| Basic gene information | Gene symbol | SFTPB |
| Gene name | surfactant protein B |
| Synonyms | PSP-B|SFTB3|SFTP3|SMDP1|SP-B |
| Cytomap | UCSC genome browser: 2p12-p11.2 |
| Type of gene | protein-coding |
| RefGenes | NM_000542.3,
NM_198843.2, |
| Description | 18 kDa pulmonary-surfactant protein6 kDa proteinpulmonary surfactant-associated protein Bpulmonary surfactant-associated proteolipid SPL(Phe) |
| Modification date | 20141207 |
| dbXrefs | MIM : 178640 |
| HGNC : HGNC |
| Ensembl : ENSG00000168878 |
| HPRD : 01523 |
| Vega : OTTHUMG00000130181 |
| Protein | UniProt:
go to UniProt's Cross Reference DB Table |
| Expression | CleanEX: HS_SFTPB |
| BioGPS: 6439 |
| Pathway | NCI Pathway Interaction Database: SFTPB |
| KEGG: SFTPB |
| REACTOME: SFTPB |
| Pathway Commons: SFTPB |
| Context | iHOP: SFTPB |
| ligand binding site mutation search in PubMed: SFTPB |
| UCL Cancer Institute: SFTPB |
| Assigned class in TissGDB* | A |
| Included tissue-specific gene expression resources | HPA,TiGER,GTEx |
| Specific-tissues in normal samples (assigned by TissGDB using HPA, TiGER, and GTEx) | Lung |
| Cancer types related to the specific-tissues in cancer samples (assigned by TissGDB using TCGA) | LUAD,LUSC |
| Reference showing the relevant tissue of SFTPB | Clinical and ultrastructural spectrum of diffuse lung disease associated with surfactant protein C mutations. Peca D, Boldrini R, Johannson J, Shieh JT, Citti A, Petrini S, Salerno T, Cazzato S, Testa R, Messina F, Onofri A, Cenacchi G, Westermark P, Ullmann N, Cogo P, Cutrera R, Danhaive O. Eur J Hum Genet. 2015 Aug;23(8):1033-41. doi: 10.1038/ejhg.2015.45. Epub 2015 Mar 18 (pmid:25782673)
go to article |
| Description by TissGene annotations | Cancer gene
|
| BRCA (tumor) | BRCA (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| COAD (tumor) | COAD (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| KICH (tumor) | KICH (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| KIRC (tumor) | KIRC (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| KIRP (tumor) | KIRP (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| LUAD (tumor) | LUAD (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| LUSC (tumor) | LUSC (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| PRAD (tumor) | PRAD (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| STAD (tumor) | STAD (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| THCA (tumor) | THCA (normal) |
| SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (tumor) | SFTPB, RARA, NKX2-1, RARG, FOXF2, CTSH (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0035220 | Respiratory Distress Syndrome, Newborn | 31 | BeFree,GAD,RGD |
| umls:C0035222 | Respiratory Distress Syndrome, Adult | 28 | BeFree,GAD,LHGDN |
| umls:C0024115 | Lung diseases | 26 | BeFree,CTD_human,GAD,LHGDN,RGD |
| umls:C1145670 | Respiratory Failure | 16 | BeFree |
| umls:C0024117 | Chronic Obstructive Airway Disease | 14 | BeFree,GAD,LHGDN |
| umls:C0476273 | Respiratory distress | 9 | BeFree |
| umls:C0206062 | Lung Diseases, Interstitial | 8 | BeFree,GAD |
| umls:C0001418 | Adenocarcinoma | 6 | BeFree |
| umls:C0006287 | Bronchopulmonary Dysplasia | 6 | BeFree,GAD |
| umls:C0006012 | Borderline Personality Disorder | 5 | BeFree |
| umls:C0032285 | Pneumonia | 5 | BeFree,GAD |
| umls:C0034050 | Pulmonary Alveolar Proteinosis | 5 | BeFree |
| umls:C0004096 | Asthma | 4 | BeFree,GAD,RGD |
| umls:C0242379 | Malignant neoplasm of lung | 4 | BeFree,GAD |
| umls:C0152013 | Adenocarcinoma of lung (disorder) | 3 | BeFree |
| umls:C0521648 | Neonatal respiratory failure | 3 | BeFree |
| umls:C0684249 | Carcinoma of lung | 3 | BeFree |
| umls:C0746102 | Chronic lung disease | 3 | BeFree |
| umls:C0852283 | Respiratory Distress Syndrome | 3 | GAD |
| umls:C0007131 | Non-Small Cell Lung Carcinoma | 2 | BeFree |
| umls:C0019284 | Diaphragmatic Hernia | 2 | CTD_human |
| umls:C0024121 | Lung Neoplasms | 2 | CTD_human,GAD |
| umls:C0035204 | Respiration Disorders | 2 | BeFree |
| umls:C0035235 | Respiratory Syncytial Virus Infections | 2 | BeFree,GAD |
| umls:C0035242 | Respiratory Tract Diseases | 2 | BeFree |
| umls:C0243026 | Sepsis | 2 | GAD |
| umls:C0694549 | Community acquired pneumonia | 2 | BeFree |
| umls:C3714514 | Infection | 2 | LHGDN |
| umls:C0000768 | Congenital Abnormality | 1 | BeFree |
| umls:C0003130 | Anoxia | 1 | GAD |
| umls:C0004626 | Pneumonia, Bacterial | 1 | RGD |
| umls:C0006274 | Bronchiolitis, Viral | 1 | GAD |
| umls:C0007121 | Bronchogenic Carcinoma | 1 | BeFree |
| umls:C0010674 | Cystic Fibrosis | 1 | LHGDN |
| umls:C0013990 | Pathological accumulation of air in tissues | 1 | GAD |
| umls:C0020440 | Hypercapnia | 1 | GAD |
| umls:C0020459 | Hyperinsulinism | 1 | BeFree |
| umls:C0020542 | Pulmonary Hypertension | 1 | GAD |
| umls:C0020630 | Hypophosphatasia | 1 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 1 | BeFree |
| umls:C0028754 | Obesity | 1 | RGD |
| umls:C0034063 | Pulmonary Edema | 1 | BeFree |
| umls:C0034067 | Pulmonary Emphysema | 1 | BeFree |
| umls:C0034069 | Pulmonary Fibrosis | 1 | GAD |
| umls:C0036421 | Systemic Scleroderma | 1 | BeFree,GAD,LHGDN |
| umls:C0149782 | Squamous cell carcinoma of lung | 1 | BeFree |
| umls:C0152021 | Congenital heart disease | 1 | BeFree |
| umls:C0206682 | Follicular thyroid carcinoma | 1 | BeFree |
| umls:C0220810 | Congenital defects | 1 | BeFree |
| umls:C0221757 | alpha 1-Antitrypsin Deficiency | 1 | GAD |
| umls:C0235833 | Congenital diaphragmatic hernia | 1 | BeFree |
| umls:C0242706 | Hyperoxia | 1 | RGD |
| umls:C0264936 | Secondary pulmonary hypertension | 1 | BeFree |
| umls:C0265783 | Congenital hypoplasia of lung | 1 | BeFree |
| umls:C0376358 | Malignant neoplasm of prostate | 1 | BeFree |
| umls:C0600139 | Prostate carcinoma | 1 | BeFree |
| umls:C0600260 | Lung Diseases, Obstructive | 1 | BeFree |
| umls:C0876973 | Infectious disease of lung | 1 | BeFree |
| umls:C1135361 | Persistent pulmonary hypertension | 1 | BeFree |
| umls:C1334455 | Pulmonary Sclerosing Hemangioma | 1 | LHGDN |
| umls:C1800706 | Idiopathic Pulmonary Fibrosis | 1 | BeFree |
| umls:C1968602 | Surfactant Metabolism Dysfunction, Pulmonary, 1 | 1 | CLINVAR,CTD_human,ORPHANET,UNIPROT |
| umls:C2931035 | Pulmonary alveolar proteinosis, congenital | 1 | BeFree |
| umls:C2931829 | RDS - infants | 1 | BeFree |
| umls:C3179539 | Congenital Deficiency of Pulmonary Surfactant Protein B | 1 | BeFree |
| umls:C3711368 | Surfactant Dysfunction | 1 | BeFree |
| umls:C1968593 | Respiratory Distress Syndrome In Premature Infants | 0 | ORPHANET |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))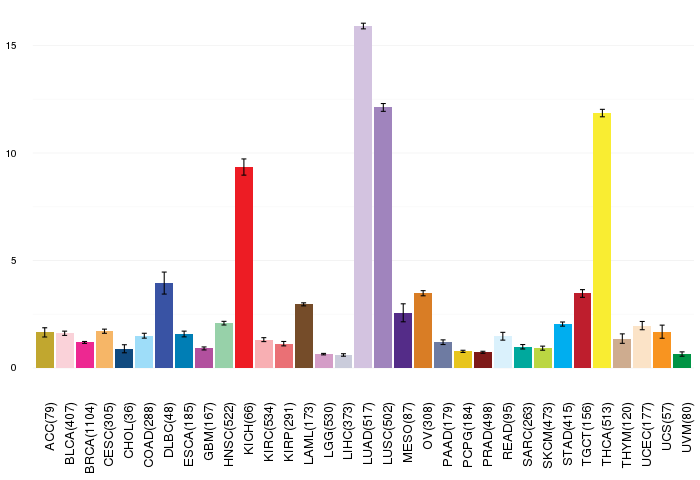
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))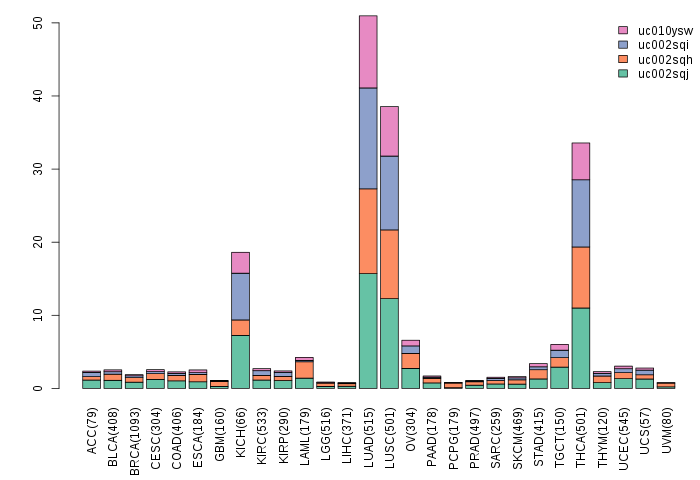
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data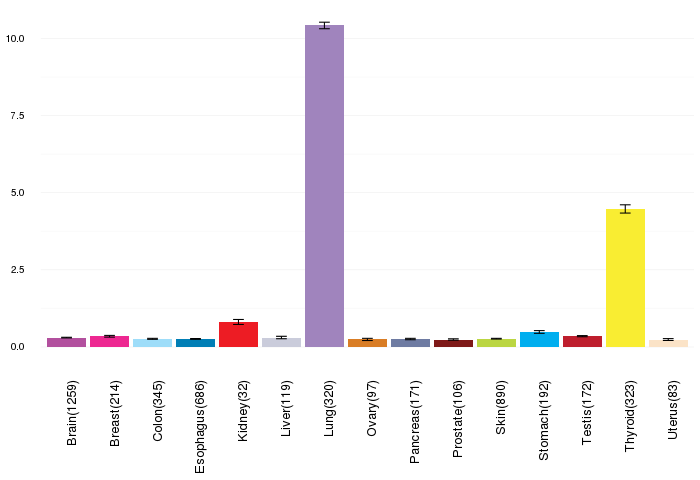
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))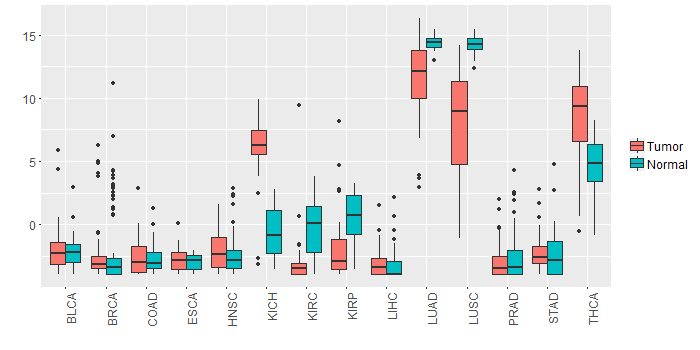
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 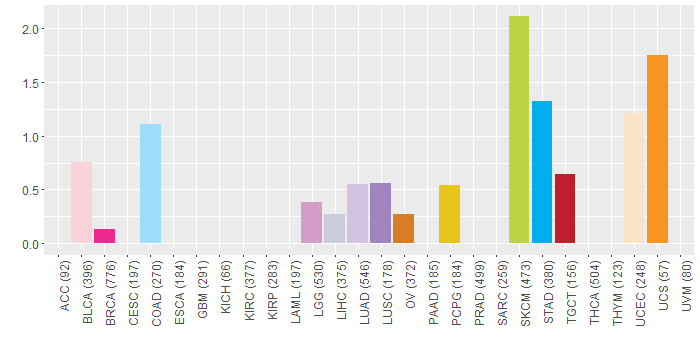
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)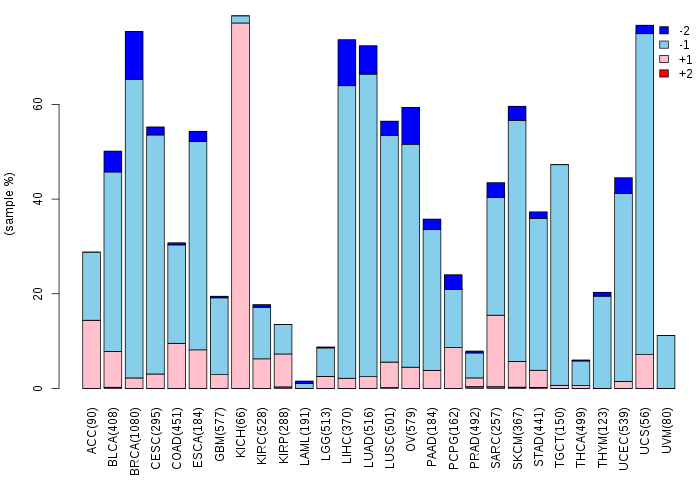
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























