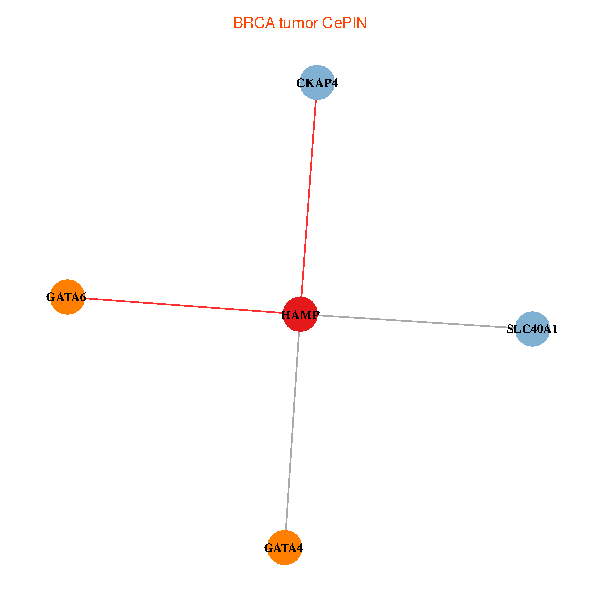
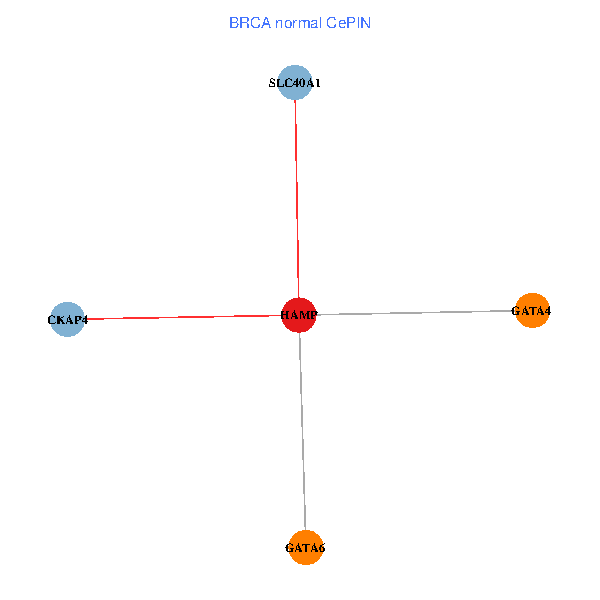
| BRCA (tumor) | BRCA (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
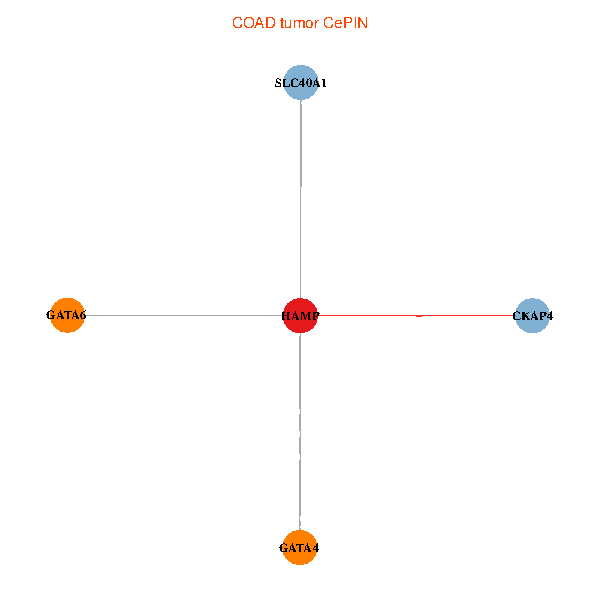
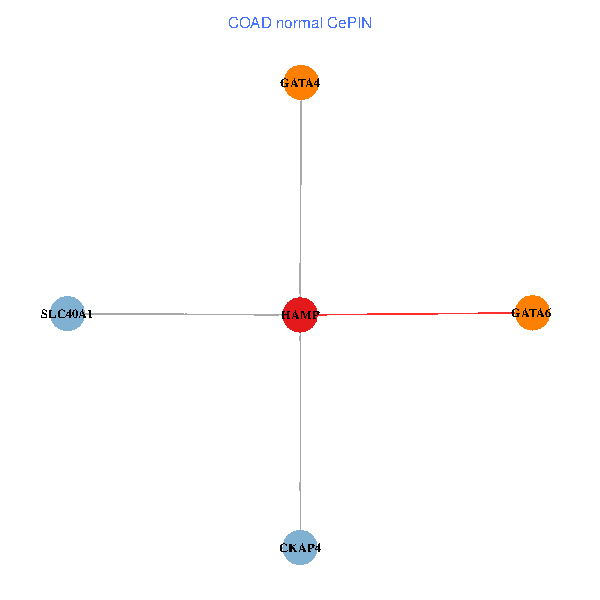
| COAD (tumor) | COAD (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
| KICH (tumor) | KICH (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
| KIRC (tumor) | KIRC (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
| KIRP (tumor) | KIRP (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
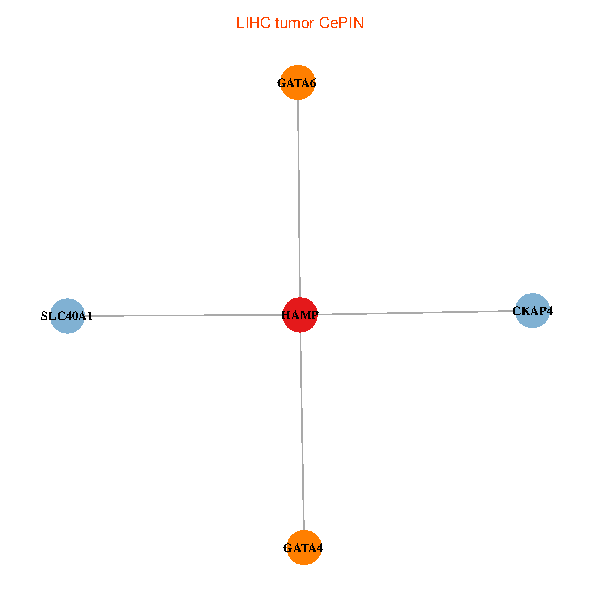
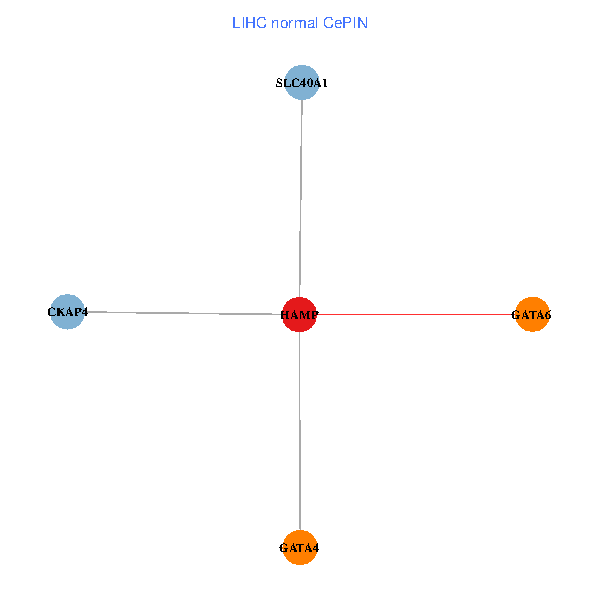
| LIHC (tumor) | LIHC (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
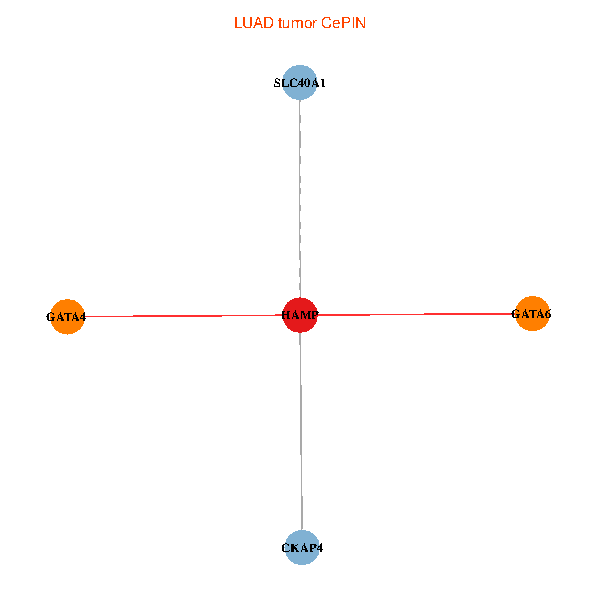
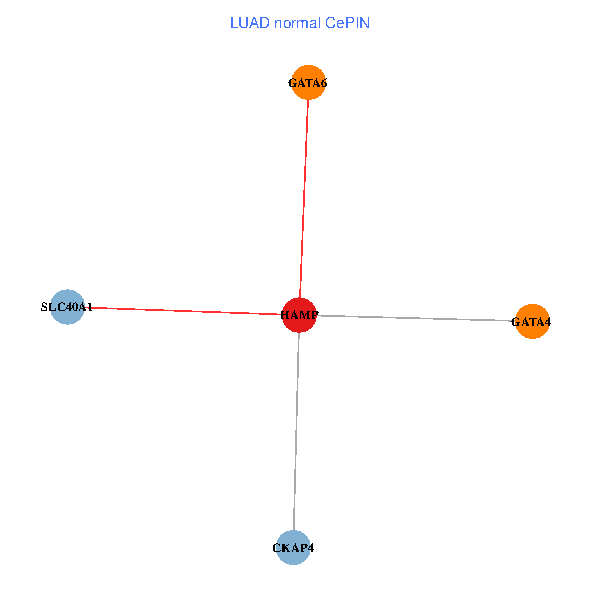
| LUAD (tumor) | LUAD (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
| LUSC (tumor) | LUSC (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
| PRAD (tumor) | PRAD (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
| STAD (tumor) | STAD (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
| THCA (tumor) | THCA (normal) |
| HAMP, CKAP4, GATA4, SLC40A1, GATA6 (tumor) | HAMP, CKAP4, GATA4, SLC40A1, GATA6 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0282193 | Iron Overload | 104 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0018995 | Hemochromatosis | 73 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0002871 | Anemia | 65 | BeFree,CTD_human,LHGDN |
| umls:C0392514 | Hereditary hemochromatosis | 48 | BeFree,CLINVAR |
| umls:C0268060 | Juvenile hemochromatosis | 27 | BeFree,CTD_human,ORPHANET |
| umls:C0240066 | Iron deficiency | 23 | BeFree |
| umls:C0002873 | Anemia of chronic disease | 18 | BeFree |
| umls:C2239176 | Liver carcinoma | 18 | BeFree |
| umls:C0005283 | beta Thalassemia | 14 | BeFree,CTD_human,GAD |
| umls:C0023903 | Liver neoplasms | 13 | BeFree |
| umls:C0039730 | Thalassemia | 11 | BeFree,LHGDN |
| umls:C0085576 | Iron-Refractory Iron Deficiency Anemia | 11 | BeFree |
| umls:C0162316 | Iron deficiency anemia | 10 | BeFree |
| umls:C0019196 | Hepatitis C | 9 | BeFree,LHGDN |
| umls:C0002875 | Cooley's anemia | 8 | BeFree |
| umls:C0028754 | Obesity | 8 | BeFree |
| umls:C0012715 | Iron Metabolism Disorders | 7 | BeFree,GAD |
| umls:C0524910 | Hepatitis C, Chronic | 6 | BeFree |
| umls:C0023896 | Alcoholic Liver Diseases | 5 | BeFree |
| umls:C0004623 | Bacterial Infections | 4 | BeFree |
| umls:C0022661 | Kidney Failure, Chronic | 4 | BeFree,CTD_human,LHGDN |
| umls:C0023895 | Liver diseases | 4 | BeFree |
| umls:C0472767 | Beta thalassemia intermedia | 4 | BeFree |
| umls:C0524620 | Metabolic Syndrome X | 4 | BeFree |
| umls:C0878682 | Ceruloplasmin deficiency | 4 | BeFree |
| umls:C0002895 | Anemia, Sickle Cell | 3 | BeFree |
| umls:C0003873 | Rheumatoid Arthritis | 3 | BeFree,LHGDN |
| umls:C0011847 | Diabetes | 3 | BeFree |
| umls:C0011849 | Diabetes Mellitus | 3 | BeFree |
| umls:C0023890 | Liver Cirrhosis | 3 | BeFree |
| umls:C0024530 | Malaria | 3 | BeFree,LHGDN |
| umls:C0162309 | Adrenoleukodystrophy | 3 | BeFree |
| umls:C0596263 | Carcinogenesis | 3 | BeFree |
| umls:C1865616 | HEMOCHROMATOSIS, TYPE 2B | 3 | CLINVAR,CTD_human,UNIPROT |
| umls:C0003850 | Arteriosclerosis | 2 | BeFree |
| umls:C0004153 | Atherosclerosis | 2 | BeFree |
| umls:C0005411 | Biliary Atresia | 2 | BeFree,CTD_human |
| umls:C0006142 | Malignant neoplasm of breast | 2 | BeFree |
| umls:C0009319 | Colitis | 2 | BeFree |
| umls:C0014130 | Endocrine System Diseases | 2 | BeFree |
| umls:C0019693 | HIV Infections | 2 | BeFree |
| umls:C0021368 | Inflammation | 2 | LHGDN |
| umls:C0024535 | Malaria, Falciparum | 2 | BeFree,LHGDN |
| umls:C0026764 | Multiple Myeloma | 2 | BeFree |
| umls:C0029928 | Ovarian Diseases | 2 | BeFree |
| umls:C0035078 | Kidney Failure | 2 | BeFree |
| umls:C0037061 | Siderosis | 2 | BeFree,LHGDN |
| umls:C0042769 | Virus Diseases | 2 | BeFree |
| umls:C0239946 | Fibrosis, Liver | 2 | BeFree |
| umls:C0271901 | Microcytic hypochromic anemia (disorder) | 2 | BeFree |
| umls:C0341439 | Chronic liver disease | 2 | BeFree |
| umls:C0400966 | Non-alcoholic Fatty Liver Disease | 2 | BeFree |
| umls:C0403447 | Chronic Kidney Insufficiency | 2 | BeFree |
| umls:C0497406 | Overweight | 2 | BeFree |
| umls:C0678222 | Breast Carcinoma | 2 | BeFree |
| umls:C0878521 | Beta thalassemia trait | 2 | BeFree |
| umls:C1561643 | Chronic Kidney Diseases | 2 | BeFree |
| umls:C1623038 | Cirrhosis | 2 | BeFree |
| umls:C1858664 | HEMOCHROMATOSIS, TYPE 3 | 2 | BeFree |
| umls:C2316810 | Chronic kidney disease stage 5 | 2 | BeFree |
| umls:C3241937 | Nonalcoholic Steatohepatitis | 2 | BeFree |
| umls:C0000786 | Spontaneous abortion | 1 | GAD |
| umls:C0001430 | Adenoma | 1 | BeFree,LHGDN |
| umls:C0002893 | Refractory anemias | 1 | BeFree |
| umls:C0004114 | Astrocytoma | 1 | BeFree |
| umls:C0008313 | Cholangitis, Sclerosing | 1 | BeFree |
| umls:C0009404 | Colorectal Neoplasms | 1 | LHGDN |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | BeFree |
| umls:C0017152 | Gastritis | 1 | BeFree |
| umls:C0019163 | Hepatitis B | 1 | BeFree |
| umls:C0019202 | Hepatolenticular Degeneration | 1 | CTD_human |
| umls:C0021390 | Inflammatory Bowel Diseases | 1 | BeFree |
| umls:C0022658 | Kidney Diseases | 1 | BeFree |
| umls:C0024141 | Lupus Erythematosus, Systemic | 1 | LHGDN |
| umls:C0024299 | Lymphoma | 1 | BeFree |
| umls:C0028756 | Obesity, Morbid | 1 | BeFree |
| umls:C0031154 | Peritonitis | 1 | BeFree |
| umls:C0032460 | Polycystic Ovary Syndrome | 1 | BeFree |
| umls:C0032461 | Polycythemia | 1 | BeFree |
| umls:C0032708 | Disorders of Porphyrin Metabolism | 1 | LHGDN |
| umls:C0032962 | Pregnancy Complications | 1 | GAD |
| umls:C0033027 | Preleukemia | 1 | BeFree |
| umls:C0036117 | Salmonella infections | 1 | BeFree |
| umls:C0041296 | Tuberculosis | 1 | BeFree |
| umls:C0149520 | Acute Cholecystitis | 1 | BeFree |
| umls:C0162566 | Porphyria Cutanea Tarda | 1 | BeFree,LHGDN |
| umls:C0238644 | Anemia, severe | 1 | BeFree |
| umls:C0242379 | Malignant neoplasm of lung | 1 | BeFree |
| umls:C0242723 | Parasitemia | 1 | BeFree |
| umls:C0268070 | Hypocupremia | 1 | BeFree |
| umls:C0271979 | Thalassemia Intermedia | 1 | BeFree |
| umls:C0275553 | Bacterial cholangitis | 1 | BeFree |
| umls:C0342793 | Malonic aciduria | 1 | BeFree |
| umls:C0376358 | Malignant neoplasm of prostate | 1 | BeFree |
| umls:C0472777 | Hemoglobin E/beta thalassemia disease | 1 | BeFree |
| umls:C0524909 | Hepatitis B, Chronic | 1 | BeFree |
| umls:C0566602 | Primary sclerosing cholangitis | 1 | BeFree |
| umls:C0600139 | Prostate carcinoma | 1 | BeFree |
| umls:C0684249 | Carcinoma of lung | 1 | BeFree |
| umls:C0740340 | Amyloidosis, Familial | 1 | LHGDN |
| umls:C0746102 | Chronic lung disease | 1 | BeFree |
| umls:C1142276 | Renal anemia | 1 | BeFree |
| umls:C1290884 | Inflammatory disorder | 1 | BeFree |
| umls:C1334815 | Multi-centric Castleman's Disease | 1 | BeFree |
| umls:C1458155 | Mammary Neoplasms | 1 | BeFree |
| umls:C1565489 | Renal Insufficiency | 1 | BeFree |
| umls:C1636149 | Macular dystrophy, corneal type 1 | 1 | BeFree |
| umls:C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 1 | GAD |
| umls:C2981142 | Refractory anemia, without ringed sideroblasts, without excess blasts | 1 | BeFree |
| umls:C3463824 | MYELODYSPLASTIC SYNDROME | 1 | BeFree |
| umls:C3714514 | Infection | 1 | LHGDN |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))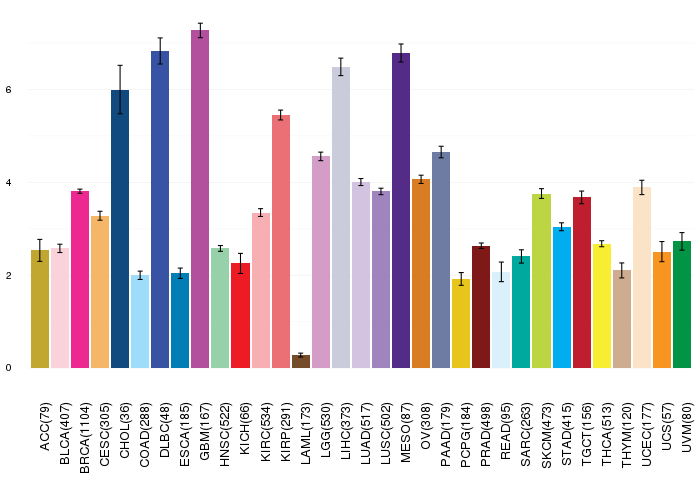
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))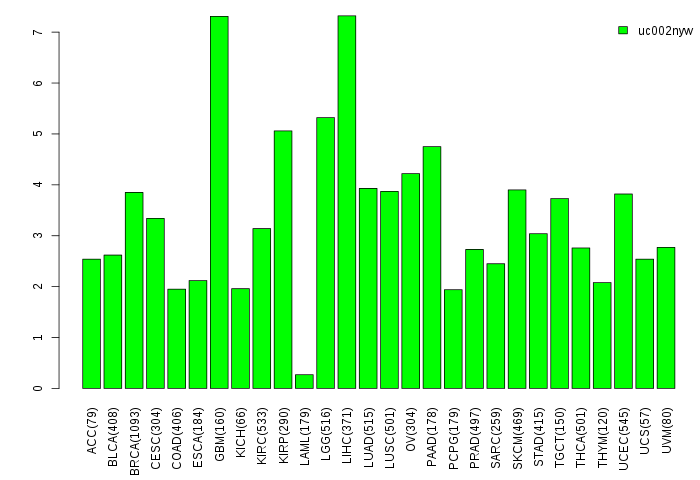
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data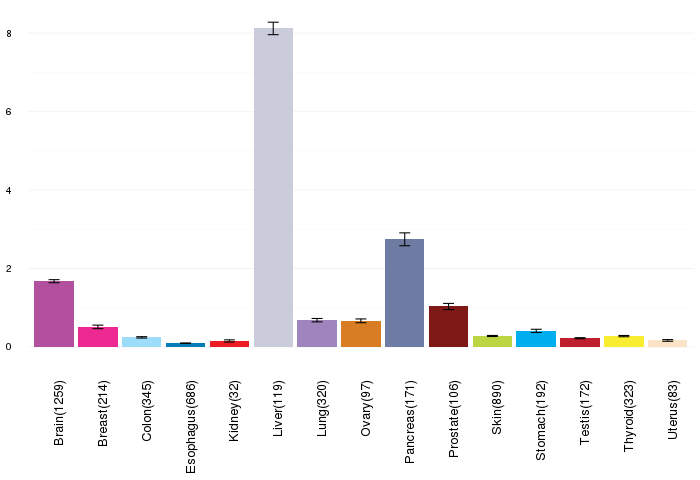
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))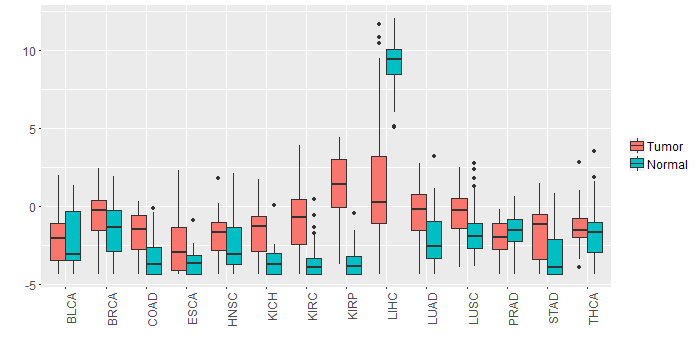
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.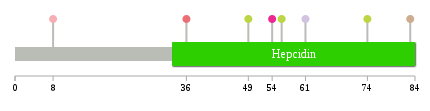

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 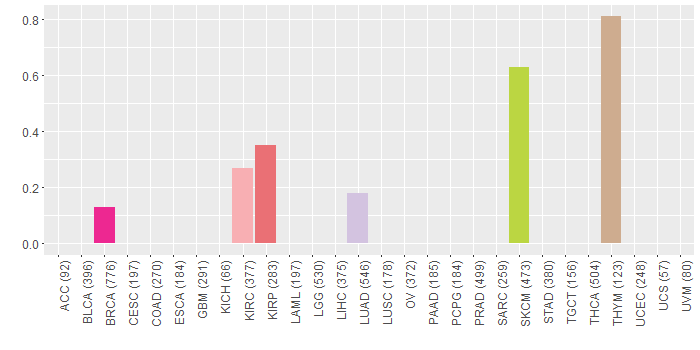
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)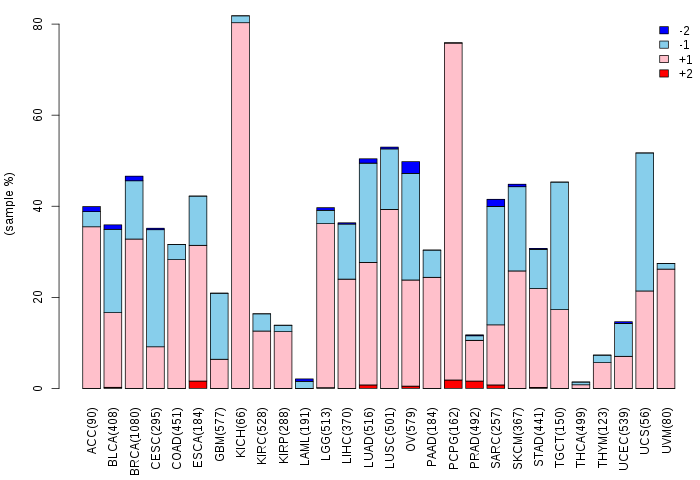
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 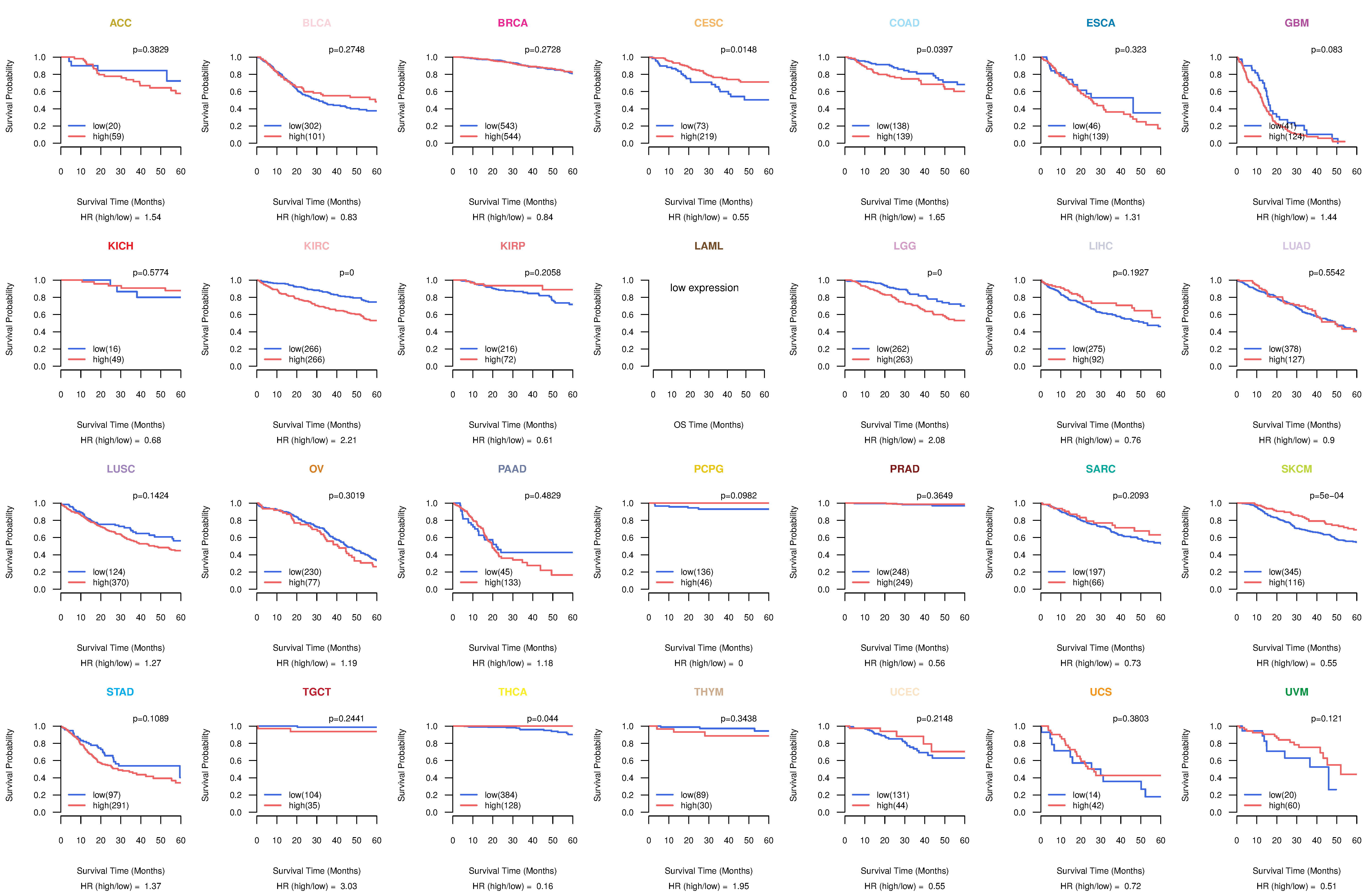
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 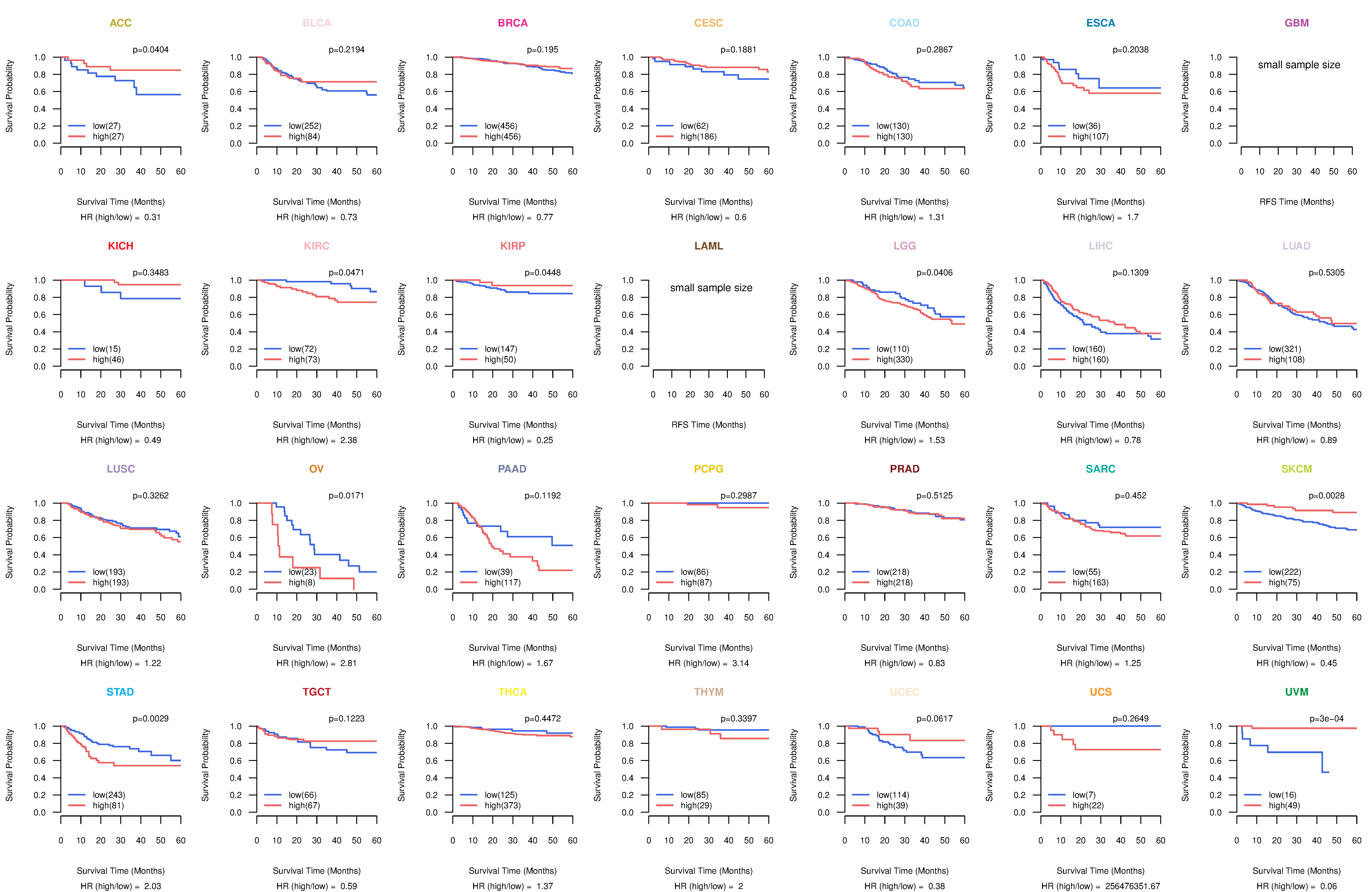
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 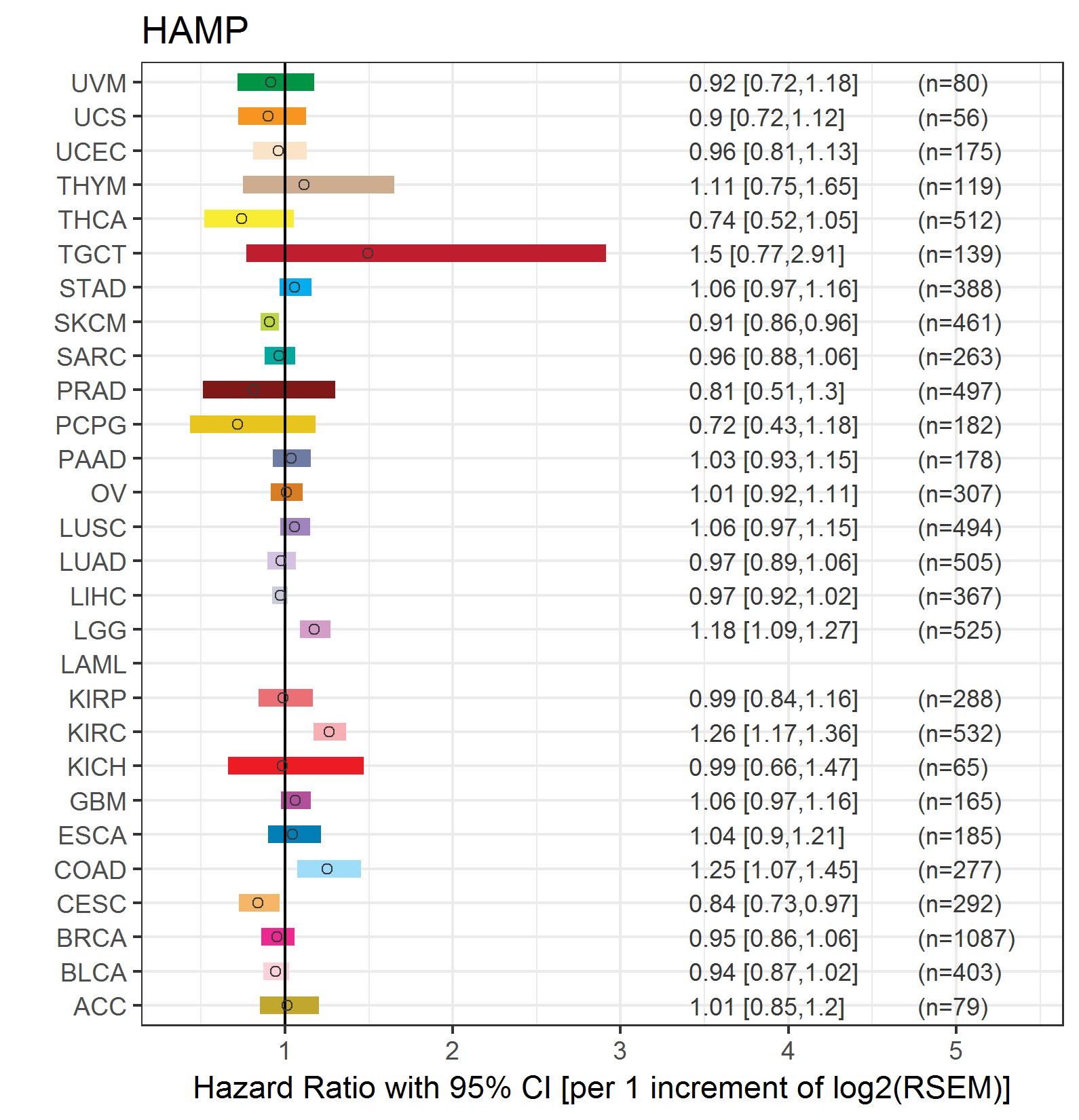
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 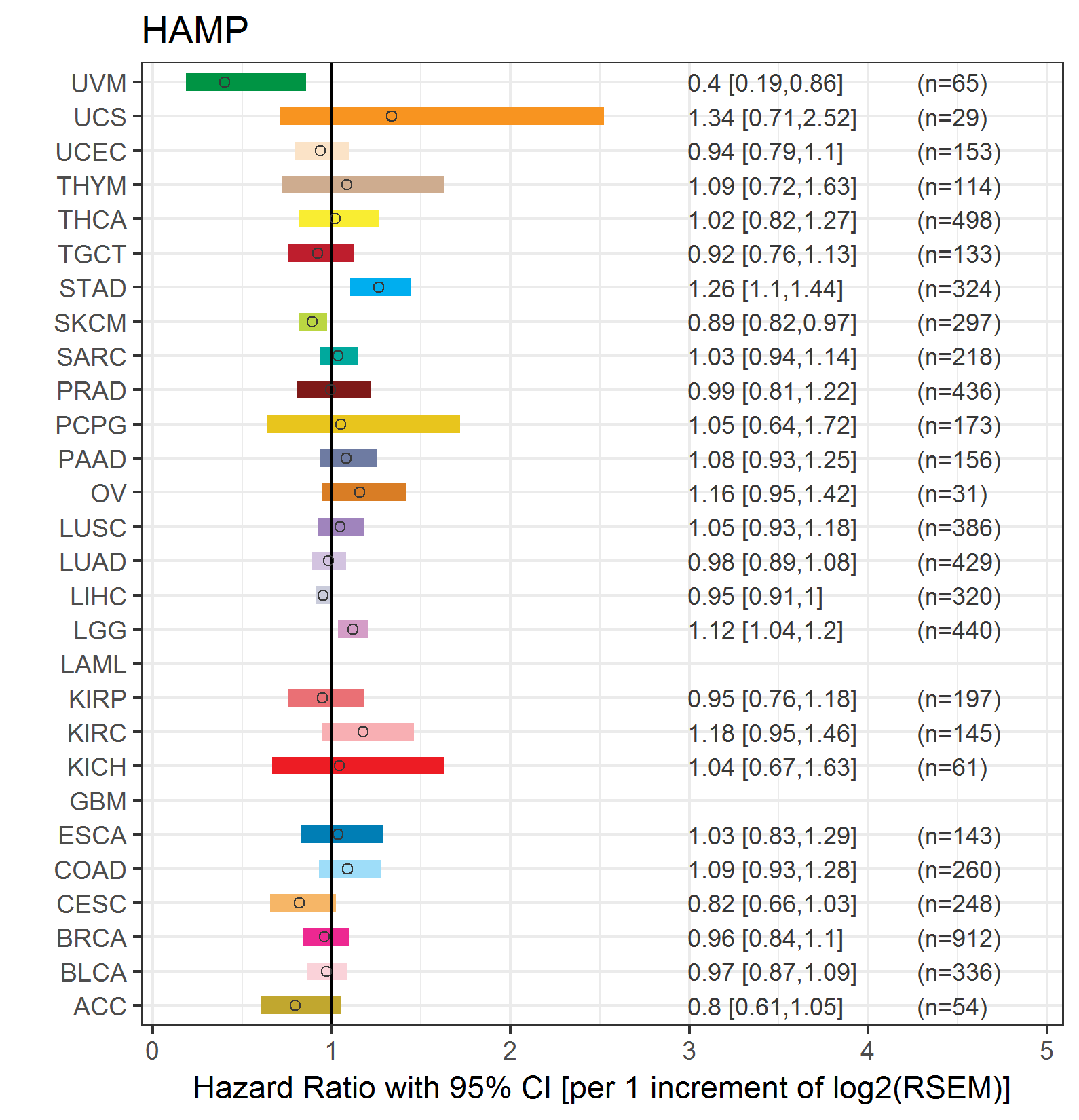
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























