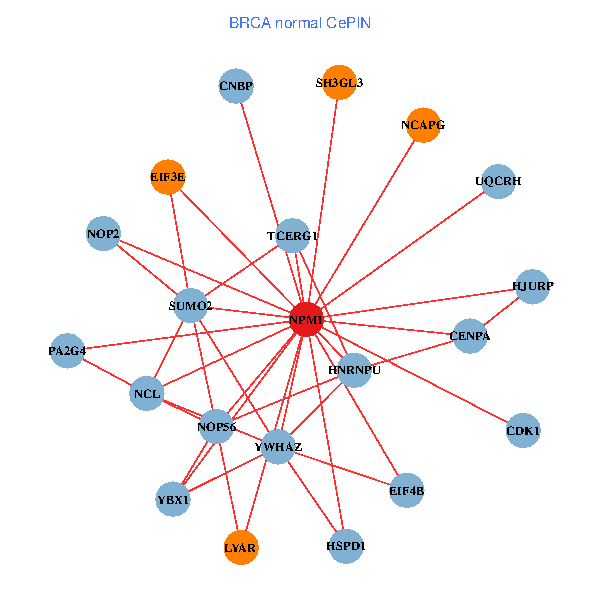
| BRCA (tumor) | BRCA (normal) |
| NPM1, TP53, TFAP2A, HMGA1, SH3GL3, HDAC2, YWHAQ, HDAC1, NCL, CTCF, CDK2, HNRNPU, GRB7, EIF3E, CDT1, EPHA1, PA2G4, RRP1B, HABP4, CASKIN1, EPHB3 (tumor) | NPM1, YBX1, YWHAZ, CNBP, SH3GL3, SUMO2, LYAR, NCL, CDK1, TCERG1, HNRNPU, NOP56, CENPA, HJURP, EIF3E, NCAPG, HSPD1, EIF4B, PA2G4, UQCRH, NOP2 (normal) |
 |  |
|
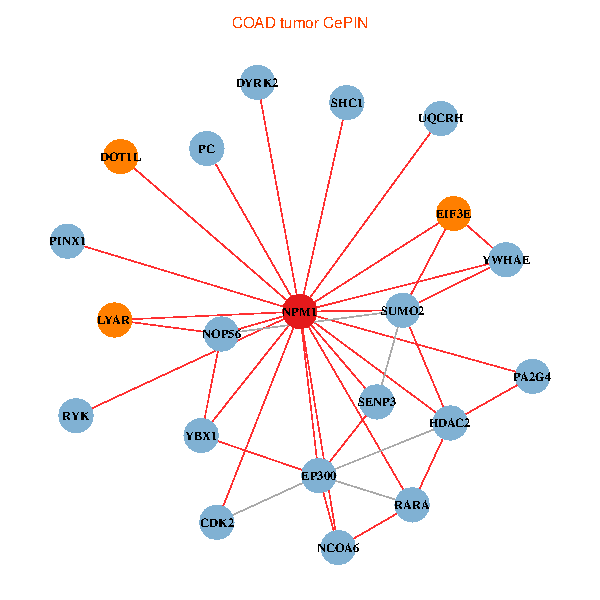
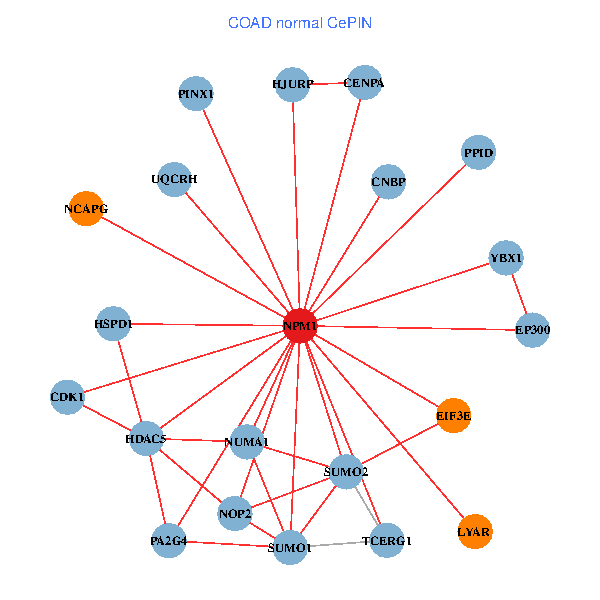
| COAD (tumor) | COAD (normal) |
| NPM1, YBX1, SUMO2, HDAC2, SHC1, RARA, LYAR, YWHAE, CDK2, NOP56, EP300, PINX1, NCOA6, DYRK2, DOT1L, EIF3E, SENP3, PC, RYK, PA2G4, UQCRH (tumor) | NPM1, YBX1, CNBP, SUMO2, LYAR, SUMO1, CDK1, HDAC5, TCERG1, CENPA, EP300, PINX1, HJURP, EIF3E, NCAPG, HSPD1, NUMA1, PA2G4, PPID, UQCRH, NOP2 (normal) |
 |  |
|
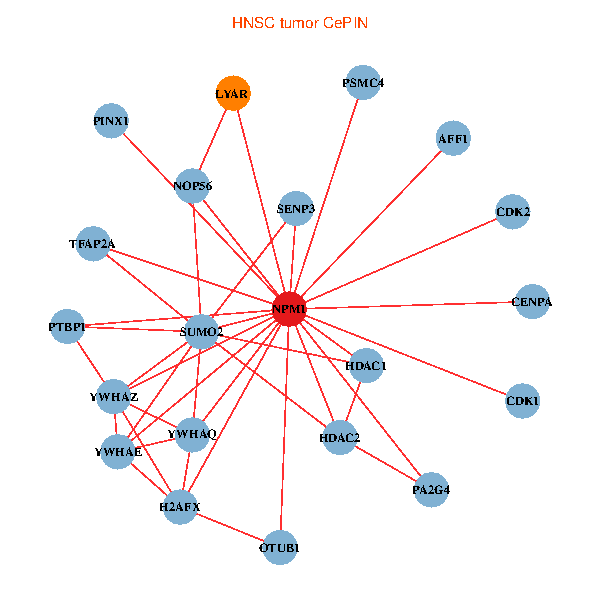
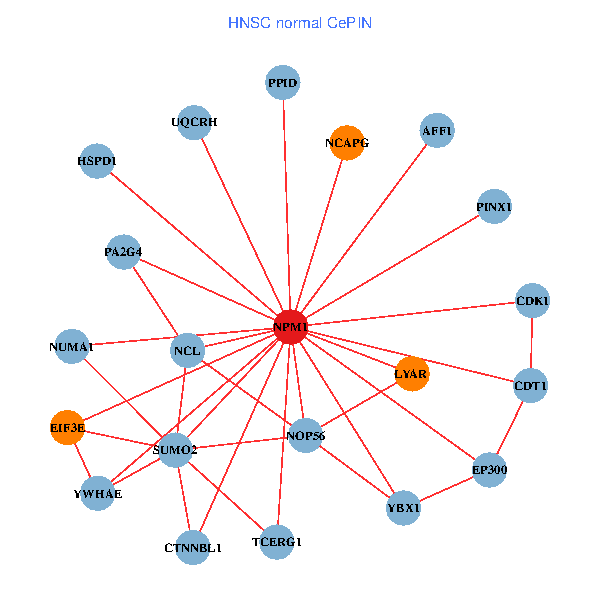
| HNSC (tumor) | HNSC (normal) |
| NPM1, TFAP2A, YWHAZ, SUMO2, HDAC2, LYAR, YWHAQ, HDAC1, YWHAE, CDK1, CDK2, NOP56, CENPA, PSMC4, PINX1, SENP3, OTUB1, PTBP1, H2AFX, AFF1, PA2G4 (tumor) | NPM1, YBX1, SUMO2, LYAR, NCL, YWHAE, CDK1, TCERG1, NOP56, EP300, PINX1, EIF3E, NCAPG, CDT1, HSPD1, AFF1, CTNNBL1, NUMA1, PA2G4, PPID, UQCRH (normal) |
 |  |
|
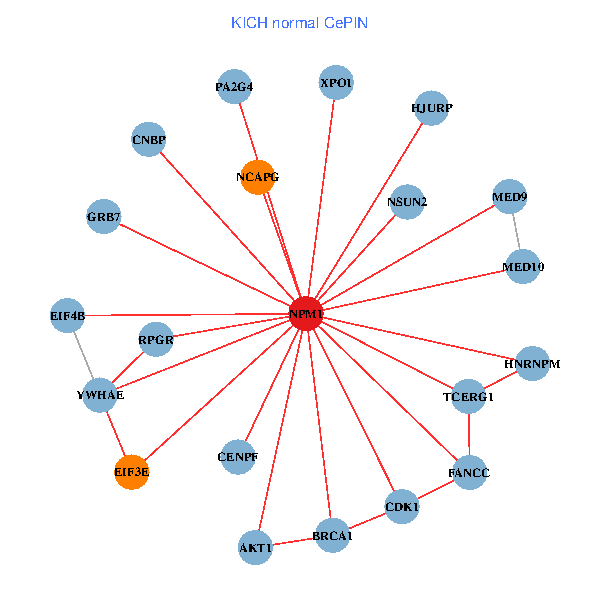
| KICH (tumor) | KICH (normal) |
| NPM1, SMARCA4, CNBP, PLCG1, YWHAQ, NCL, SUMO1, YWHAE, CTCF, HNRNPU, EP300, AKT1, PSMC4, NCOA6, EIF3E, PARP1, AFF1, ACACA, ABCC1, NUMA1, RRP1B (tumor) | NPM1, BRCA1, CNBP, MED9, YWHAE, CDK1, TCERG1, XPO1, GRB7, RPGR, AKT1, HJURP, EIF3E, NCAPG, HNRNPM, MED10, EIF4B, FANCC, CENPF, PA2G4, NSUN2 (normal) |
 |  |
|
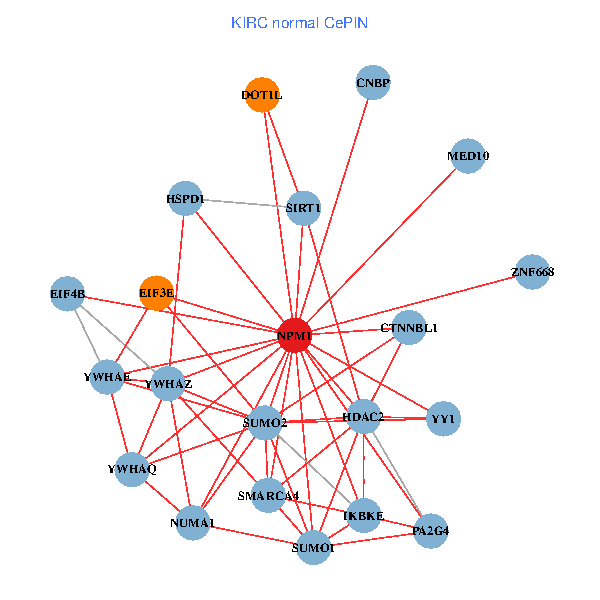
| KIRC (tumor) | KIRC (normal) |
| NPM1, SMARCA4, YWHAZ, CNBP, SUMO2, HDAC2, CDKN2A, YWHAQ, SUMO1, CDK1, CDK2, CENPA, MYL12A, DOT1L, HJURP, EIF3E, NCAPG, MED10, CENPF, NUMA1, PA2G4 (tumor) | NPM1, IKBKE, SMARCA4, YWHAZ, CNBP, SUMO2, HDAC2, YWHAQ, SUMO1, YWHAE, YY1, SIRT1, DOT1L, EIF3E, HSPD1, MED10, EIF4B, CTNNBL1, NUMA1, PA2G4, ZNF668 (normal) |
 |  |
|
| KIRP (tumor) | KIRP (normal) |
| NPM1, CNBP, SUMO2, HDAC2, LYAR, YWHAQ, HDAC1, SUMO1, CDK2, XPO1, EP300, MYL12A, NCOA6, DOT1L, EIF3E, RYK, MED10, PTBP1, EIF4B, NUMA1, PA2G4 (tumor) | NPM1, YBX1, CNBP, LYAR, NCL, SUMO1, CDK1, CENPA, AKT1, DOT1L, HJURP, EIF3E, SENP3, PARP1, NCAPG, EPHA1, HSPD1, MED10, EIF4B, CENPF, PA2G4 (normal) |
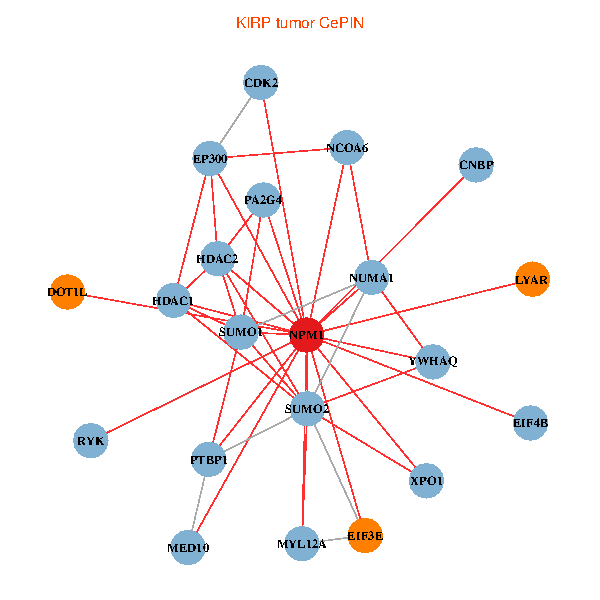 | 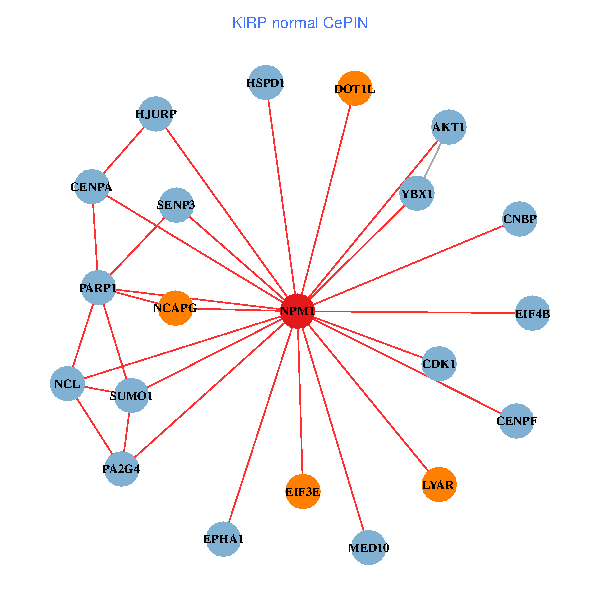 |
|
| LIHC (tumor) | LIHC (normal) |
| NPM1, YBX1, CNBP, SUMO2, HDAC2, LYAR, HDAC1, YWHAE, MED19, NOP56, RPGR, AKT1, MYL12A, PINX1, EIF3E, EPHA1, CDC14A, RYK, MED10, PA2G4, HABP4 (tumor) | NPM1, YBX1, HMGA1, YWHAZ, HDAC2, ESR1, RARA, LYAR, NCL, TCERG1, PIK3R1, HNRNPU, NOP56, EIF3E, HNRNPM, HSPD1, MED10, GADD45A, EIF4B, PA2G4, NOP2 (normal) |
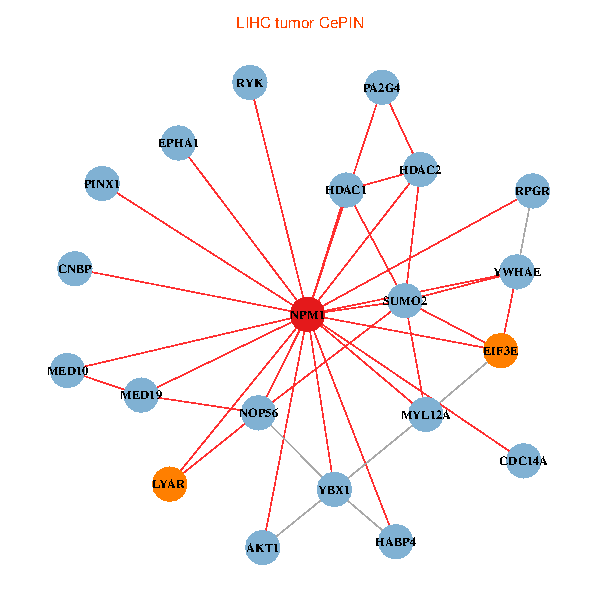 | 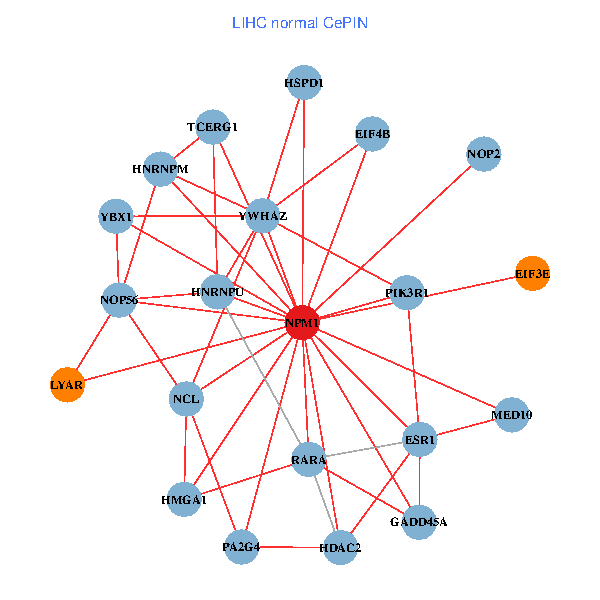 |
|
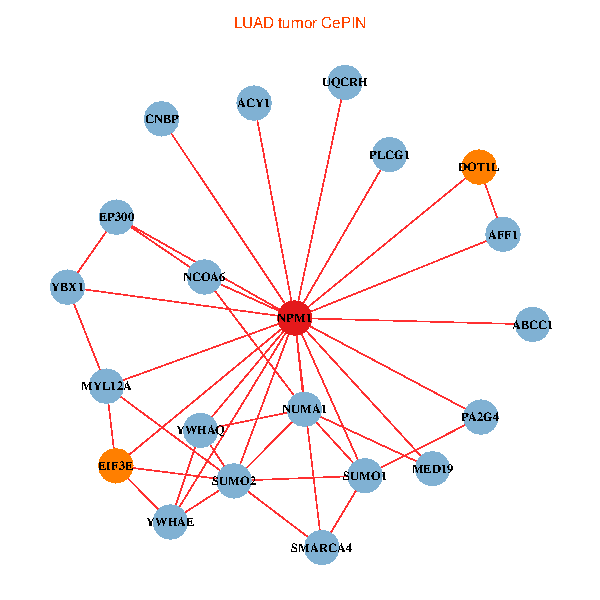
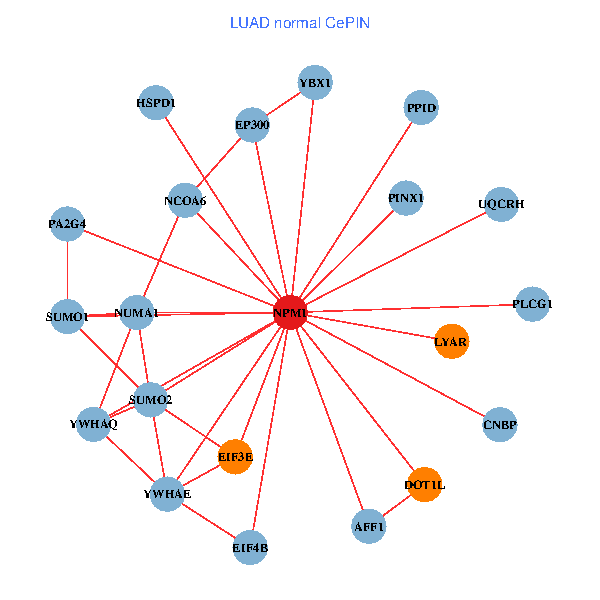
| LUAD (tumor) | LUAD (normal) |
| NPM1, YBX1, SMARCA4, ACY1, CNBP, SUMO2, PLCG1, YWHAQ, SUMO1, YWHAE, MED19, EP300, MYL12A, NCOA6, DOT1L, EIF3E, AFF1, ABCC1, NUMA1, PA2G4, UQCRH (tumor) | NPM1, YBX1, CNBP, SUMO2, PLCG1, LYAR, YWHAQ, SUMO1, YWHAE, EP300, PINX1, NCOA6, DOT1L, EIF3E, HSPD1, AFF1, EIF4B, NUMA1, PA2G4, PPID, UQCRH (normal) |
 |  |
|
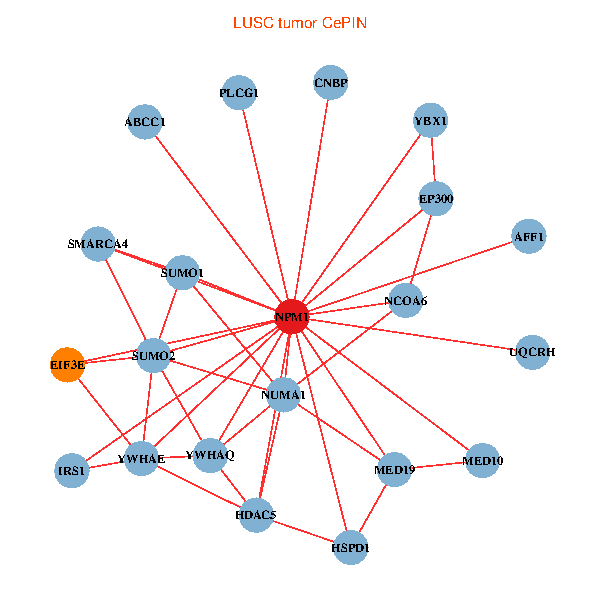
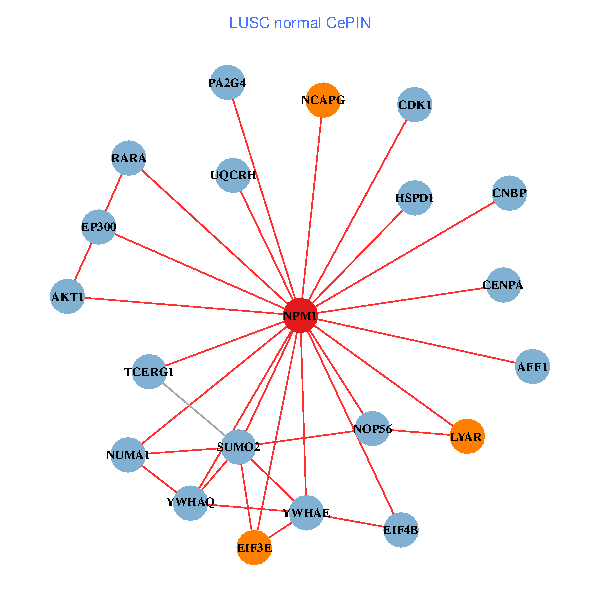
| LUSC (tumor) | LUSC (normal) |
| NPM1, YBX1, SMARCA4, CNBP, SUMO2, PLCG1, YWHAQ, SUMO1, YWHAE, HDAC5, MED19, EP300, NCOA6, IRS1, EIF3E, HSPD1, MED10, AFF1, ABCC1, NUMA1, UQCRH (tumor) | NPM1, CNBP, SUMO2, RARA, LYAR, YWHAQ, YWHAE, CDK1, TCERG1, NOP56, CENPA, EP300, AKT1, EIF3E, NCAPG, HSPD1, AFF1, EIF4B, NUMA1, PA2G4, UQCRH (normal) |
 |  |
|
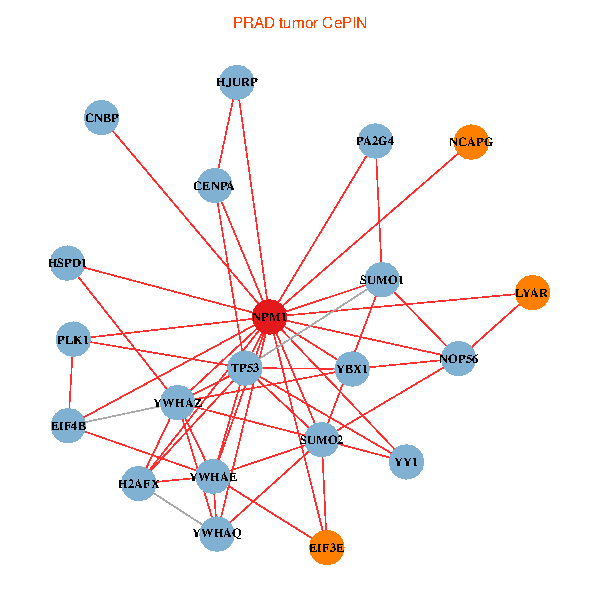
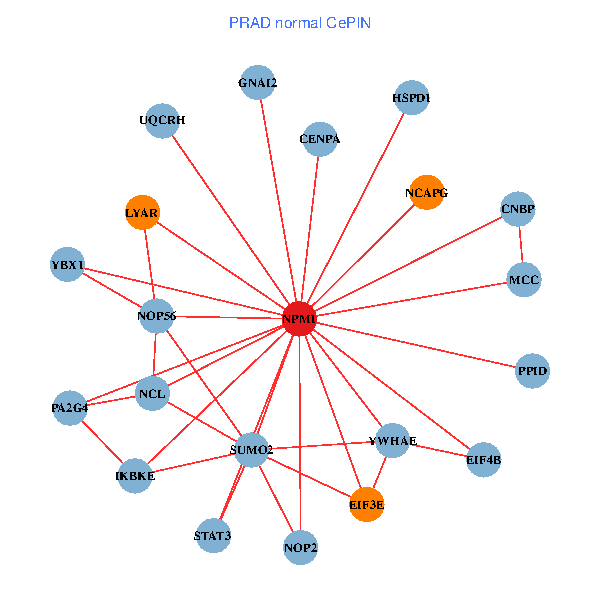
| PRAD (tumor) | PRAD (normal) |
| NPM1, YBX1, TP53, YWHAZ, CNBP, SUMO2, PLK1, LYAR, YWHAQ, SUMO1, YWHAE, YY1, NOP56, CENPA, HJURP, EIF3E, NCAPG, HSPD1, H2AFX, EIF4B, PA2G4 (tumor) | NPM1, YBX1, IKBKE, CNBP, SUMO2, LYAR, STAT3, NCL, YWHAE, NOP56, CENPA, MCC, GNAI2, EIF3E, NCAPG, HSPD1, EIF4B, PA2G4, PPID, UQCRH, NOP2 (normal) |
 |  |
|
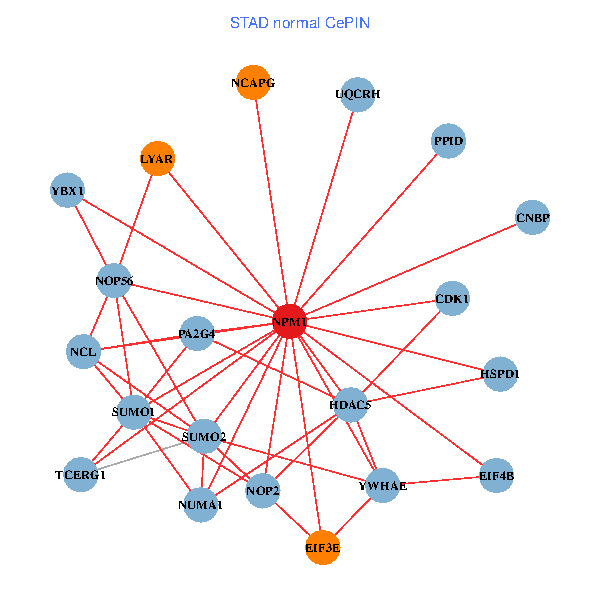
| STAD (tumor) | STAD (normal) |
| NPM1, YBX1, TRAF6, SUMO2, PLCG1, HDAC2, SHC1, LYAR, STAT3, SUMO1, MED19, NOP56, EP300, PINX1, NCOA6, DYRK2, EIF3E, MED10, NUMA1, PA2G4, UQCRH (tumor) | NPM1, YBX1, CNBP, SUMO2, LYAR, NCL, SUMO1, YWHAE, CDK1, HDAC5, TCERG1, NOP56, EIF3E, NCAPG, HSPD1, EIF4B, NUMA1, PA2G4, PPID, UQCRH, NOP2 (normal) |
 |  |
|
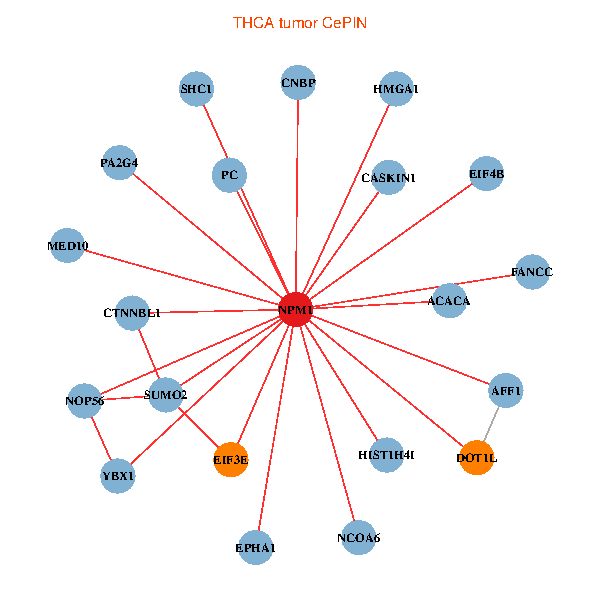
| THCA (tumor) | THCA (normal) |
| NPM1, YBX1, HMGA1, CNBP, SUMO2, SHC1, NOP56, NCOA6, HIST1H4I, DOT1L, EIF3E, EPHA1, PC, MED10, AFF1, ACACA, EIF4B, FANCC, CTNNBL1, PA2G4, CASKIN1 (tumor) | NPM1, SMARCA4, TRAF6, HMGA1, CNBP, SUMO2, PLCG1, HDAC2, NCL, GRB7, MCC, MYL12A, IRS1, DOT1L, EIF3E, CDT1, PC, HSPD1, GADD45A, EIF4B, PA2G4 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0023467 | Leukemia, Myelocytic, Acute | 298 | BeFree,CLINVAR,CTD_human,GAD,LHGDN |
| umls:C0206180 | Ki-1+ Anaplastic Large Cell Lymphoma | 128 | BeFree,LHGDN |
| umls:C0023418 | leukemia | 41 | BeFree,GAD,LHGDN |
| umls:C0242596 | Neoplasm, Residual | 35 | BeFree,GAD |
| umls:C3839868 | Cytogenetically normal acute myeloid leukemia | 33 | BeFree |
| umls:C0024299 | Lymphoma | 32 | BeFree,LHGDN |
| umls:C0596263 | Carcinogenesis | 25 | BeFree |
| umls:C0598766 | Leukemogenesis | 25 | BeFree |
| umls:C0023487 | Acute Promyelocytic Leukemia | 22 | BeFree,CTD_human,ORPHANET |
| umls:C0023470 | Myeloid Leukemia | 17 | BeFree,GAD |
| umls:C0033027 | Preleukemia | 17 | BeFree,GAD |
| umls:C0220615 | Adult Acute Myeloblastic Leukemia | 17 | BeFree |
| umls:C3463824 | MYELODYSPLASTIC SYNDROME | 16 | BeFree,GAD,LHGDN |
| umls:C1332079 | Anaplastic Large Cell Lymphoma, ALK-Positive | 14 | BeFree |
| umls:C0008626 | Congenital chromosomal disease | 12 | BeFree |
| umls:C1840451 | MULTICYSTIC RENAL DYSPLASIA, BILATERAL | 10 | BeFree |
| umls:C0220621 | pediatric acute myeloblastic leukemia | 9 | BeFree |
| umls:C0376545 | Hematologic Neoplasms | 8 | BeFree |
| umls:C0027708 | Nephroblastoma | 6 | BeFree |
| umls:C0079772 | T-Cell Lymphoma | 6 | BeFree,LHGDN |
| umls:C0022660 | Kidney Failure, Acute | 5 | BeFree |
| umls:C0023480 | Leukemia, Myelomonocytic, Chronic | 5 | BeFree,GAD |
| umls:C0026985 | Myelodysplasia | 5 | BeFree |
| umls:C0264490 | Acute respiratory failure | 5 | BeFree |
| umls:C2239176 | Liver carcinoma | 5 | BeFree |
| umls:C0024623 | Malignant neoplasm of stomach | 4 | BeFree |
| umls:C0027022 | Myeloproliferative disease | 4 | BeFree,LHGDN |
| umls:C0027627 | Neoplasm Metastasis | 4 | BeFree |
| umls:C0152276 | Granulocytic Sarcoma | 4 | BeFree,LHGDN |
| umls:C1321546 | Anaplastic large B-cell lymphoma | 4 | BeFree |
| umls:C0005684 | Malignant neoplasm of urinary bladder | 3 | BeFree |
| umls:C0005699 | Blast Phase | 3 | BeFree,GAD |
| umls:C0006142 | Malignant neoplasm of breast | 3 | BeFree,GAD |
| umls:C0008625 | Chromosome Aberrations | 3 | GAD |
| umls:C0015625 | Fanconi Anemia | 3 | BeFree,LHGDN |
| umls:C0023473 | Myeloid Leukemia, Chronic | 3 | BeFree |
| umls:C0033578 | Prostatic Neoplasms | 3 | BeFree,LHGDN |
| umls:C0178874 | Tumor Progression | 3 | BeFree |
| umls:C0280100 | Solid tumour | 3 | BeFree |
| umls:C0280449 | secondary acute myeloid leukemia | 3 | BeFree |
| umls:C0521158 | Recurrent tumor | 3 | BeFree |
| umls:C0699791 | Stomach Carcinoma | 3 | BeFree |
| umls:C0699885 | Carcinoma of bladder | 3 | BeFree |
| umls:C2745900 | Promyelocytic leukemia | 3 | BeFree |
| umls:C2826177 | Acute myeloid leukemia with mutated NPM1 | 3 | BeFree |
| umls:C3469521 | FANCONI ANEMIA, COMPLEMENTATION GROUP A (disorder) | 3 | BeFree |
| umls:C0001430 | Adenoma | 2 | BeFree |
| umls:C0001815 | Primary Myelofibrosis | 2 | BeFree,LHGDN |
| umls:C0004114 | Astrocytoma | 2 | BeFree |
| umls:C0007131 | Non-Small Cell Lung Carcinoma | 2 | BeFree |
| umls:C0009402 | Colorectal Carcinoma | 2 | BeFree |
| umls:C0019693 | HIV Infections | 2 | BeFree |
| umls:C0023449 | Acute lymphocytic leukemia | 2 | BeFree |
| umls:C0026764 | Multiple Myeloma | 2 | BeFree |
| umls:C0027819 | Neuroblastoma | 2 | BeFree |
| umls:C0038356 | Stomach Neoplasms | 2 | BeFree,CTD_human |
| umls:C0079731 | B-Cell Lymphomas | 2 | BeFree |
| umls:C0085110 | Severe Combined Immunodeficiency | 2 | BeFree |
| umls:C0085669 | Acute leukemia | 2 | BeFree |
| umls:C0153381 | Malignant neoplasm of mouth | 2 | BeFree |
| umls:C0206182 | Lymphomatoid Papulosis | 2 | BeFree,ORPHANET |
| umls:C0220641 | Lip and Oral Cavity Carcinoma | 2 | BeFree |
| umls:C0376358 | Malignant neoplasm of prostate | 2 | BeFree |
| umls:C0600139 | Prostate carcinoma | 2 | BeFree |
| umls:C0678222 | Breast Carcinoma | 2 | BeFree |
| umls:C0700095 | Central neuroblastoma | 2 | BeFree |
| umls:C1261473 | Sarcoma | 2 | BeFree |
| umls:C1318544 | M5b Acute differentiated monocytic leukemia | 2 | BeFree |
| umls:C1336548 | Systemic Anaplastic Large Cell Lymphoma | 2 | BeFree |
| umls:C1336735 | Treatment related acute myeloid leukaemia | 2 | BeFree |
| umls:C1709246 | Non-Neoplastic Disorder | 2 | BeFree |
| umls:C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 2 | BeFree |
| umls:C0007102 | Malignant tumor of colon | 1 | BeFree |
| umls:C0007103 | Malignant neoplasm of endometrium | 1 | BeFree |
| umls:C0007112 | Adenocarcinoma of prostate | 1 | BeFree |
| umls:C0007134 | Renal Cell Carcinoma | 1 | BeFree |
| umls:C0007137 | Squamous cell carcinoma | 1 | BeFree |
| umls:C0007621 | Neoplastic Cell Transformation | 1 | GAD |
| umls:C0014057 | Japanese Encephalitis | 1 | LHGDN |
| umls:C0015230 | Exanthema | 1 | BeFree |
| umls:C0017636 | Glioblastoma | 1 | BeFree |
| umls:C0018133 | Graft-vs-Host Disease | 1 | BeFree |
| umls:C0019163 | Hepatitis B | 1 | BeFree |
| umls:C0019208 | Hepatoma, Novikoff | 1 | BeFree |
| umls:C0019623 | Malignant histiocytosis | 1 | BeFree |
| umls:C0019829 | Hodgkin Disease | 1 | BeFree |
| umls:C0021051 | Immunologic Deficiency Syndromes | 1 | BeFree |
| umls:C0021400 | Influenza | 1 | BeFree |
| umls:C0021943 | Chromosome inversion | 1 | GAD |
| umls:C0023434 | Chronic Lymphocytic Leukemia | 1 | BeFree |
| umls:C0023465 | Acute monocytic leukemia | 1 | BeFree |
| umls:C0023474 | Leukemia, Myeloid, Chronic-Phase | 1 | BeFree |
| umls:C0023479 | Acute myelomonocytic leukemia | 1 | BeFree |
| umls:C0023890 | Liver Cirrhosis | 1 | BeFree |
| umls:C0023903 | Liver neoplasms | 1 | BeFree |
| umls:C0024121 | Lung Neoplasms | 1 | LHGDN |
| umls:C0024305 | Lymphoma, Non-Hodgkin | 1 | BeFree |
| umls:C0024899 | Mastocytosis | 1 | BeFree |
| umls:C0026499 | Monosomy | 1 | BeFree |
| umls:C0029463 | Osteosarcoma | 1 | LHGDN |
| umls:C0035335 | Retinoblastoma | 1 | BeFree |
| umls:C0036220 | Kaposi Sarcoma | 1 | BeFree |
| umls:C0037286 | Skin Neoplasms | 1 | BeFree |
| umls:C0040715 | Chromosomal translocation | 1 | GAD |
| umls:C0042111 | Urticaria Pigmentosa | 1 | BeFree |
| umls:C0043346 | Xeroderma Pigmentosum | 1 | BeFree |
| umls:C0043379 | XYY Karyotype | 1 | GAD |
| umls:C0079744 | Diffuse Large B-Cell Lymphoma | 1 | LHGDN |
| umls:C0079773 | Lymphoma, T-Cell, Cutaneous | 1 | BeFree |
| umls:C0085702 | Monocytosis | 1 | BeFree |
| umls:C0153886 | Acute myeloid leukemia in remission | 1 | BeFree |
| umls:C0153888 | Chronic myeloid leukemia in remission | 1 | BeFree |
| umls:C0235974 | Pancreatic carcinoma | 1 | BeFree |
| umls:C0238246 | Hemangioma of liver | 1 | BeFree |
| umls:C0242379 | Malignant neoplasm of lung | 1 | BeFree |
| umls:C0275524 | Coinfection | 1 | BeFree |
| umls:C0278484 | Malignant neoplasm of colon stage IV | 1 | BeFree |
| umls:C0279000 | Liver and Intrahepatic Biliary Tract Carcinoma | 1 | BeFree |
| umls:C0333980 | Focal Nodular Hyperplasia | 1 | BeFree |
| umls:C0334121 | Inflammatory Myofibroblastic Tumor | 1 | BeFree |
| umls:C0334634 | Malignant lymphoma, lymphocytic, intermediate differentiation, diffuse | 1 | BeFree |
| umls:C0345904 | Malignant neoplasm of liver | 1 | BeFree |
| umls:C0346647 | Malignant neoplasm of pancreas | 1 | BeFree |
| umls:C0349667 | Sarcoma of breast | 1 | BeFree |
| umls:C0476089 | Endometrial Carcinoma | 1 | BeFree |
| umls:C0684249 | Carcinoma of lung | 1 | BeFree |
| umls:C0686619 | Secondary malignant neoplasm of lymph node | 1 | BeFree |
| umls:C0699790 | Colon Carcinoma | 1 | BeFree |
| umls:C0795824 | Chromosome 8, monosomy 8p | 1 | BeFree |
| umls:C0848332 | Spots on skin | 1 | BeFree |
| umls:C0850572 | Adenomatous polyp of colon | 1 | BeFree |
| umls:C0854917 | Rhabdoid Tumor of the Kidney | 1 | BeFree |
| umls:C1292753 | Primary Effusion Lymphoma | 1 | BeFree |
| umls:C1292773 | Acute myeloid leukemia with multilineage dysplasia | 1 | BeFree |
| umls:C1292778 | Chronic myeloproliferative disorder | 1 | BeFree |
| umls:C1292780 | Therapy-related myelodysplastic syndrome | 1 | BeFree |
| umls:C1301362 | Lymphoma, Primary Cutaneous Anaplastic Large Cell | 1 | BeFree,ORPHANET |
| umls:C1332078 | Anaplastic large cell lymphoma, ALK negative | 1 | BeFree |
| umls:C1458155 | Mammary Neoplasms | 1 | BeFree |
| umls:C1527249 | Colorectal Cancer | 1 | BeFree |
| umls:C1621958 | Glioblastoma Multiforme | 1 | BeFree |
| umls:C1856689 | FRIEDREICH ATAXIA 1 | 1 | BeFree |
| umls:C1883486 | Uterine Corpus Cancer | 1 | BeFree |
| umls:C2613439 | Extramedullary Hematopoiesis (disorder) | 1 | BeFree |
| umls:C2826178 | Acute myeloid leukemia with mutated CEBPA | 1 | BeFree |
| umls:C2931822 | Nasopharyngeal carcinoma | 1 | BeFree |
| umls:C2939461 | Myeloid neoplasm | 1 | BeFree |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))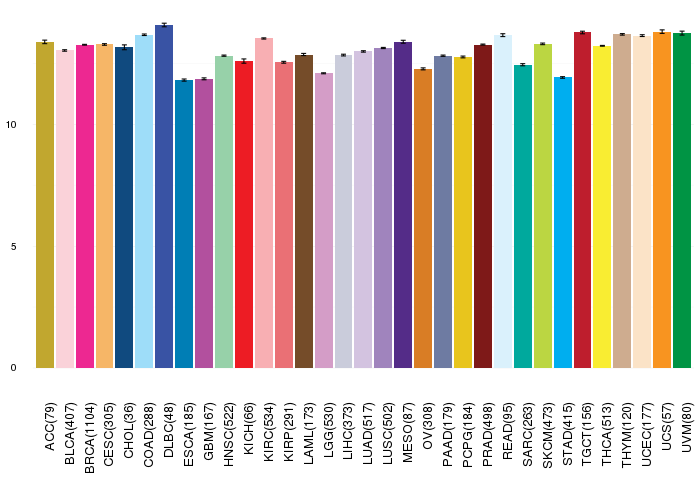
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))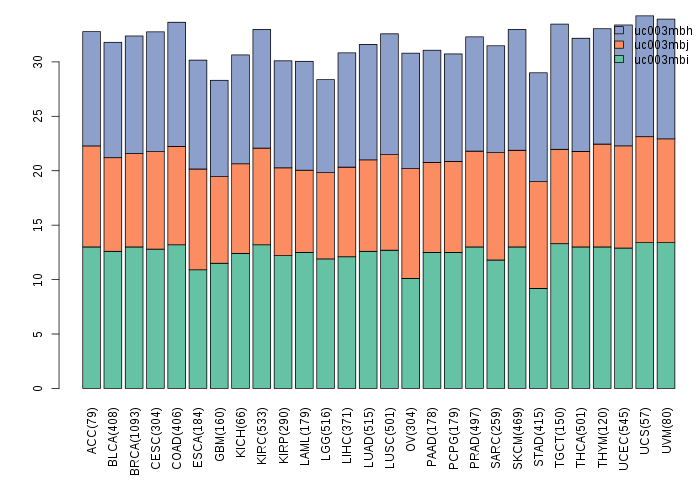
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data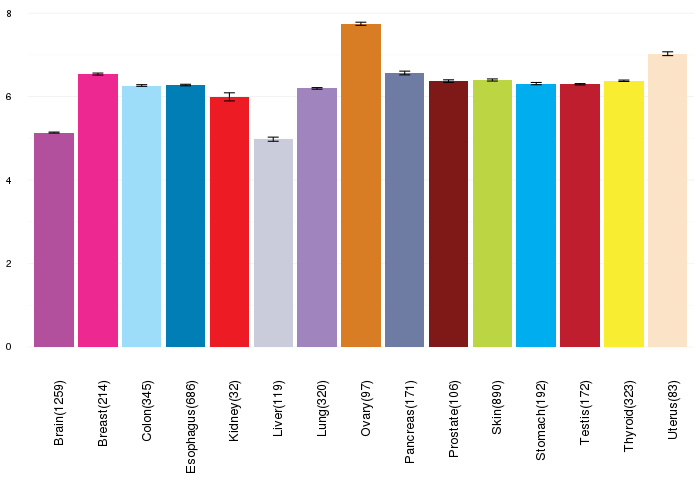
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))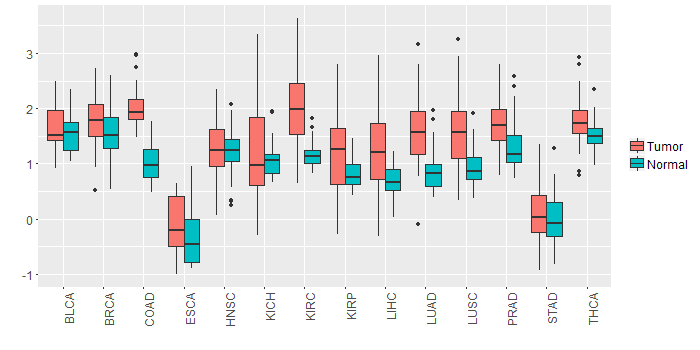
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.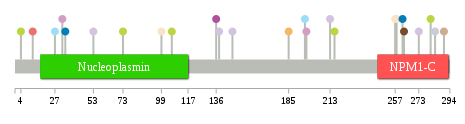

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 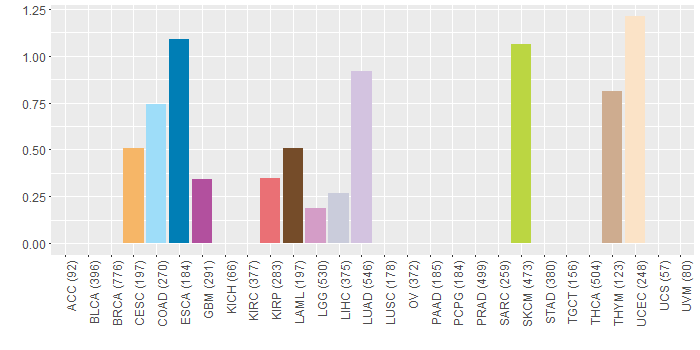
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)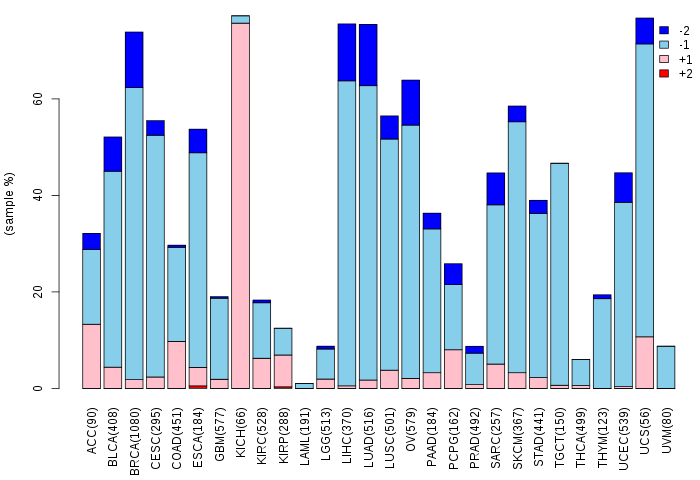
 Fusion genes including TissGene
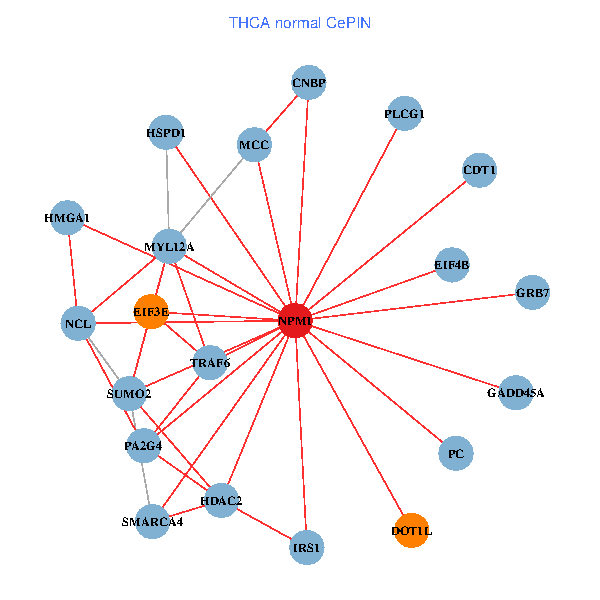
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 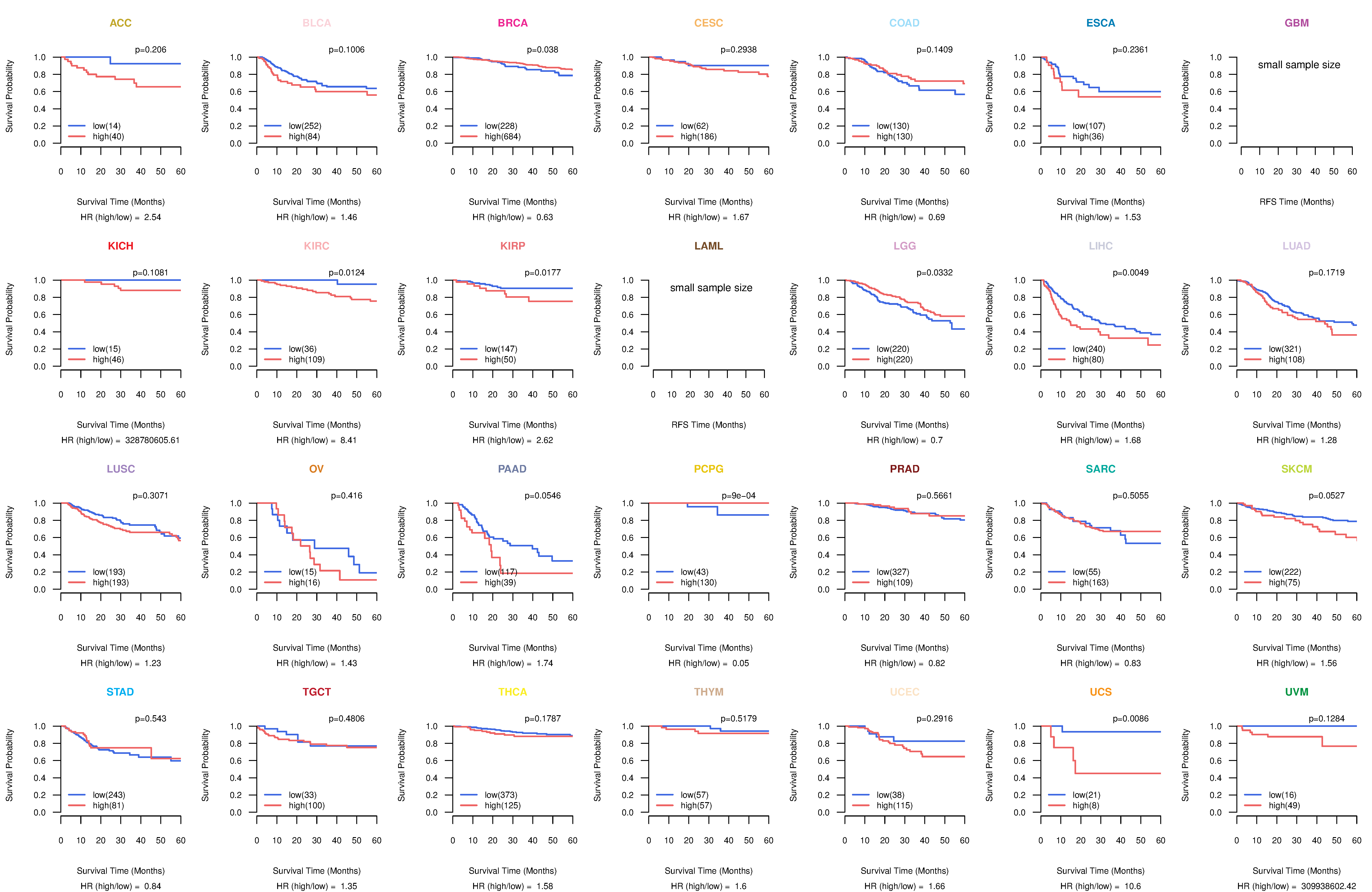
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 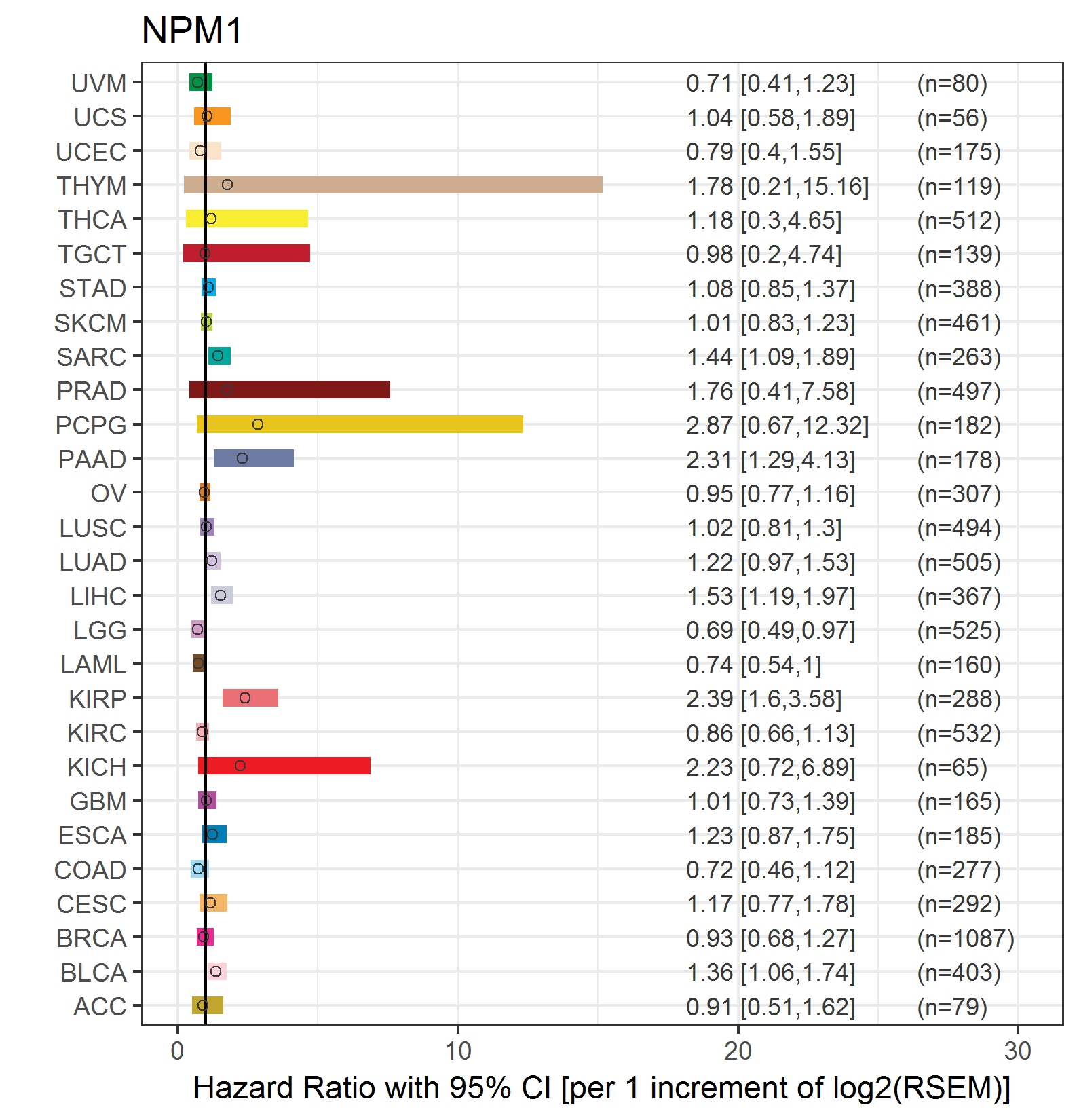
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 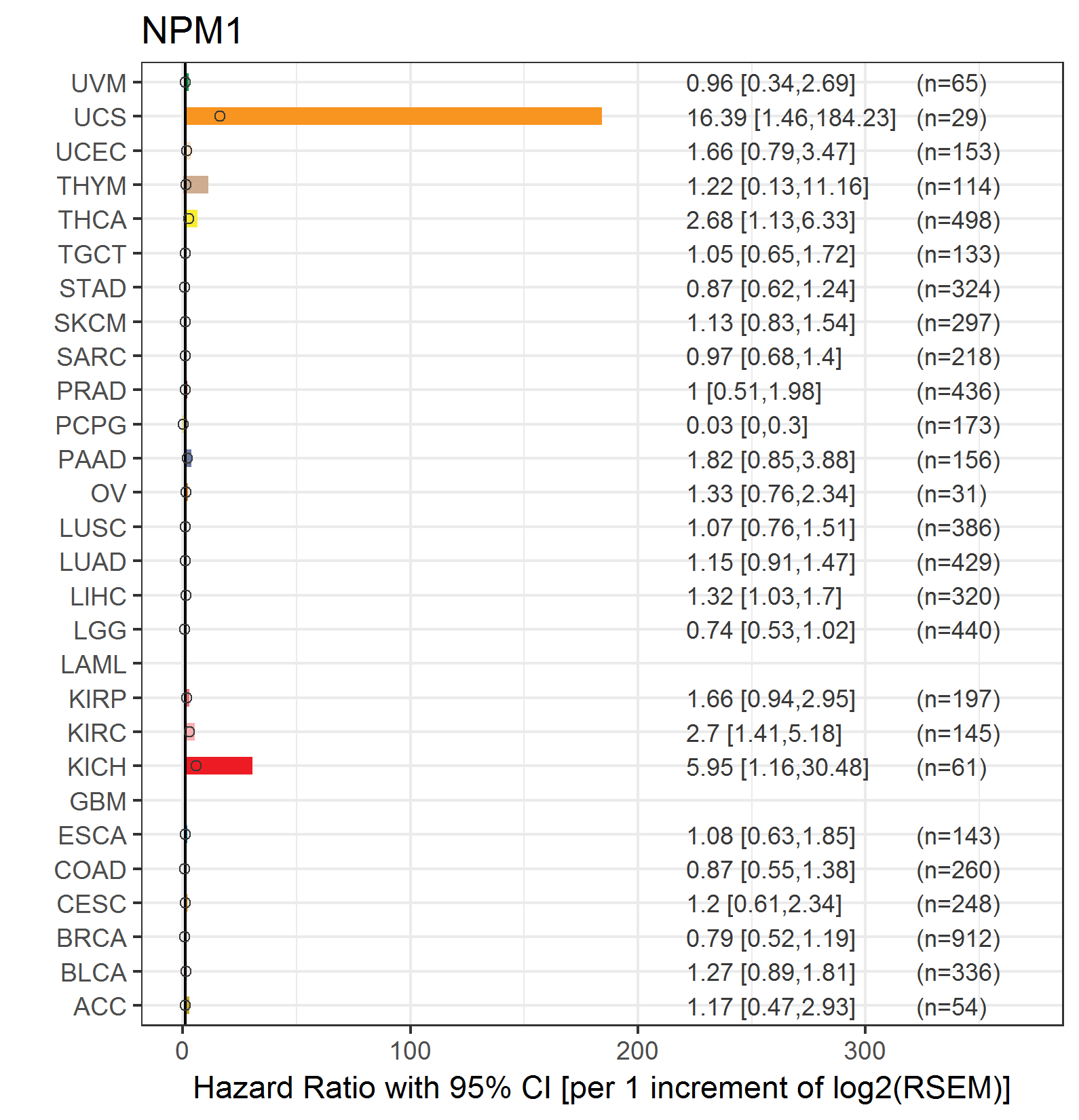
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























