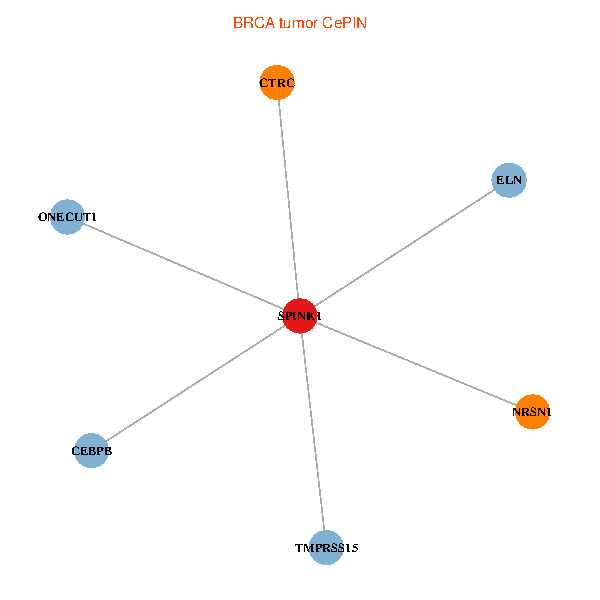
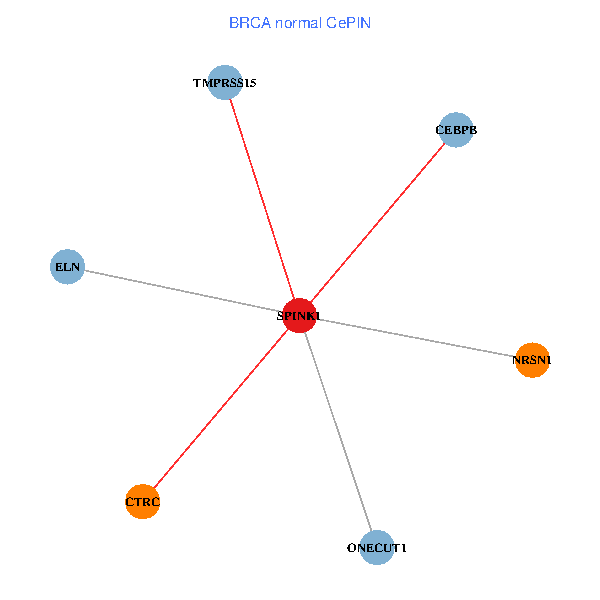
| BRCA (tumor) | BRCA (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
 |  |
|
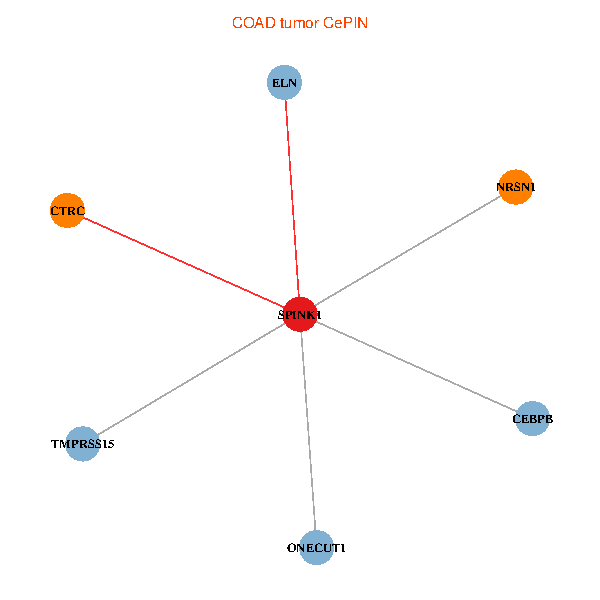
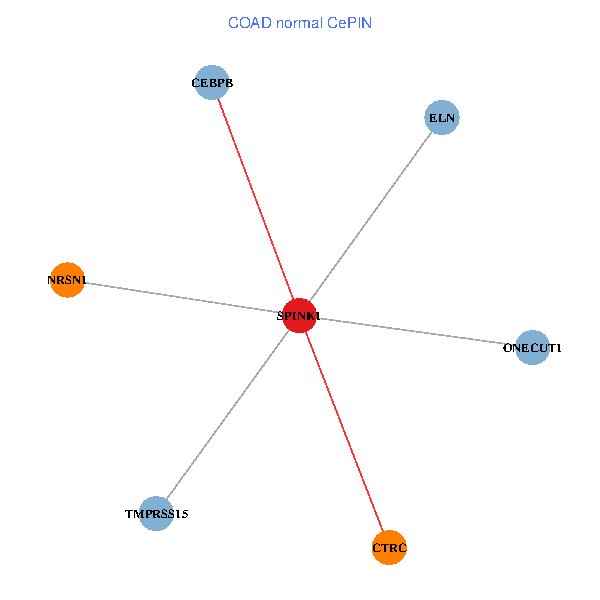
| COAD (tumor) | COAD (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
 |  |
|
| KICH (tumor) | KICH (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
 |  |
|
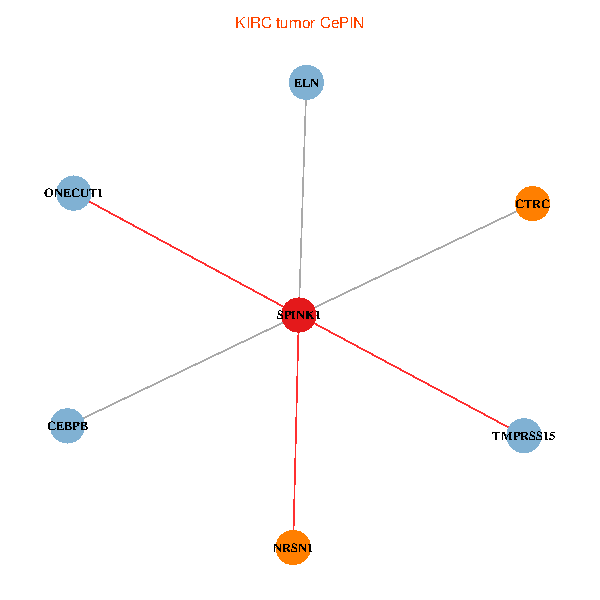
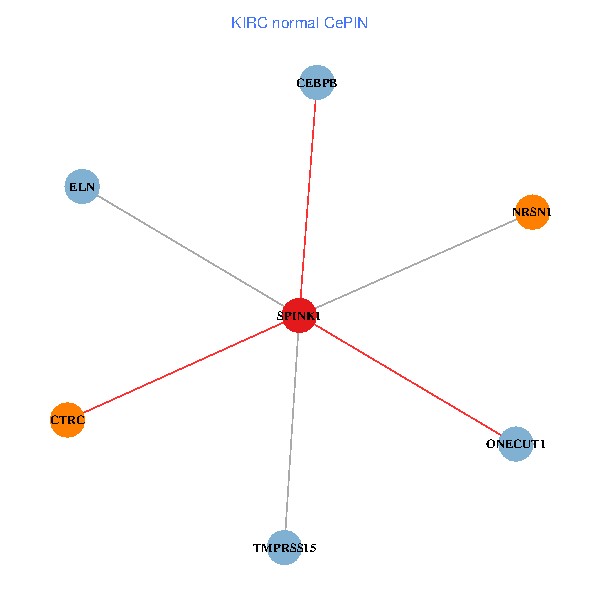
| KIRC (tumor) | KIRC (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
 |  |
|
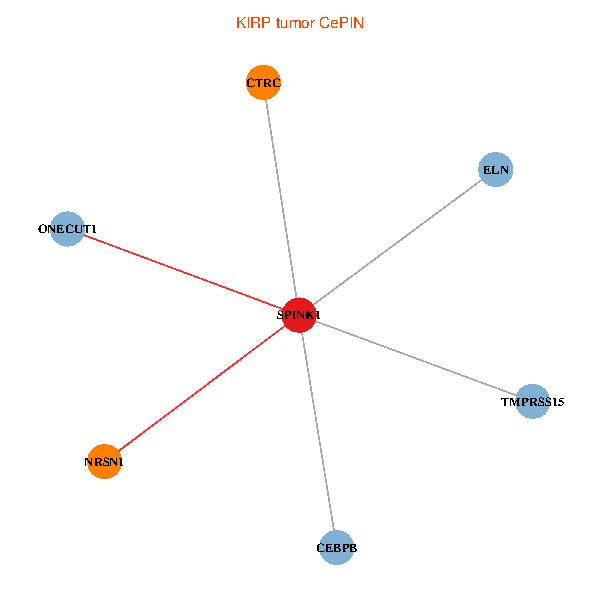
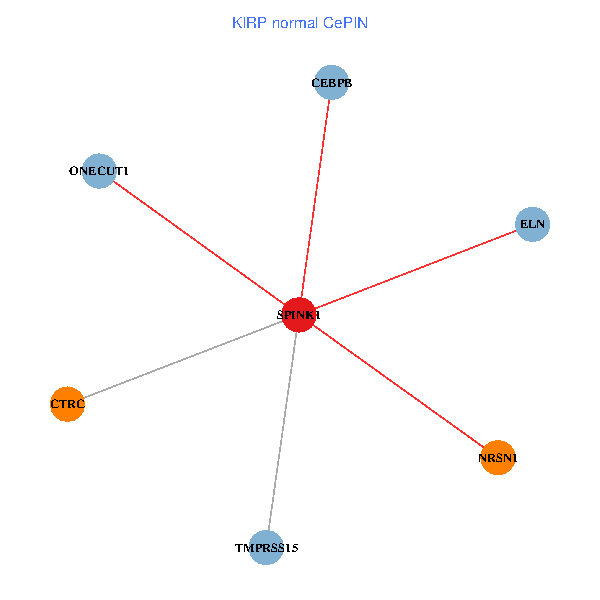
| KIRP (tumor) | KIRP (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
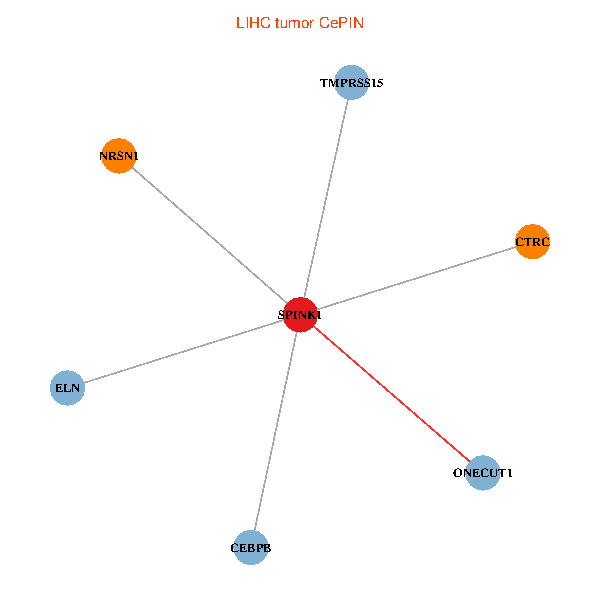 | 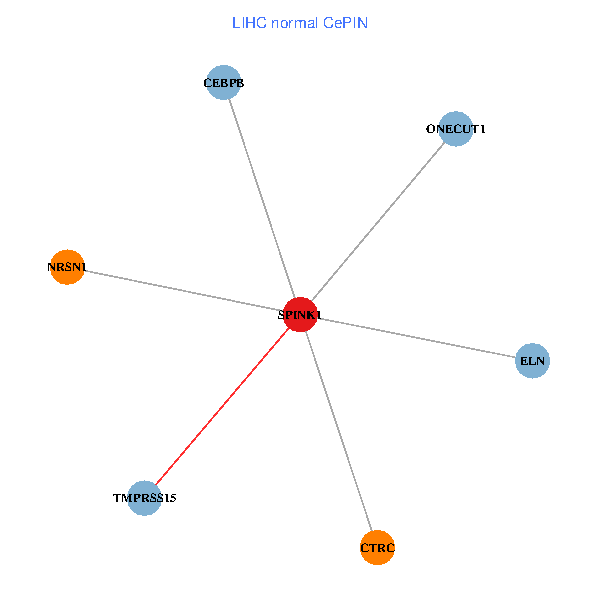 |
|
| LUAD (tumor) | LUAD (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
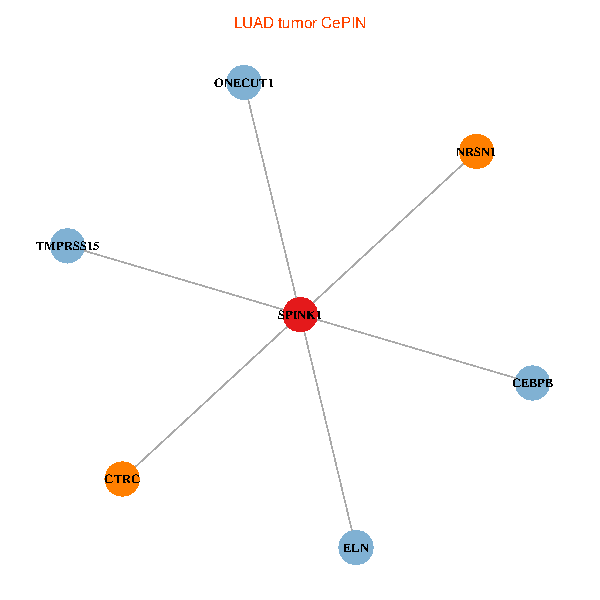 | 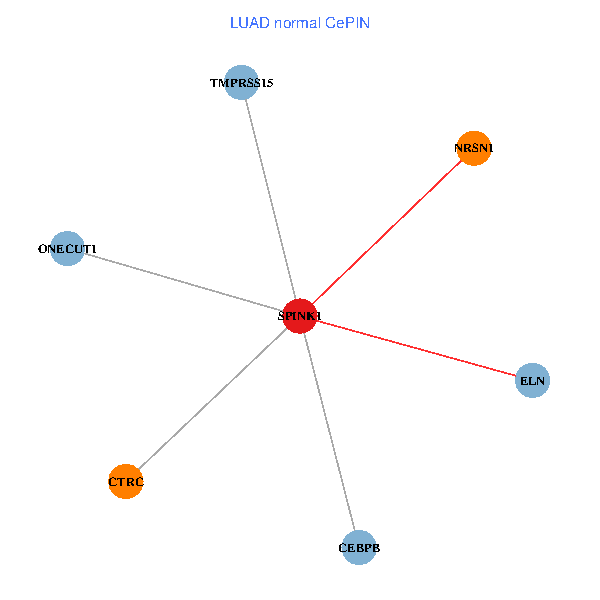 |
|
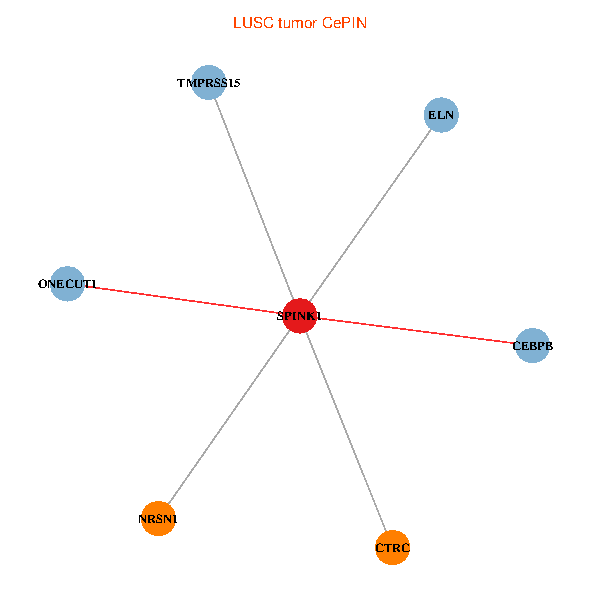
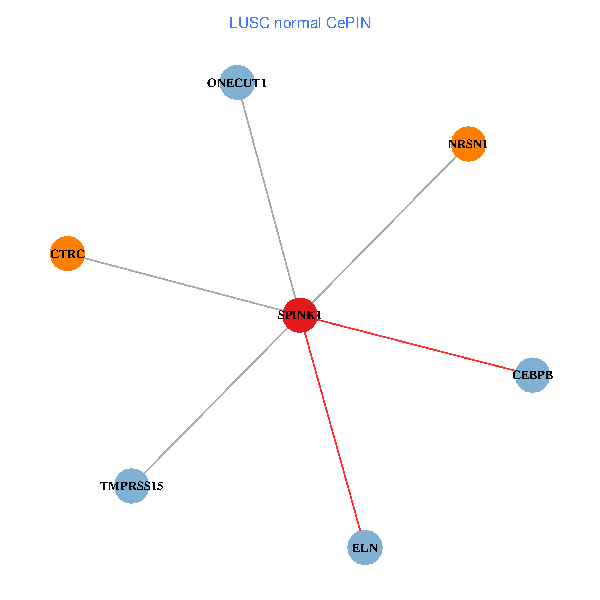
| LUSC (tumor) | LUSC (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
 |  |
|
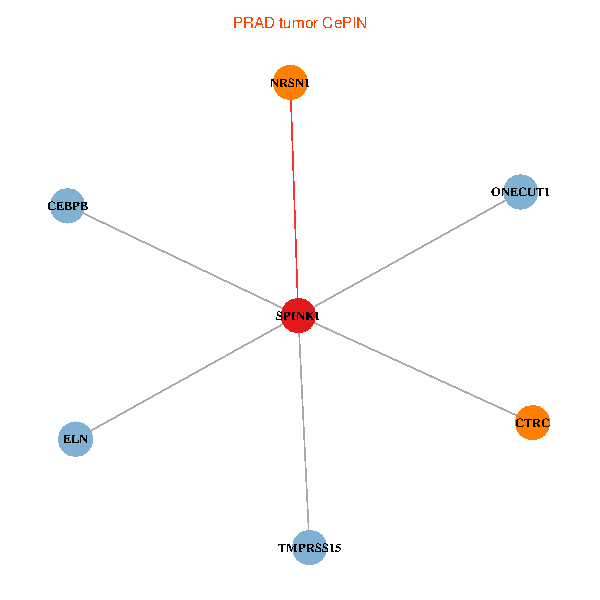
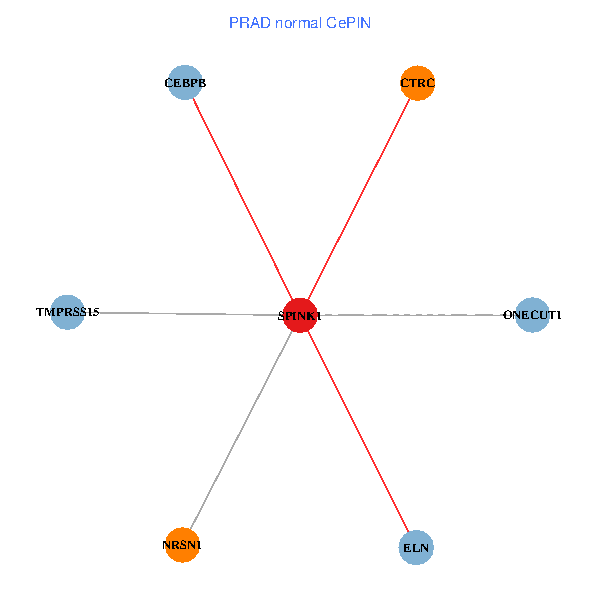
| PRAD (tumor) | PRAD (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
 |  |
|
| STAD (tumor) | STAD (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
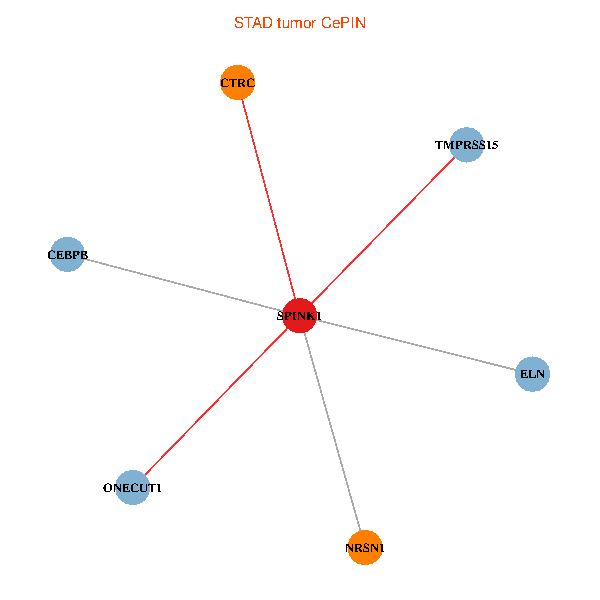 | 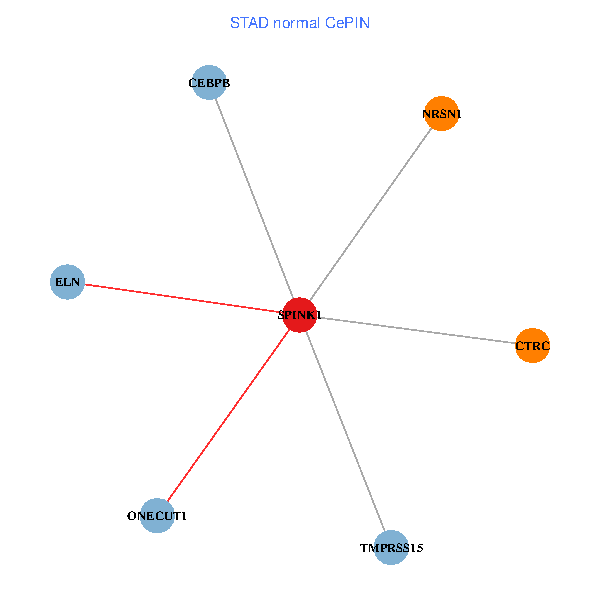 |
|
| THCA (tumor) | THCA (normal) |
| SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (tumor) | SPINK1, ONECUT1, CEBPB, ELN, TMPRSS15, CTRC, NRSN1 (normal) |
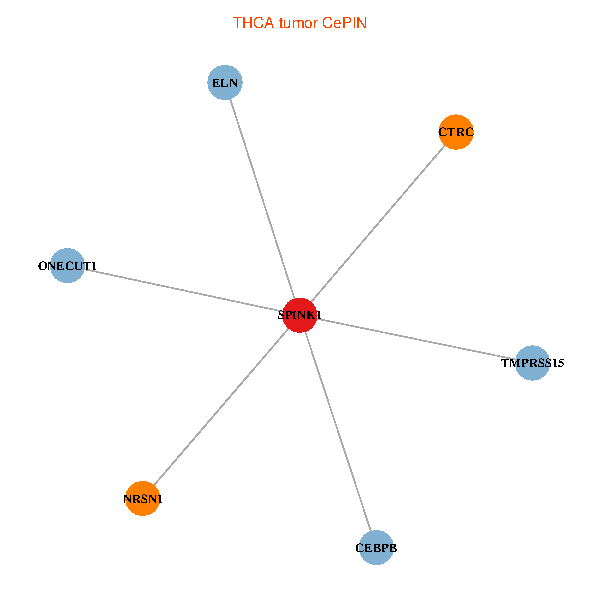 | 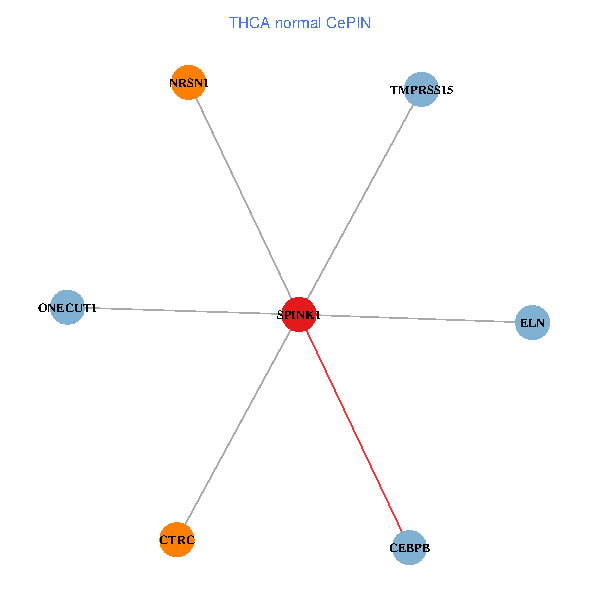 |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0149521 | Pancreatitis, Chronic | 103 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0030305 | Pancreatitis | 76 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0238339 | Hereditary pancreatitis | 21 | BeFree,CLINVAR,UNIPROT |
| umls:C0001339 | Acute pancreatitis | 18 | BeFree,GAD |
| umls:C0341471 | Idiopathic chronic pancreatitis | 18 | BeFree,GAD |
| umls:C0376358 | Malignant neoplasm of prostate | 13 | BeFree |
| umls:C0600139 | Prostate carcinoma | 12 | BeFree |
| umls:C0010674 | Cystic Fibrosis | 11 | BeFree,GAD,LHGDN |
| umls:C0376670 | Pancreatitis, Alcoholic | 11 | BeFree,GAD,LHGDN |
| umls:C1842402 | TROPICAL CALCIFIC PANCREATITIS | 11 | BeFree,CLINVAR,CTD_human,GAD,ORPHANET,UNIPROT |
| umls:C0235974 | Pancreatic carcinoma | 10 | BeFree |
| umls:C0346647 | Malignant neoplasm of pancreas | 10 | BeFree,GAD |
| umls:C0747198 | pancreatitis idiopathic | 10 | BeFree |
| umls:C2239176 | Liver carcinoma | 6 | BeFree |
| umls:C0008354 | Cholera | 5 | BeFree |
| umls:C0011849 | Diabetes Mellitus | 5 | BeFree,LHGDN |
| umls:C0030286 | Pancreatic Diseases | 5 | BeFree,GAD |
| umls:C0267937 | Acute recurrent pancreatitis | 5 | BeFree |
| umls:C0011847 | Diabetes | 4 | BeFree |
| umls:C0271642 | Fibrocalculous pancreatic diabetes | 4 | BeFree |
| umls:C0001418 | Adenocarcinoma | 3 | BeFree |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 3 | BeFree,GAD |
| umls:C0029925 | Ovarian Carcinoma | 3 | BeFree |
| umls:C0030297 | Pancreatic Neoplasm | 3 | CTD_human,GAD,LHGDN |
| umls:C0162565 | Acute intermittent porphyria | 3 | BeFree |
| umls:C0266270 | Pancreas divisum | 3 | BeFree,GAD |
| umls:C0281361 | Adenocarcinoma of pancreas | 3 | BeFree |
| umls:C0005684 | Malignant neoplasm of urinary bladder | 2 | BeFree |
| umls:C0006663 | Calcinosis | 2 | GAD |
| umls:C0007102 | Malignant tumor of colon | 2 | BeFree |
| umls:C0007570 | Celiac Disease | 2 | GAD |
| umls:C0009404 | Colorectal Neoplasms | 2 | BeFree,LHGDN |
| umls:C0015695 | Fatty Liver | 2 | BeFree,GAD |
| umls:C0023903 | Liver neoplasms | 2 | BeFree |
| umls:C0033578 | Prostatic Neoplasms | 2 | BeFree,LHGDN |
| umls:C0175683 | Citrullinemia | 2 | BeFree |
| umls:C0221002 | Hyperparathyroidism, Primary | 2 | BeFree,GAD |
| umls:C0267963 | Exocrine pancreatic insufficiency | 2 | BeFree,GAD |
| umls:C0400966 | Non-alcoholic Fatty Liver Disease | 2 | BeFree |
| umls:C0677886 | Epithelial ovarian cancer | 2 | BeFree |
| umls:C0699790 | Colon Carcinoma | 2 | BeFree |
| umls:C0699885 | Carcinoma of bladder | 2 | BeFree |
| umls:C0810032 | Pancreatic disorders (not diabetes) | 2 | BeFree |
| umls:C1140680 | Malignant neoplasm of ovary | 2 | BeFree |
| umls:C2609129 | Autoimmune pancreatitis | 2 | BeFree |
| umls:C0001430 | Adenoma | 1 | BeFree |
| umls:C0005699 | Blast Phase | 1 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 1 | BeFree |
| umls:C0007134 | Renal Cell Carcinoma | 1 | BeFree |
| umls:C0007137 | Squamous cell carcinoma | 1 | BeFree |
| umls:C0009375 | Colonic Neoplasms | 1 | LHGDN |
| umls:C0009376 | Colonic Polyps | 1 | BeFree |
| umls:C0009402 | Colorectal Carcinoma | 1 | BeFree |
| umls:C0010068 | Coronary heart disease | 1 | BeFree |
| umls:C0011854 | Diabetes Mellitus, Insulin-Dependent | 1 | BeFree |
| umls:C0011881 | Diabetic Nephropathy | 1 | LHGDN |
| umls:C0017185 | Gastrointestinal Neoplasms | 1 | LHGDN |
| umls:C0019163 | Hepatitis B | 1 | BeFree |
| umls:C0019189 | Hepatitis, Chronic | 1 | BeFree |
| umls:C0019196 | Hepatitis C | 1 | BeFree |
| umls:C0019693 | HIV Infections | 1 | BeFree |
| umls:C0020437 | Hypercalcemia | 1 | BeFree |
| umls:C0020473 | Hyperlipidemia | 1 | GAD |
| umls:C0021051 | Immunologic Deficiency Syndromes | 1 | BeFree |
| umls:C0021368 | Inflammation | 1 | RGD |
| umls:C0021655 | Insulin Resistance | 1 | GAD |
| umls:C0022661 | Kidney Failure, Chronic | 1 | GAD |
| umls:C0023467 | Leukemia, Myelocytic, Acute | 1 | BeFree |
| umls:C0023473 | Myeloid Leukemia, Chronic | 1 | BeFree |
| umls:C0023890 | Liver Cirrhosis | 1 | BeFree |
| umls:C0024299 | Lymphoma | 1 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 1 | BeFree |
| umls:C0028259 | Nodule | 1 | BeFree |
| umls:C0030293 | Pancreatic Insufficiency | 1 | BeFree |
| umls:C0030583 | Parotitis | 1 | BeFree |
| umls:C0032927 | Precancerous Conditions | 1 | BeFree |
| umls:C0035078 | Kidney Failure | 1 | BeFree |
| umls:C0038356 | Stomach Neoplasms | 1 | LHGDN |
| umls:C0041234 | Chagas Disease | 1 | BeFree |
| umls:C0085694 | Chronic cholecystitis | 1 | BeFree |
| umls:C0153676 | Secondary malignant neoplasm of lung | 1 | BeFree |
| umls:C0162309 | Adrenoleukodystrophy | 1 | BeFree |
| umls:C0178874 | Tumor Progression | 1 | BeFree |
| umls:C0205645 | Adenocarcinoma, Tubular | 1 | BeFree |
| umls:C0272138 | Erythroblastosis | 1 | BeFree |
| umls:C0280100 | Solid tumour | 1 | BeFree |
| umls:C0339985 | Idiopathic bronchiectasis | 1 | BeFree |
| umls:C0345905 | Intrahepatic Cholangiocarcinoma | 1 | BeFree |
| umls:C0524620 | Metabolic Syndrome X | 1 | BeFree |
| umls:C0678222 | Breast Carcinoma | 1 | BeFree |
| umls:C0740457 | Malignant neoplasm of kidney | 1 | BeFree |
| umls:C0878500 | Intraepithelial Neoplasia | 1 | BeFree |
| umls:C1168327 | High-Grade Prostatic Intraepithelial Neoplasia | 1 | BeFree |
| umls:C1290807 | Diarrheal disorder | 1 | BeFree |
| umls:C1290884 | Inflammatory disorder | 1 | BeFree |
| umls:C1378703 | Renal carcinoma | 1 | BeFree |
| umls:C1512409 | Hepatocarcinogenesis | 1 | BeFree |
| umls:C1527249 | Colorectal Cancer | 1 | BeFree |
| umls:C1527349 | Ductal Breast Carcinoma | 1 | BeFree |
| umls:C1565489 | Renal Insufficiency | 1 | BeFree |
| umls:C1623038 | Cirrhosis | 1 | BeFree |
| umls:C1704272 | Benign Prostatic Hyperplasia | 1 | BeFree |
| umls:C1863844 | Adult-onset citrullinemia type 2 | 1 | BeFree |
| umls:C1997910 | Citrin deficiency | 1 | BeFree |
| umls:C2931038 | Pancreatic carcinoma, familial | 1 | BeFree |
| umls:C1969419 | PANCREATITIS, CHRONIC, SUSCEPTIBILITY TO | 0 | CLINVAR |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))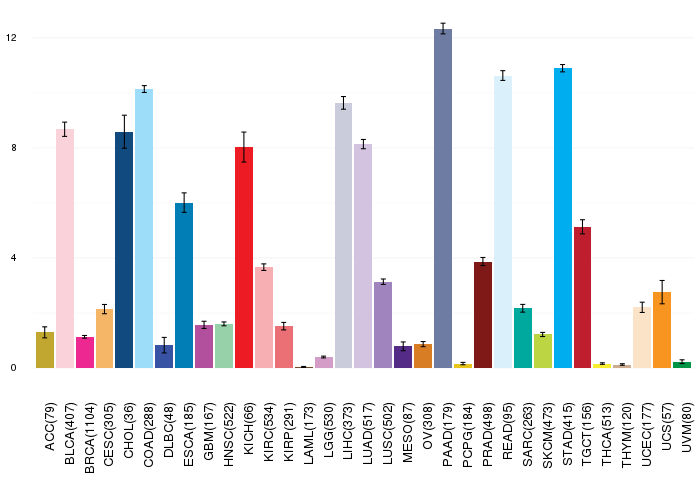
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))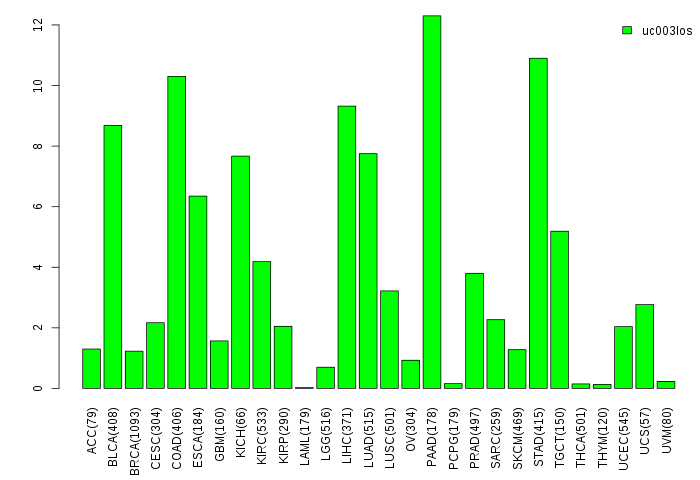
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data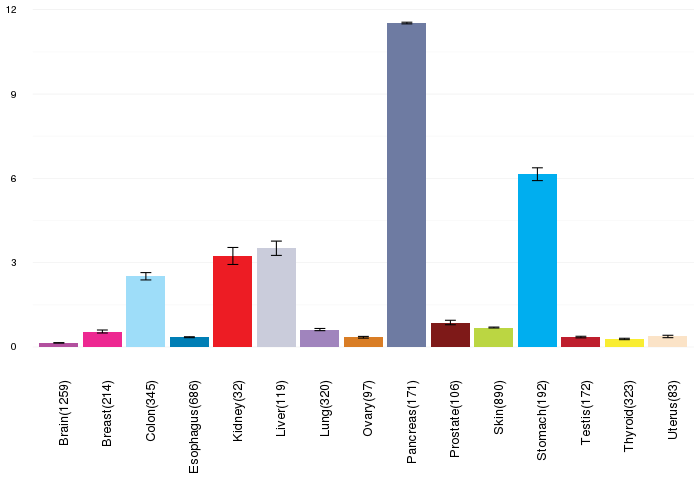
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))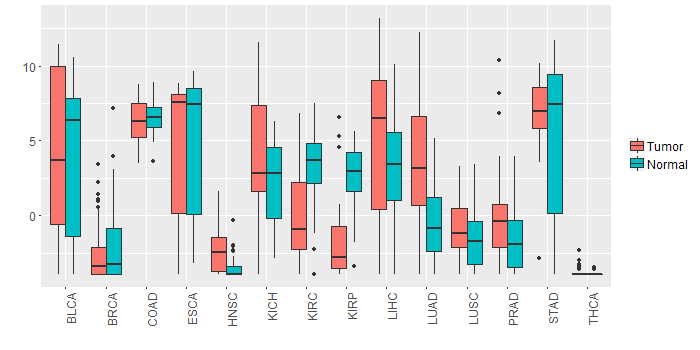
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.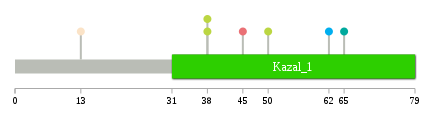

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 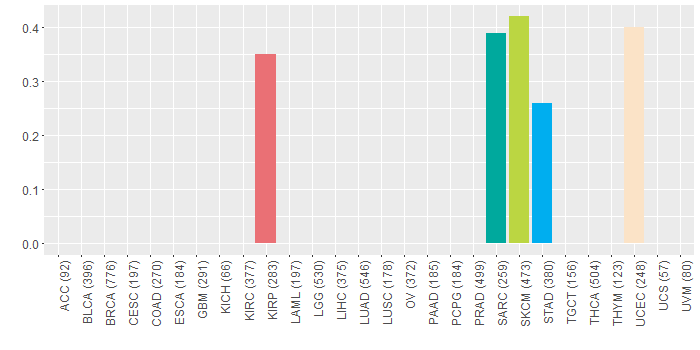
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)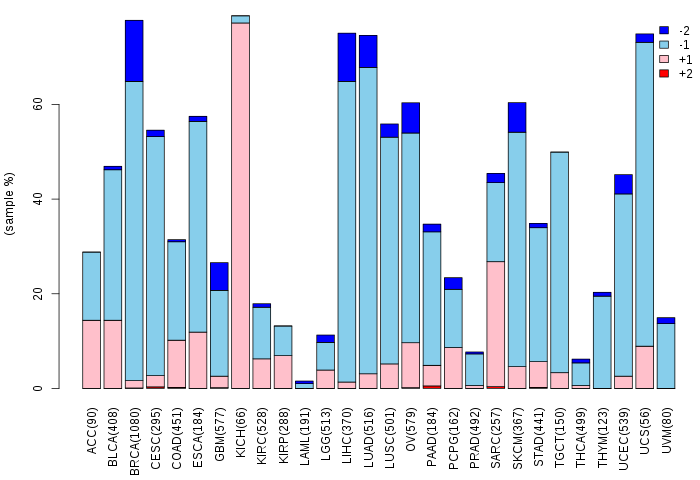
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 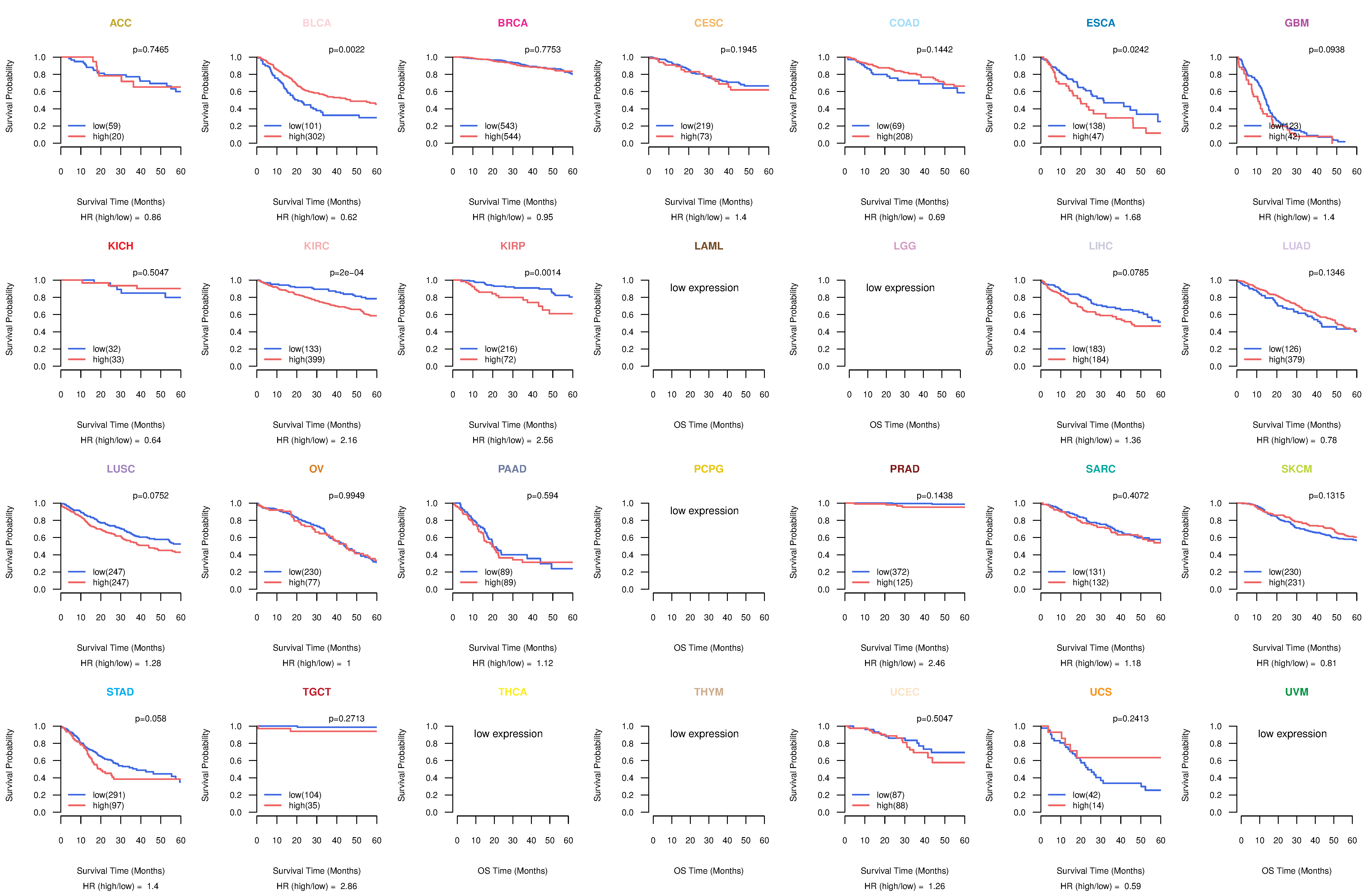
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 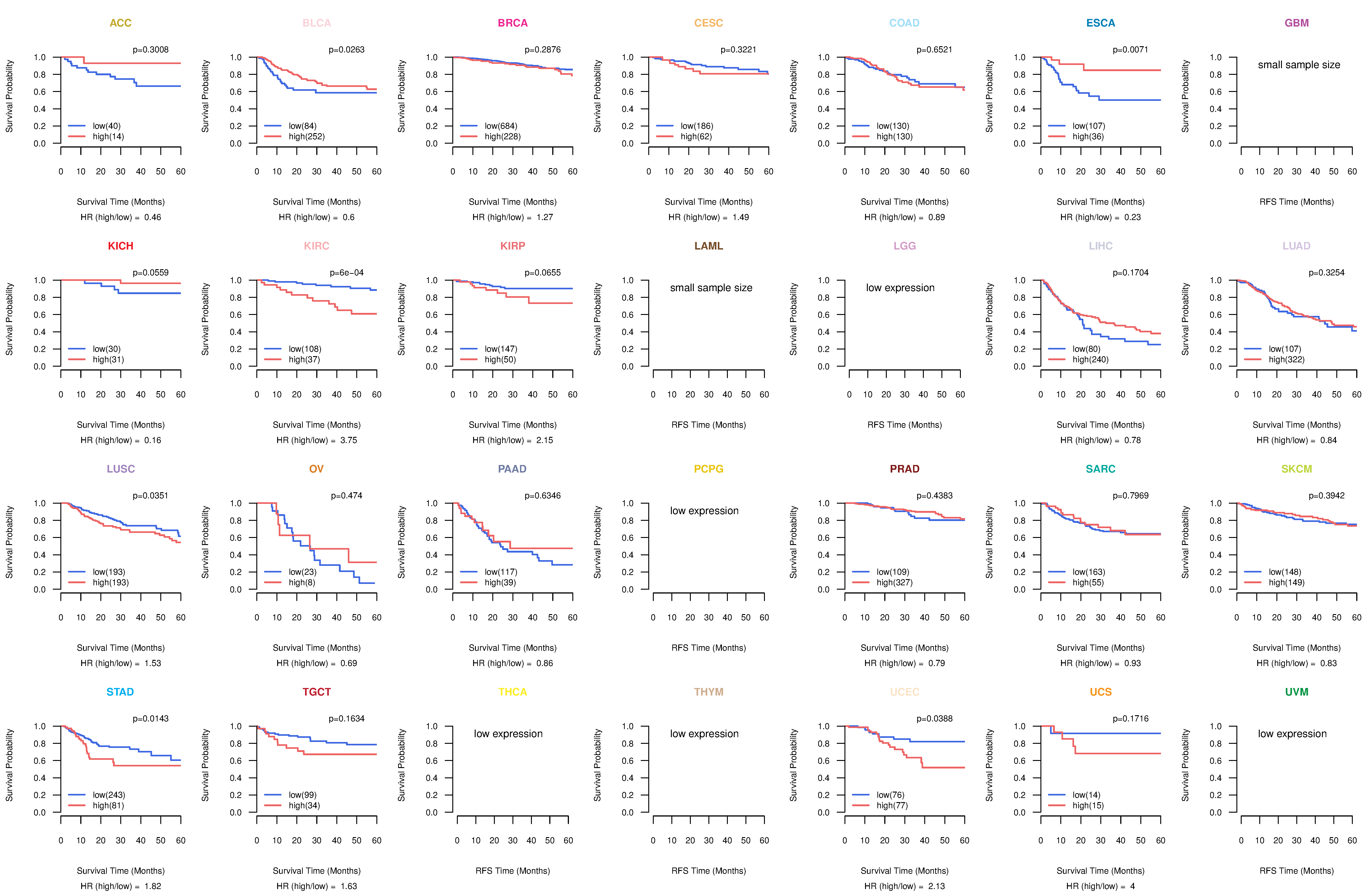
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 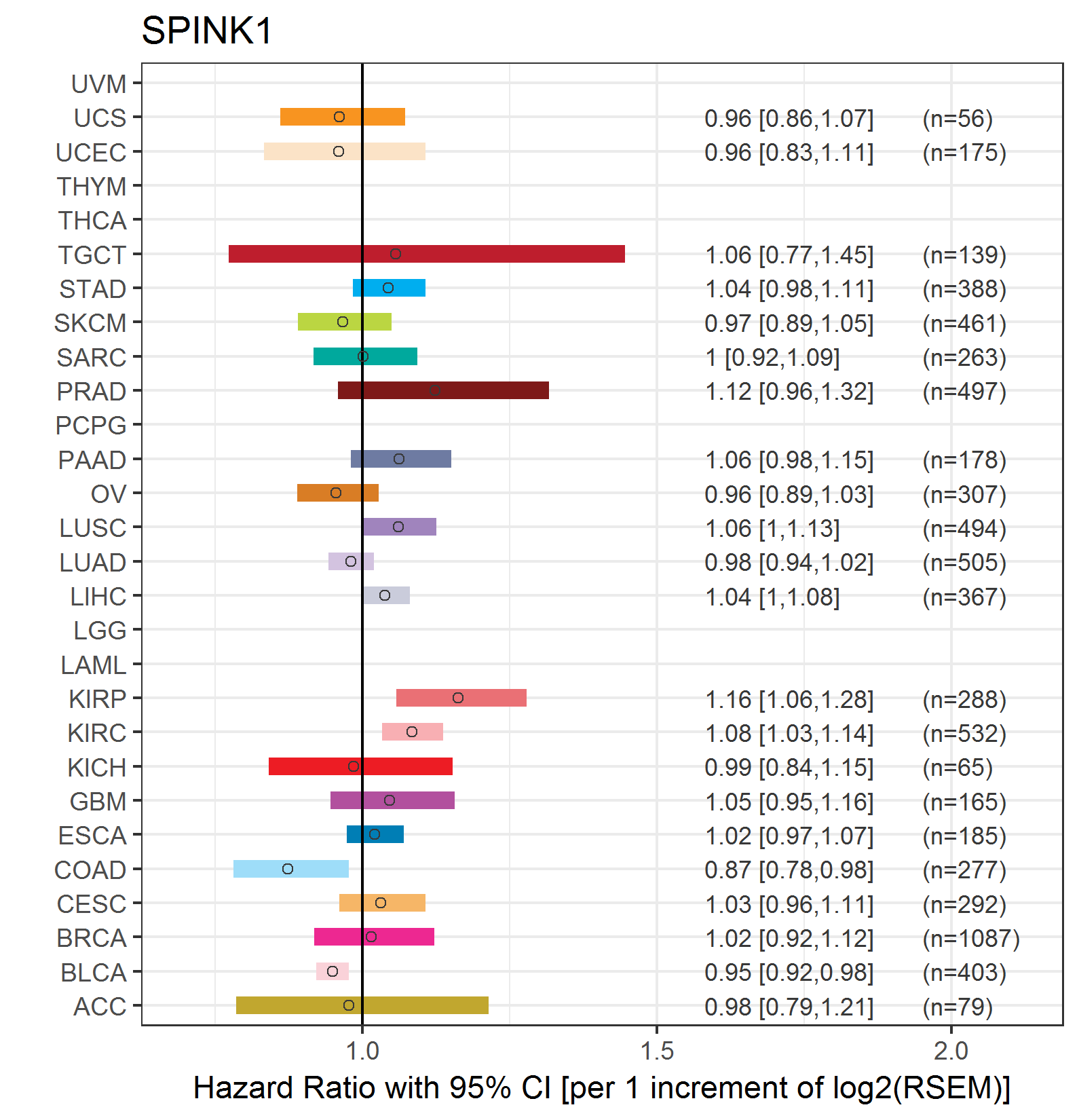
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 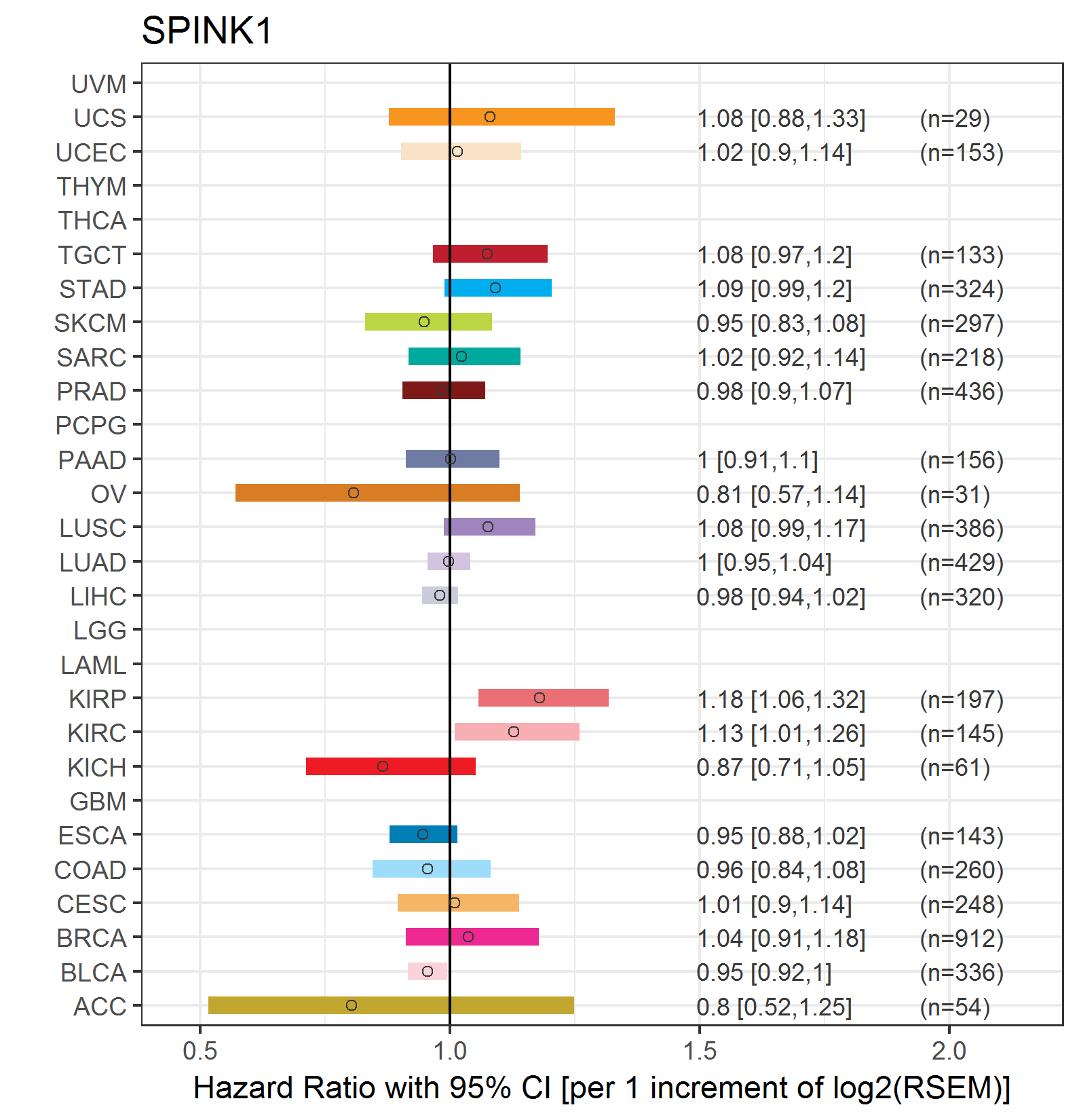
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























