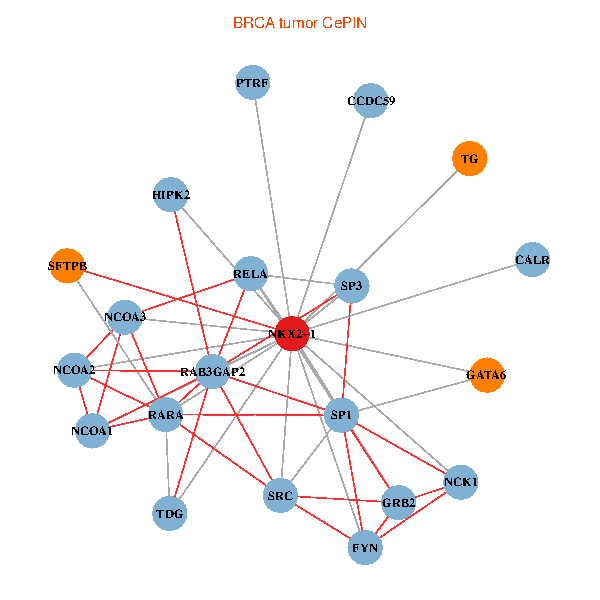
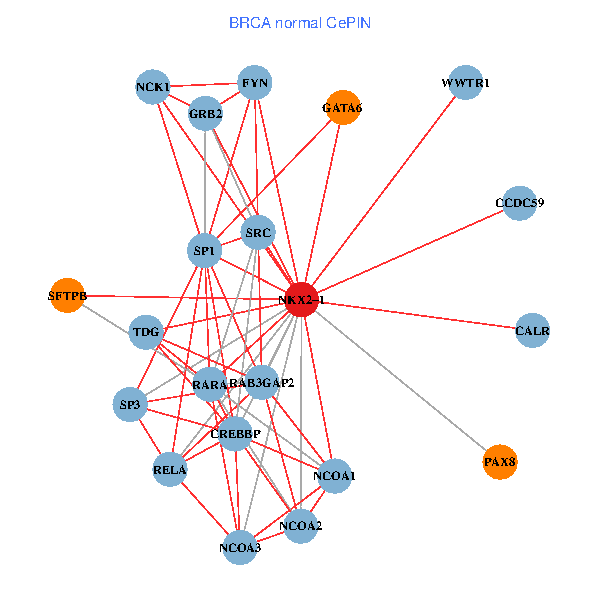
| BRCA (tumor) | BRCA (normal) |
| NKX2-1, NCK1, NCOA3, HIPK2, SP1, GRB2, RELA, SRC, RARA, NCOA1, FYN, NCOA2, RAB3GAP2, CALR, SP3, TDG, CCDC59, GATA6, PTRF, TG, SFTPB (tumor) | NKX2-1, NCK1, NCOA3, SP1, GRB2, RELA, SRC, RARA, CREBBP, NCOA1, FYN, NCOA2, RAB3GAP2, CALR, SP3, TDG, WWTR1, PAX8, CCDC59, GATA6, SFTPB (normal) |
 |  |
|
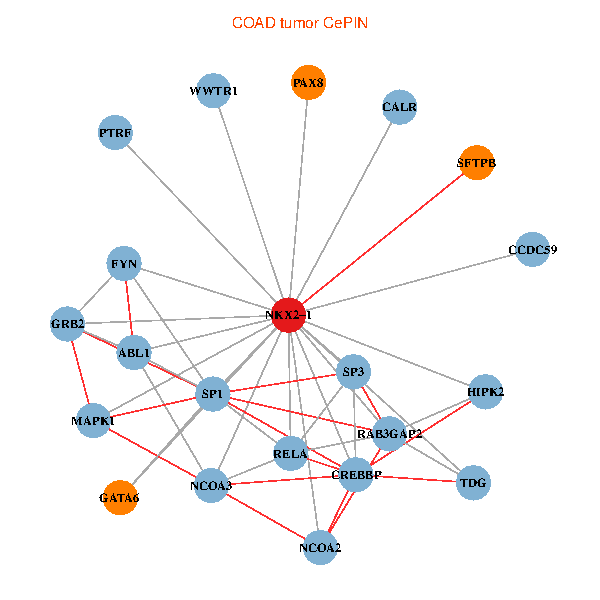
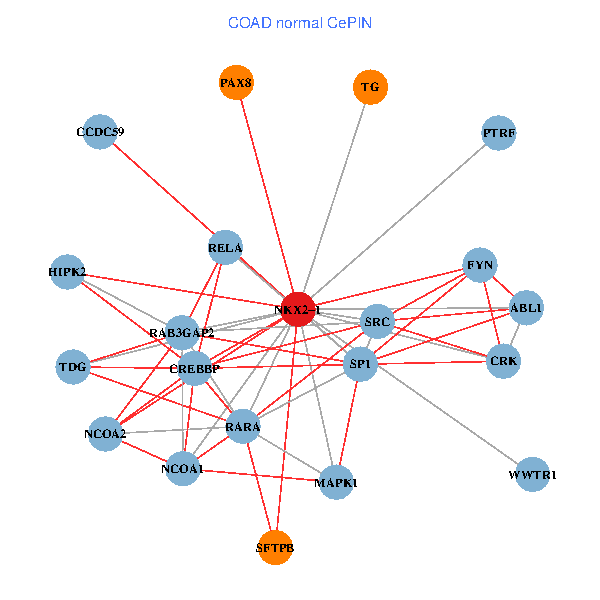
| COAD (tumor) | COAD (normal) |
| NKX2-1, NCOA3, HIPK2, SP1, GRB2, RELA, MAPK1, CREBBP, FYN, NCOA2, RAB3GAP2, ABL1, CALR, SP3, TDG, WWTR1, PAX8, CCDC59, GATA6, PTRF, SFTPB (tumor) | NKX2-1, HIPK2, SP1, RELA, SRC, MAPK1, RARA, CREBBP, NCOA1, FYN, NCOA2, RAB3GAP2, ABL1, CRK, TDG, WWTR1, PAX8, CCDC59, PTRF, TG, SFTPB (normal) |
 |  |
|
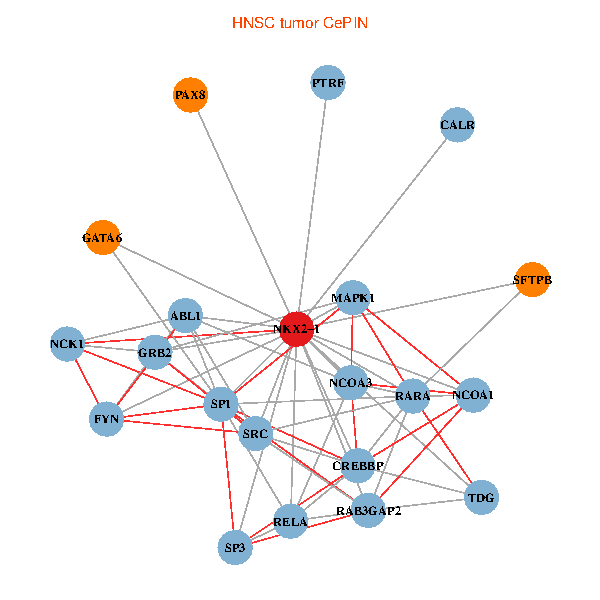
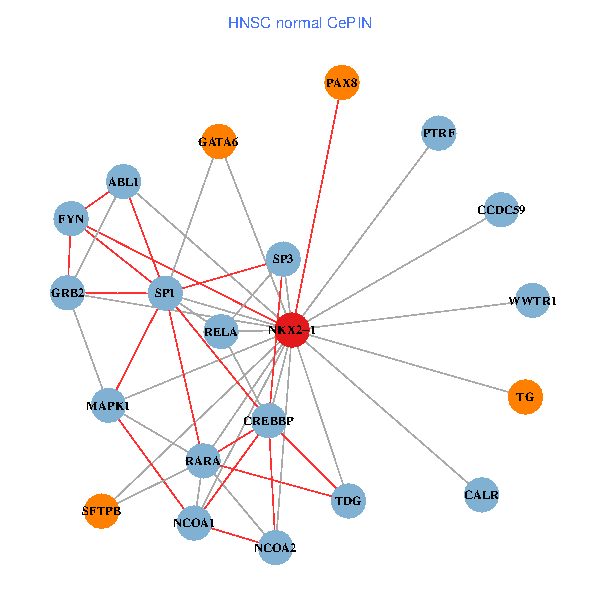
| HNSC (tumor) | HNSC (normal) |
| NKX2-1, NCK1, NCOA3, SP1, GRB2, RELA, SRC, MAPK1, RARA, CREBBP, NCOA1, FYN, RAB3GAP2, ABL1, CALR, SP3, TDG, PAX8, GATA6, PTRF, SFTPB (tumor) | NKX2-1, SP1, GRB2, RELA, MAPK1, RARA, CREBBP, NCOA1, FYN, NCOA2, ABL1, CALR, SP3, TDG, WWTR1, PAX8, CCDC59, GATA6, PTRF, TG, SFTPB (normal) |
 |  |
|
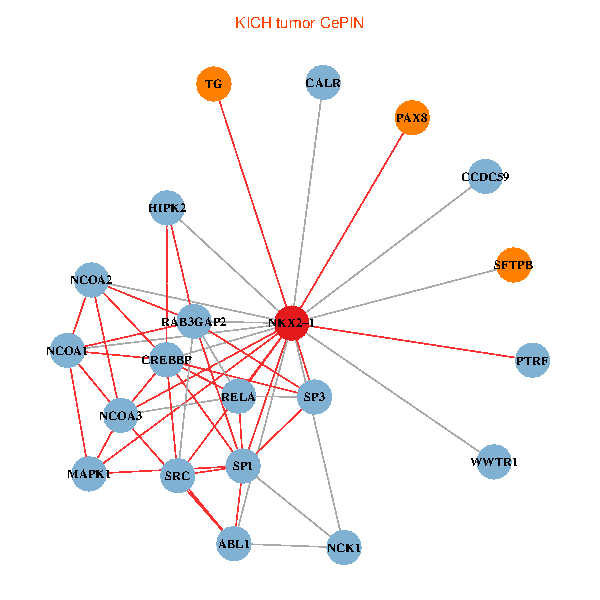
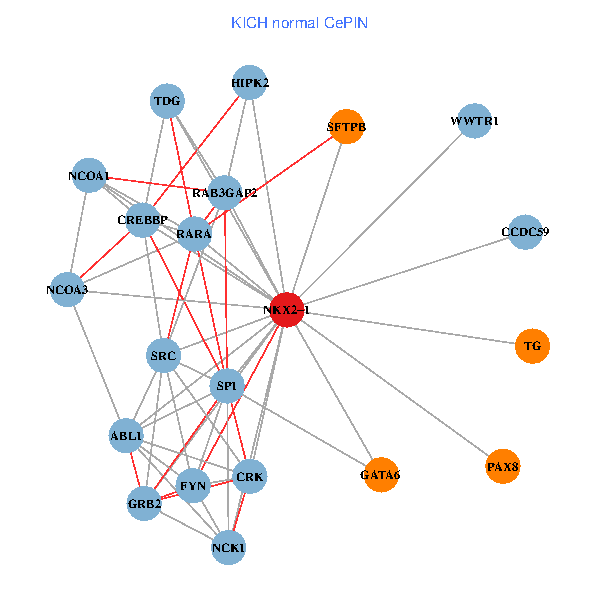
| KICH (tumor) | KICH (normal) |
| NKX2-1, NCK1, NCOA3, HIPK2, SP1, RELA, SRC, MAPK1, CREBBP, NCOA1, NCOA2, RAB3GAP2, ABL1, CALR, SP3, WWTR1, PAX8, CCDC59, PTRF, TG, SFTPB (tumor) | NKX2-1, NCK1, NCOA3, HIPK2, SP1, GRB2, SRC, RARA, CREBBP, NCOA1, FYN, RAB3GAP2, ABL1, CRK, TDG, WWTR1, PAX8, CCDC59, GATA6, TG, SFTPB (normal) |
 |  |
|
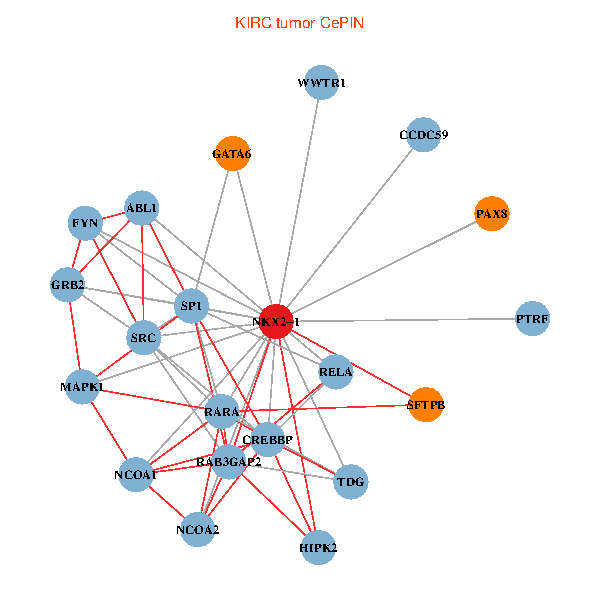
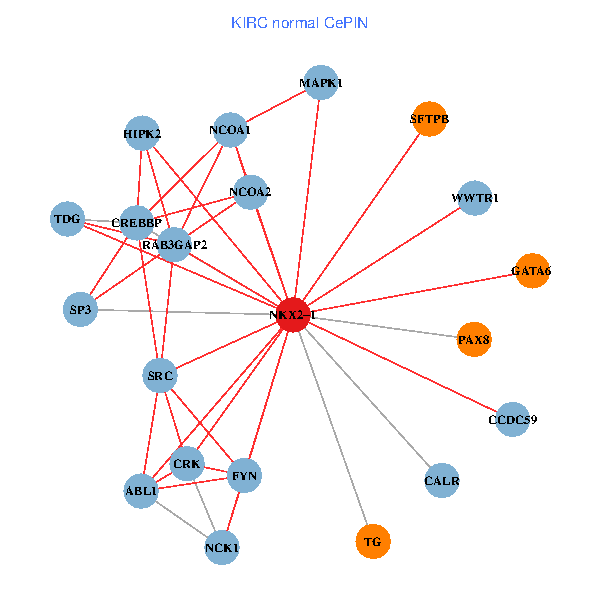
| KIRC (tumor) | KIRC (normal) |
| NKX2-1, HIPK2, SP1, GRB2, RELA, SRC, MAPK1, RARA, CREBBP, NCOA1, FYN, NCOA2, RAB3GAP2, ABL1, TDG, WWTR1, PAX8, CCDC59, GATA6, PTRF, SFTPB (tumor) | NKX2-1, NCK1, HIPK2, SRC, MAPK1, CREBBP, NCOA1, FYN, NCOA2, RAB3GAP2, ABL1, CALR, SP3, CRK, TDG, WWTR1, PAX8, CCDC59, GATA6, TG, SFTPB (normal) |
 |  |
|
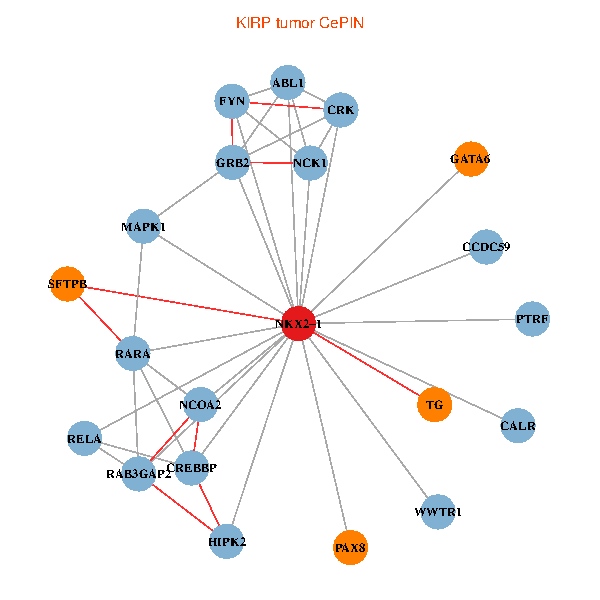
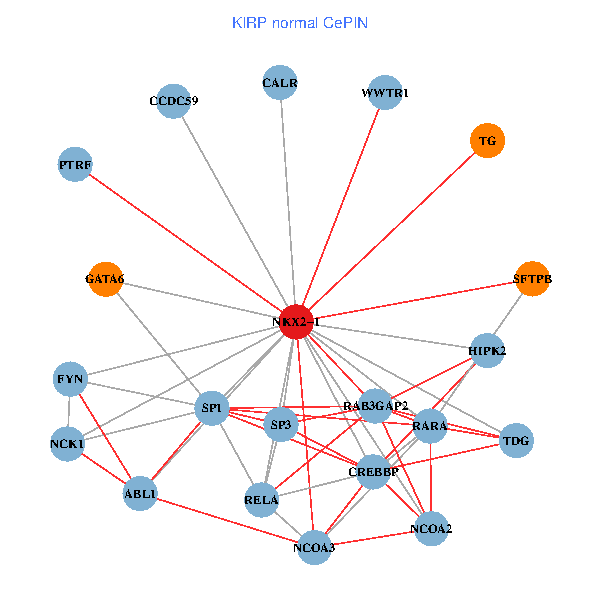
| KIRP (tumor) | KIRP (normal) |
| NKX2-1, NCK1, HIPK2, GRB2, RELA, MAPK1, RARA, CREBBP, FYN, NCOA2, RAB3GAP2, ABL1, CALR, CRK, WWTR1, PAX8, CCDC59, GATA6, PTRF, TG, SFTPB (tumor) | NKX2-1, NCK1, NCOA3, HIPK2, SP1, RELA, RARA, CREBBP, FYN, NCOA2, RAB3GAP2, ABL1, CALR, SP3, TDG, WWTR1, CCDC59, GATA6, PTRF, TG, SFTPB (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| NKX2-1, NCK1, HIPK2, SP1, GRB2, RELA, MAPK1, CREBBP, NCOA1, NCOA2, RAB3GAP2, ABL1, CALR, SP3, CRK, TDG, WWTR1, PAX8, CCDC59, TG, SFTPB (tumor) | NKX2-1, NCK1, NCOA3, HIPK2, SP1, MAPK1, RARA, CREBBP, NCOA1, FYN, RAB3GAP2, ABL1, CALR, SP3, TDG, WWTR1, PAX8, GATA6, PTRF, TG, SFTPB (normal) |
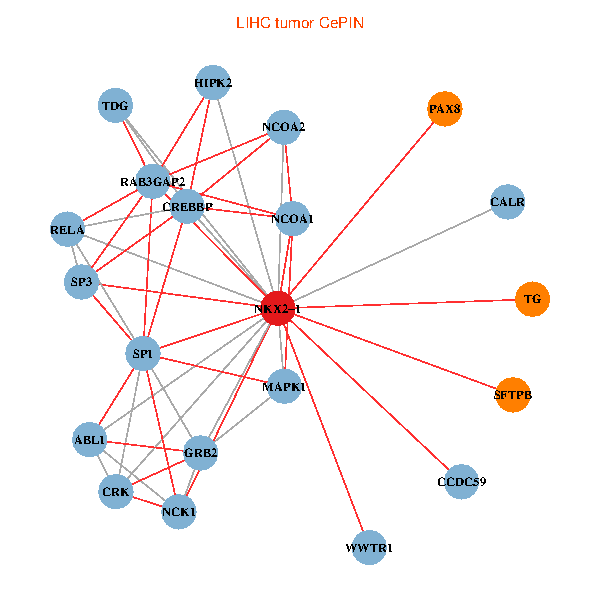 | 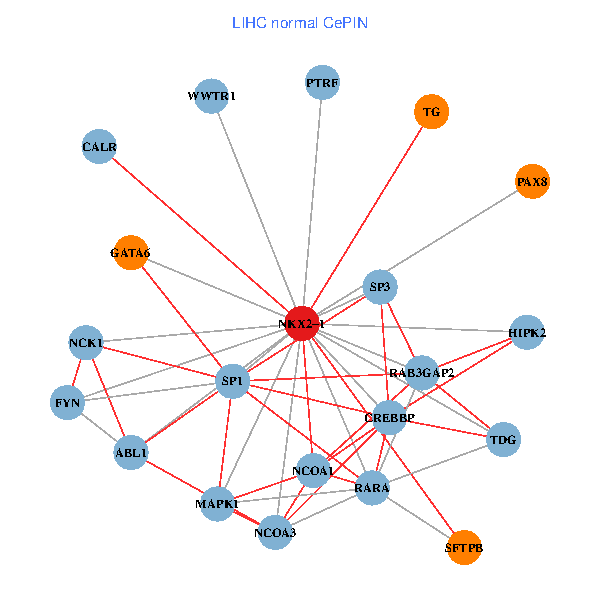 |
|
| LUAD (tumor) | LUAD (normal) |
| NKX2-1, NCK1, NCOA3, HIPK2, SP1, RELA, SRC, MAPK1, RARA, CREBBP, FYN, NCOA2, CALR, SP3, CRK, TDG, WWTR1, PAX8, CCDC59, TG, SFTPB (tumor) | NKX2-1, NCK1, NCOA3, HIPK2, SP1, GRB2, RELA, SRC, RARA, CREBBP, NCOA1, FYN, NCOA2, SP3, CRK, TDG, PAX8, CCDC59, GATA6, TG, SFTPB (normal) |
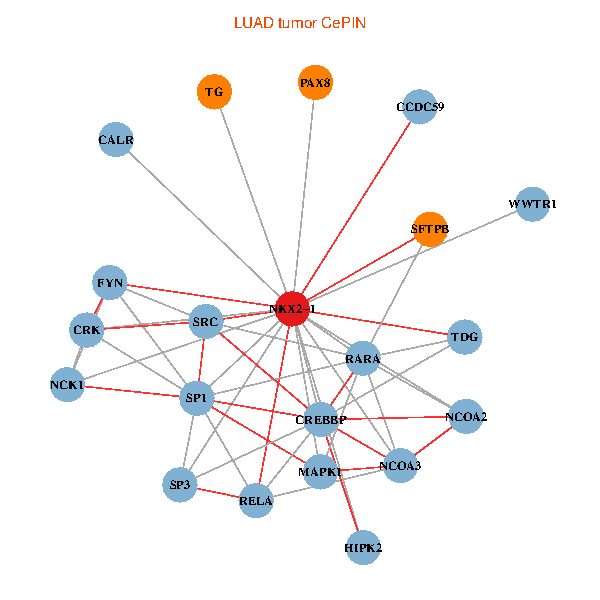 | 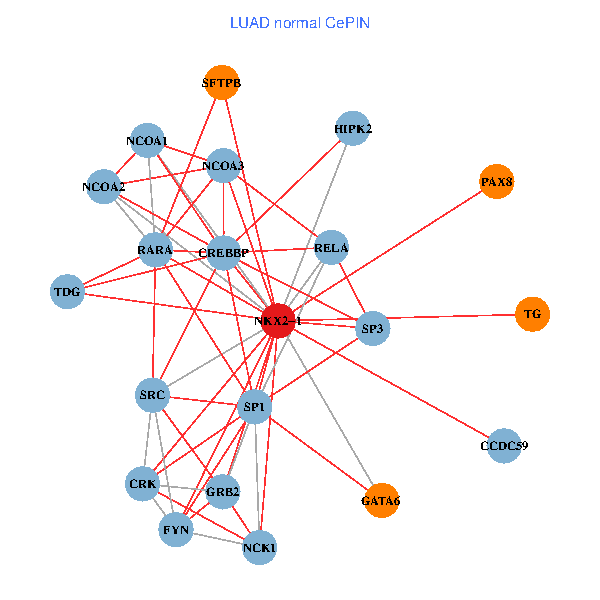 |
|
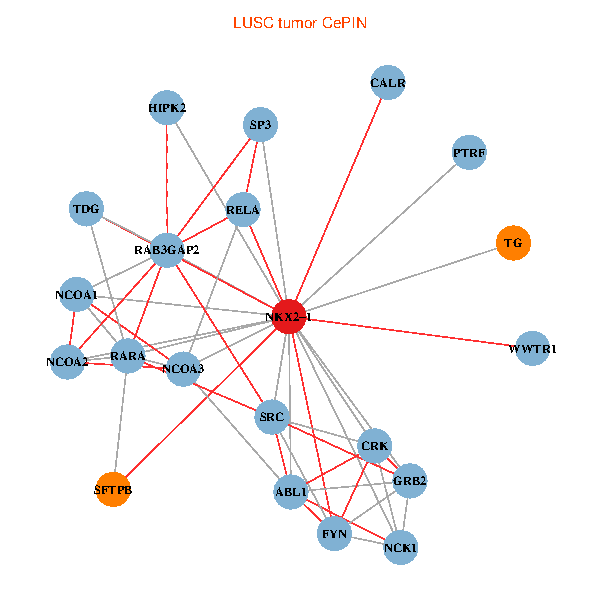
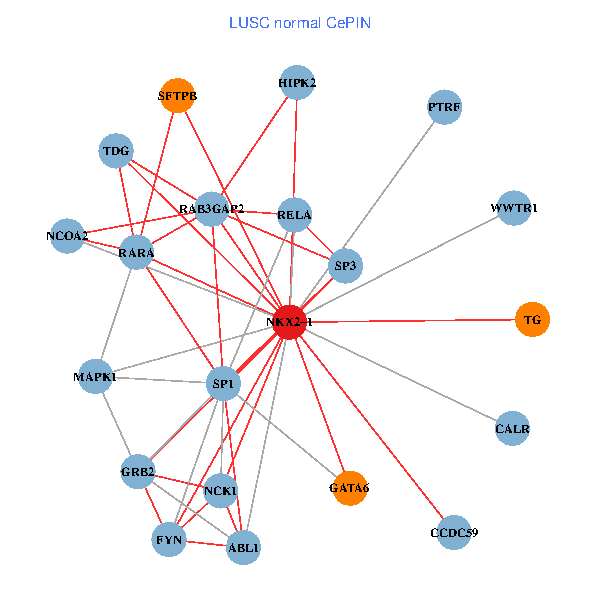
| LUSC (tumor) | LUSC (normal) |
| NKX2-1, NCK1, NCOA3, HIPK2, GRB2, RELA, SRC, RARA, NCOA1, FYN, NCOA2, RAB3GAP2, ABL1, CALR, SP3, CRK, TDG, WWTR1, PTRF, TG, SFTPB (tumor) | NKX2-1, NCK1, HIPK2, SP1, GRB2, RELA, MAPK1, RARA, FYN, NCOA2, RAB3GAP2, ABL1, CALR, SP3, TDG, WWTR1, CCDC59, GATA6, PTRF, TG, SFTPB (normal) |
 |  |
|
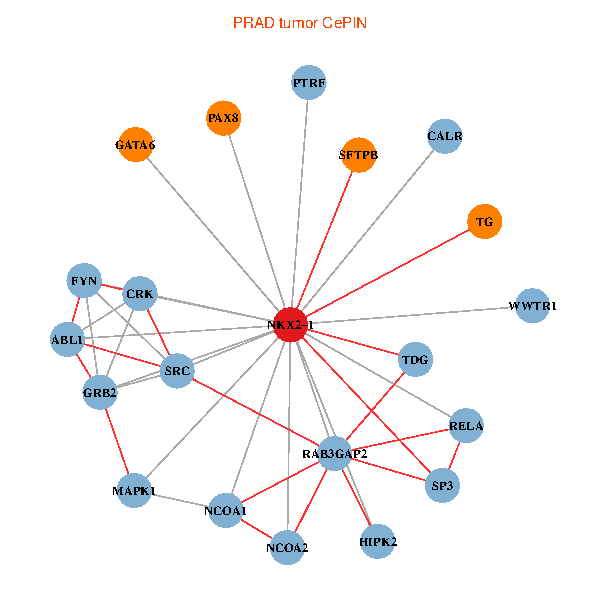
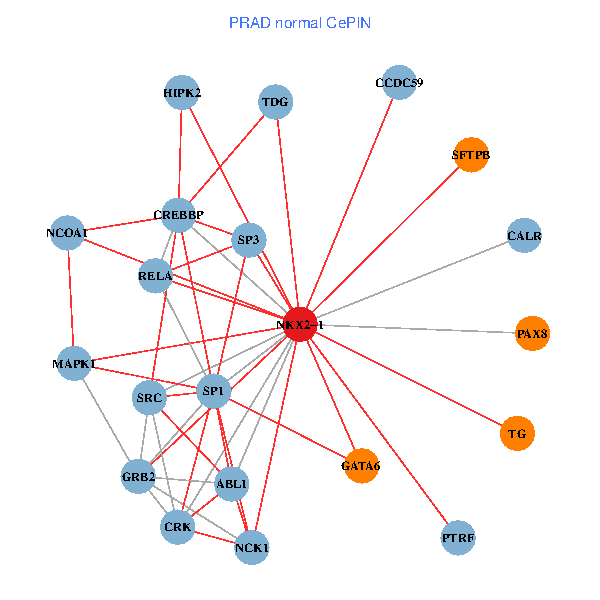
| PRAD (tumor) | PRAD (normal) |
| NKX2-1, HIPK2, GRB2, RELA, SRC, MAPK1, NCOA1, FYN, NCOA2, RAB3GAP2, ABL1, CALR, SP3, CRK, TDG, WWTR1, PAX8, GATA6, PTRF, TG, SFTPB (tumor) | NKX2-1, NCK1, HIPK2, SP1, GRB2, RELA, SRC, MAPK1, CREBBP, NCOA1, ABL1, CALR, SP3, CRK, TDG, PAX8, CCDC59, GATA6, PTRF, TG, SFTPB (normal) |
 |  |
|
| STAD (tumor) | STAD (normal) |
| NKX2-1, NCK1, GRB2, RELA, SRC, MAPK1, RARA, CREBBP, NCOA1, FYN, NCOA2, RAB3GAP2, ABL1, CALR, CRK, TDG, WWTR1, GATA6, PTRF, TG, SFTPB (tumor) | NKX2-1, NCK1, NCOA3, HIPK2, SP1, GRB2, SRC, MAPK1, RARA, FYN, NCOA2, RAB3GAP2, ABL1, CALR, SP3, CRK, TDG, GATA6, PTRF, TG, SFTPB (normal) |
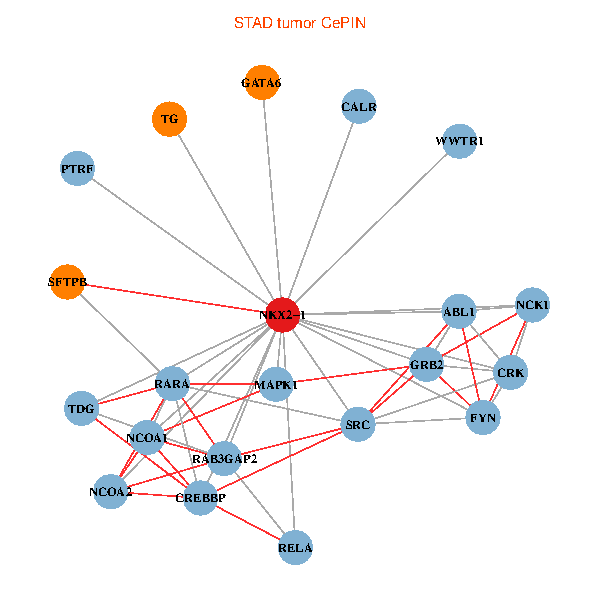 | 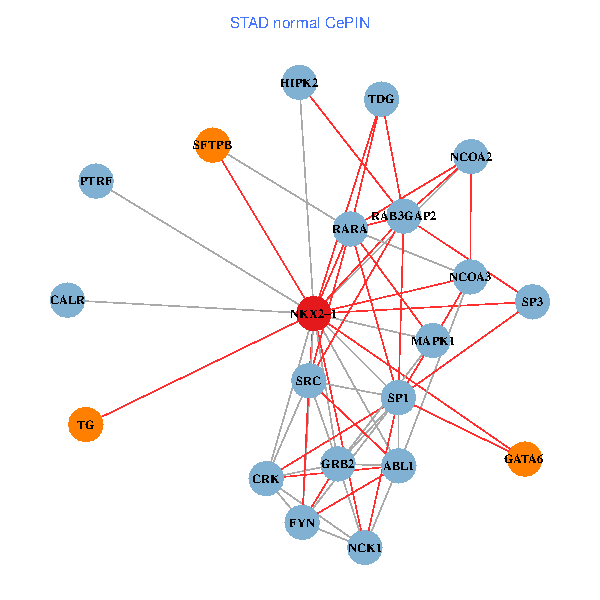 |
|
| THCA (tumor) | THCA (normal) |
| NKX2-1, NCOA3, HIPK2, GRB2, RELA, SRC, MAPK1, RARA, CREBBP, NCOA1, FYN, ABL1, CALR, SP3, CRK, TDG, WWTR1, PAX8, PTRF, TG, SFTPB (tumor) | NKX2-1, NCK1, NCOA3, SP1, GRB2, SRC, MAPK1, RARA, CREBBP, NCOA1, NCOA2, RAB3GAP2, ABL1, CALR, SP3, CRK, WWTR1, PAX8, CCDC59, GATA6, TG (normal) |
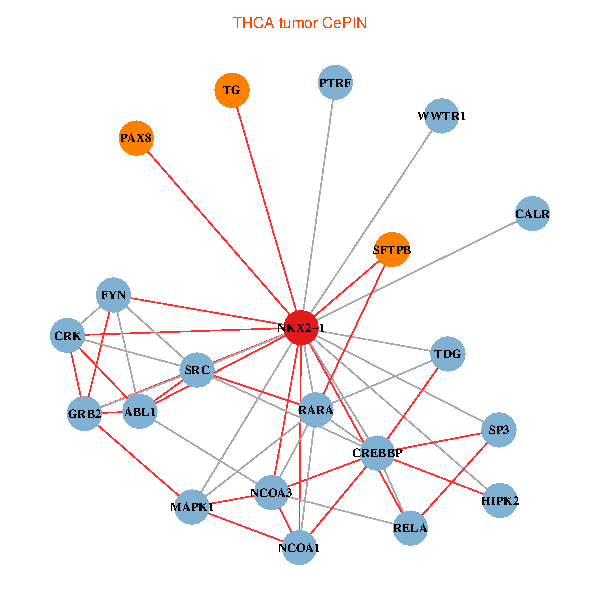 | 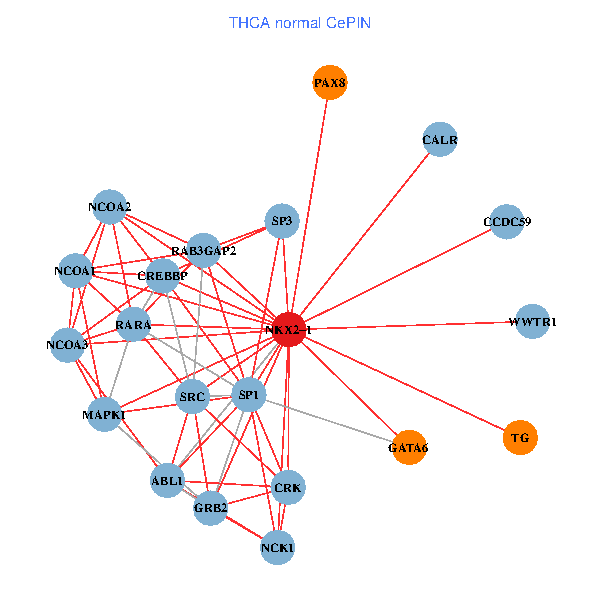 |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0001418 | Adenocarcinoma | 69 | BeFree,LHGDN |
| umls:C0152013 | Adenocarcinoma of lung (disorder) | 28 | BeFree |
| umls:C0242379 | Malignant neoplasm of lung | 24 | BeFree |
| umls:C0684249 | Carcinoma of lung | 21 | BeFree |
| umls:C0393584 | Benign Hereditary Chorea | 20 | BeFree,CLINVAR,ORPHANET,UNIPROT |
| umls:C0010308 | Congenital Hypothyroidism | 17 | BeFree,GAD,LHGDN |
| umls:C0342200 | Endemic Cretinism | 17 | BeFree |
| umls:C0549473 | Thyroid carcinoma | 15 | BeFree,GAD |
| umls:C0007131 | Non-Small Cell Lung Carcinoma | 14 | BeFree |
| umls:C0238463 | Papillary thyroid carcinoma | 14 | BeFree,ORPHANET |
| umls:C0007137 | Squamous cell carcinoma | 13 | BeFree |
| umls:C0024121 | Lung Neoplasms | 12 | BeFree,LHGDN |
| umls:C0040136 | Thyroid Neoplasm | 10 | BeFree,GAD,LHGDN |
| umls:C0020676 | Hypothyroidism | 8 | BeFree,LHGDN |
| umls:C0596263 | Carcinogenesis | 8 | BeFree |
| umls:C1563716 | Thyroid Dysgenesis | 8 | BeFree |
| umls:C0024115 | Lung diseases | 7 | BeFree,CTD_human |
| umls:C0027627 | Neoplasm Metastasis | 7 | BeFree |
| umls:C0262584 | Carcinoma, Small Cell | 7 | BeFree |
| umls:C0007133 | Carcinoma, Papillary | 6 | BeFree,GAD |
| umls:C1145670 | Respiratory Failure | 6 | BeFree |
| umls:C0007115 | Malignant neoplasm of thyroid | 5 | BeFree |
| umls:C0032002 | Pituitary Diseases | 5 | BeFree |
| umls:C0149925 | Small cell carcinoma of lung | 5 | BeFree,LHGDN |
| umls:C1970269 | Choreoathetosis, Hypothyroidism, And Neonatal Respiratory Distress | 5 | BeFree,CLINVAR,CTD_human,ORPHANET,UNIPROT |
| umls:C0007095 | Carcinoid Tumor | 4 | BeFree |
| umls:C0008489 | Chorea | 4 | BeFree,CTD_human,LHGDN |
| umls:C0678222 | Breast Carcinoma | 4 | BeFree |
| umls:C1337013 | Differentiated Thyroid Gland Carcinoma | 4 | BeFree |
| umls:C0001430 | Adenoma | 3 | BeFree,LHGDN |
| umls:C0006142 | Malignant neoplasm of breast | 3 | BeFree |
| umls:C0007130 | Mucinous Adenocarcinoma | 3 | BeFree |
| umls:C0019284 | Diaphragmatic Hernia | 3 | CTD_human,RGD |
| umls:C0019569 | Hirschsprung Disease | 3 | BeFree,GAD,LHGDN |
| umls:C0032227 | Pleural effusion disorder | 3 | BeFree |
| umls:C0178874 | Tumor Progression | 3 | BeFree |
| umls:C0238461 | Anaplastic thyroid carcinoma | 3 | BeFree |
| umls:C0852283 | Respiratory Distress Syndrome | 3 | BeFree |
| umls:C1334455 | Pulmonary Sclerosing Hemangioma | 3 | BeFree,LHGDN |
| umls:C0002793 | Anaplasia | 2 | BeFree |
| umls:C0004096 | Asthma | 2 | BeFree |
| umls:C0004134 | Ataxia | 2 | BeFree |
| umls:C0019693 | HIV Infections | 2 | BeFree |
| umls:C0032285 | Pneumonia | 2 | BeFree |
| umls:C0035204 | Respiration Disorders | 2 | BeFree |
| umls:C0035242 | Respiratory Tract Diseases | 2 | BeFree |
| umls:C0085583 | Choreoathetosis | 2 | BeFree |
| umls:C0205696 | Anaplastic carcinoma | 2 | BeFree |
| umls:C0205698 | Undifferentiated carcinoma | 2 | BeFree |
| umls:C0206704 | Carcinoma, Large Cell | 2 | BeFree |
| umls:C0220650 | Metastatic malignant neoplasm to brain | 2 | BeFree |
| umls:C0238462 | Medullary carcinoma of thyroid | 2 | BeFree |
| umls:C0265783 | Congenital hypoplasia of lung | 2 | BeFree |
| umls:C0342208 | Multinodular goiter | 2 | BeFree |
| umls:C0699734 | progressive chorea | 2 | BeFree |
| umls:C1265996 | Large cell neuroendocrine carcinoma | 2 | BeFree |
| umls:C1709246 | Non-Neoplastic Disorder | 2 | BeFree |
| umls:C1961099 | Precursor T-Cell Lymphoblastic Leukemia-Lymphoma | 2 | BeFree |
| umls:C0000768 | Congenital Abnormality | 1 | BeFree |
| umls:C0007103 | Malignant neoplasm of endometrium | 1 | BeFree |
| umls:C0007621 | Neoplastic Cell Transformation | 1 | GAD |
| umls:C0009404 | Colorectal Neoplasms | 1 | BeFree |
| umls:C0011265 | Presenile dementia | 1 | BeFree |
| umls:C0013362 | Dysarthria | 1 | BeFree |
| umls:C0013421 | Dystonia | 1 | BeFree |
| umls:C0016522 | Foramen Ovale, Patent | 1 | GAD |
| umls:C0017154 | Gastritis, Atrophic | 1 | BeFree |
| umls:C0018021 | Goiter | 1 | BeFree |
| umls:C0018023 | Nodular Goiter | 1 | GAD |
| umls:C0018213 | Graves Disease | 1 | BeFree |
| umls:C0019348 | Herpes Simplex Infections | 1 | BeFree |
| umls:C0020179 | Huntington Disease | 1 | BeFree |
| umls:C0024117 | Chronic Obstructive Airway Disease | 1 | BeFree |
| umls:C0025202 | melanoma | 1 | BeFree |
| umls:C0025267 | Multiple Endocrine Neoplasia Type 1 | 1 | BeFree |
| umls:C0026650 | Movement Disorders | 1 | BeFree |
| umls:C0026827 | Muscle hypotonia | 1 | BeFree |
| umls:C0027066 | Myoclonus | 1 | BeFree |
| umls:C0027708 | Nephroblastoma | 1 | LHGDN |
| umls:C0028754 | Obesity | 1 | GAD |
| umls:C0029925 | Ovarian Carcinoma | 1 | BeFree |
| umls:C0032460 | Polycystic Ovary Syndrome | 1 | GAD |
| umls:C0034012 | Delayed Puberty | 1 | GAD |
| umls:C0034013 | Precocious Puberty | 1 | GAD |
| umls:C0038454 | Cerebrovascular accident | 1 | GAD |
| umls:C0079744 | Diffuse Large B-Cell Lymphoma | 1 | BeFree |
| umls:C0080032 | Pleural Effusion, Malignant | 1 | BeFree |
| umls:C0085110 | Severe Combined Immunodeficiency | 1 | BeFree |
| umls:C0085167 | Granular cell tumor | 1 | BeFree |
| umls:C0085215 | Ovarian Failure, Premature | 1 | GAD |
| umls:C0206062 | Lung Diseases, Interstitial | 1 | BeFree |
| umls:C0206693 | Medullary carcinoma | 1 | LHGDN |
| umls:C0206695 | Carcinoma, Neuroendocrine | 1 | LHGDN |
| umls:C0235031 | Neurologic Symptoms | 1 | BeFree |
| umls:C0265780 | Congenital absence of lung | 1 | BeFree |
| umls:C0271829 | Pendred's syndrome | 1 | BeFree |
| umls:C0334276 | Adenocarcinoma in Situ | 1 | BeFree |
| umls:C0342196 | Thyroid Dyshormonogenesis 5 | 1 | BeFree |
| umls:C0342409 | Hypophysitis | 1 | BeFree |
| umls:C0342419 | Pituitary mass | 1 | BeFree |
| umls:C0345963 | Benign neoplasm of lung | 1 | BeFree |
| umls:C0349476 | Congenital goiter | 1 | BeFree |
| umls:C0393593 | Dystonia Disorders | 1 | BeFree |
| umls:C0424605 | Developmental delay (disorder) | 1 | BeFree |
| umls:C0456891 | Primary pulmonary hypoplasia | 1 | BeFree |
| umls:C0476089 | Endometrial Carcinoma | 1 | BeFree |
| umls:C0497327 | Dementia | 1 | BeFree |
| umls:C0520947 | Clumsiness - motor delay | 1 | BeFree |
| umls:C0521158 | Recurrent tumor | 1 | BeFree |
| umls:C0521648 | Neonatal respiratory failure | 1 | BeFree |
| umls:C0854988 | Adenocarcinoma of lung, stage IV | 1 | BeFree |
| umls:C0919267 | ovarian neoplasm | 1 | BeFree |
| umls:C1140680 | Malignant neoplasm of ovary | 1 | BeFree |
| umls:C1266050 | Poorly Differentiated Thyroid Carcinoma | 1 | BeFree |
| umls:C1333873 | Granular cell tumor of neurohypophysis | 1 | BeFree |
| umls:C1569637 | Adenocarcinoma, Endometrioid | 1 | BeFree |
| umls:C1834570 | Myoclonic dystonia | 1 | BeFree |
| umls:C1868682 | Paroxysmal kinesigenic choreoathetosis | 1 | BeFree |
| umls:C1883486 | Uterine Corpus Cancer | 1 | BeFree |
| umls:C2986550 | Pituicytoma | 1 | BeFree |
| umls:C3161105 | Neuroendocrine cell hyperplasia of infancy | 1 | BeFree |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))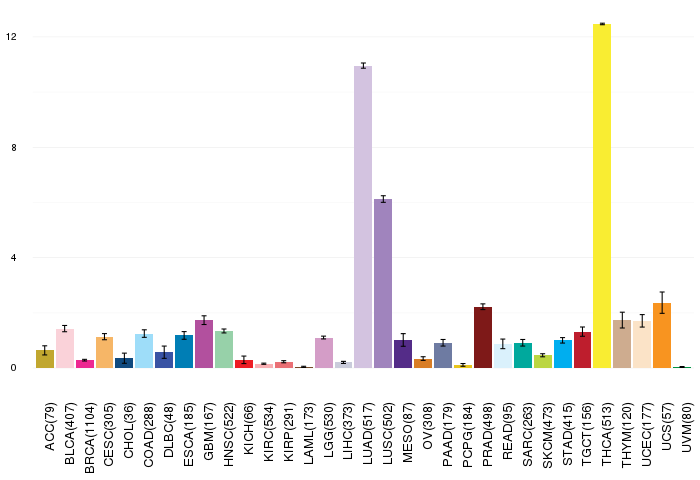
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))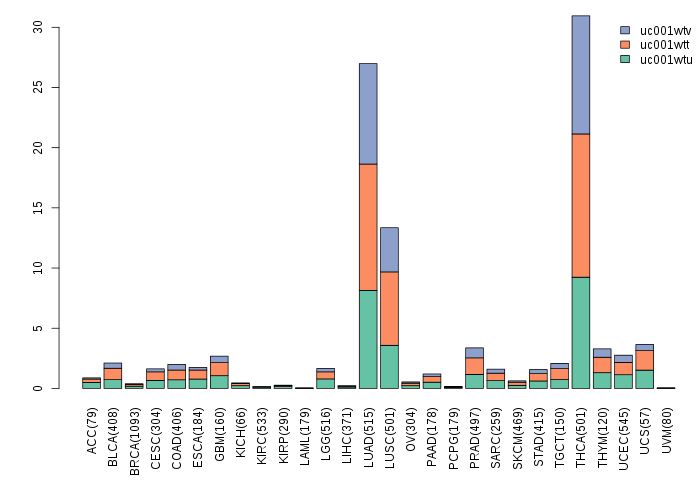
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data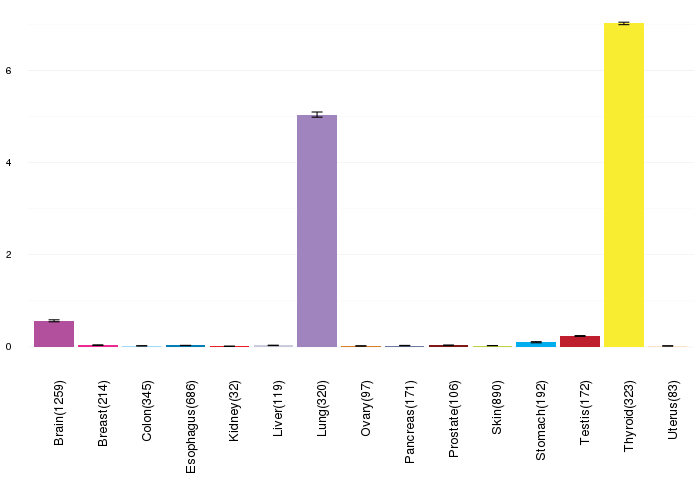
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1)) Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.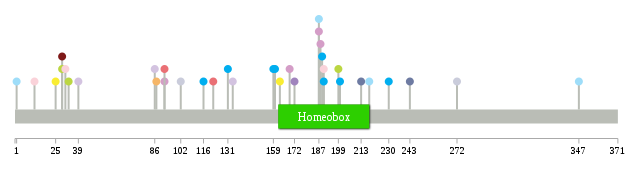

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 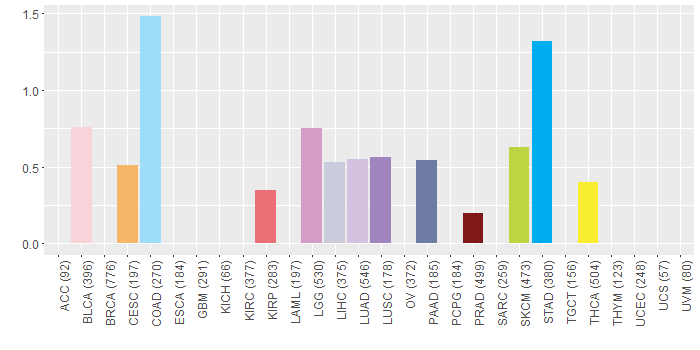
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)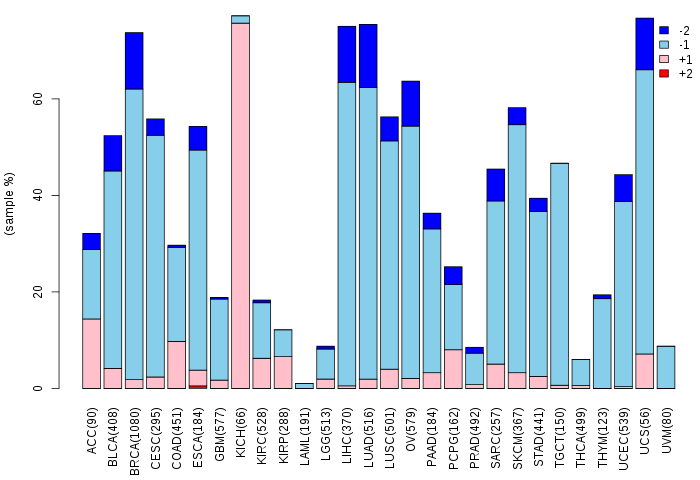
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 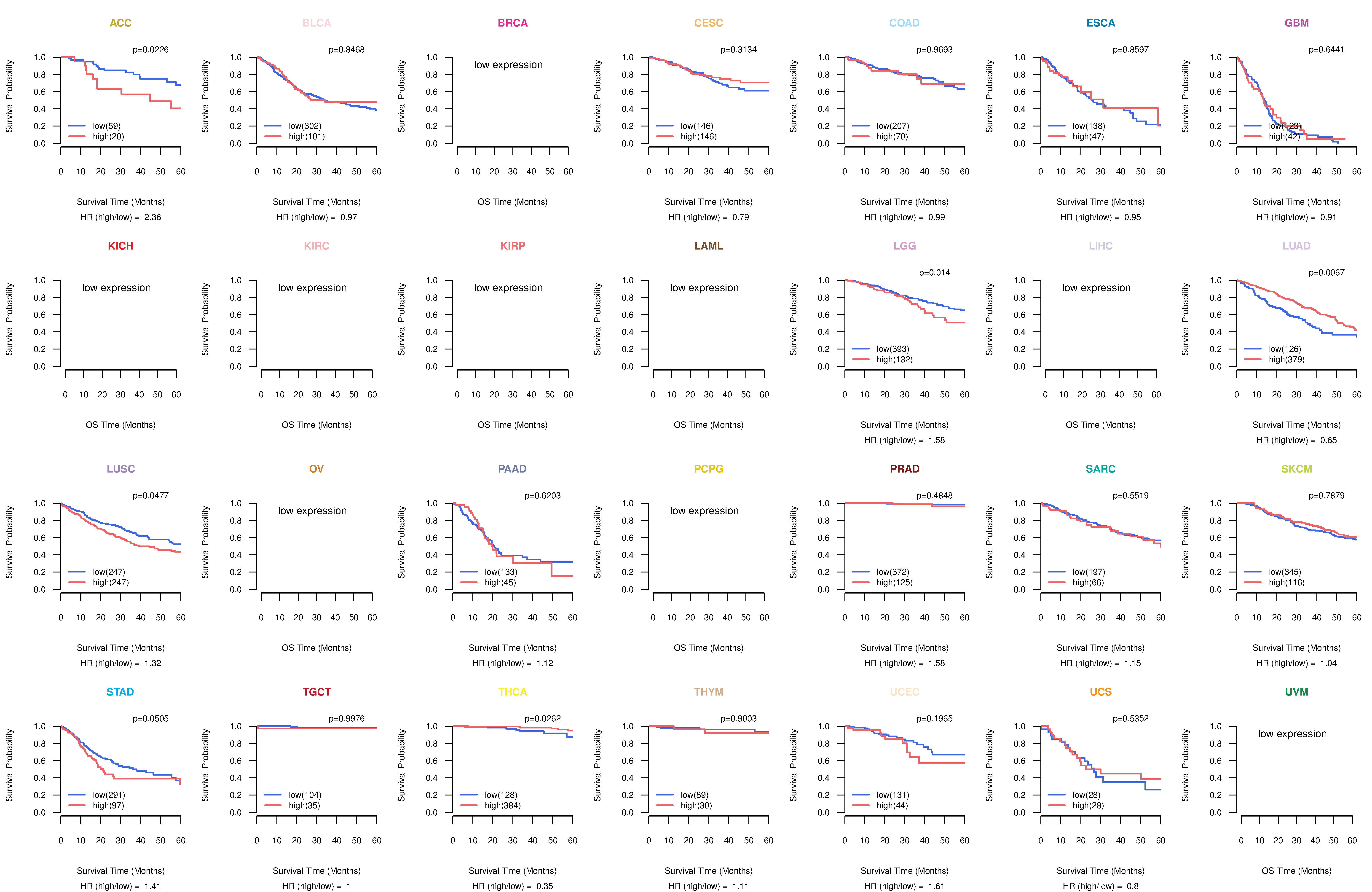
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 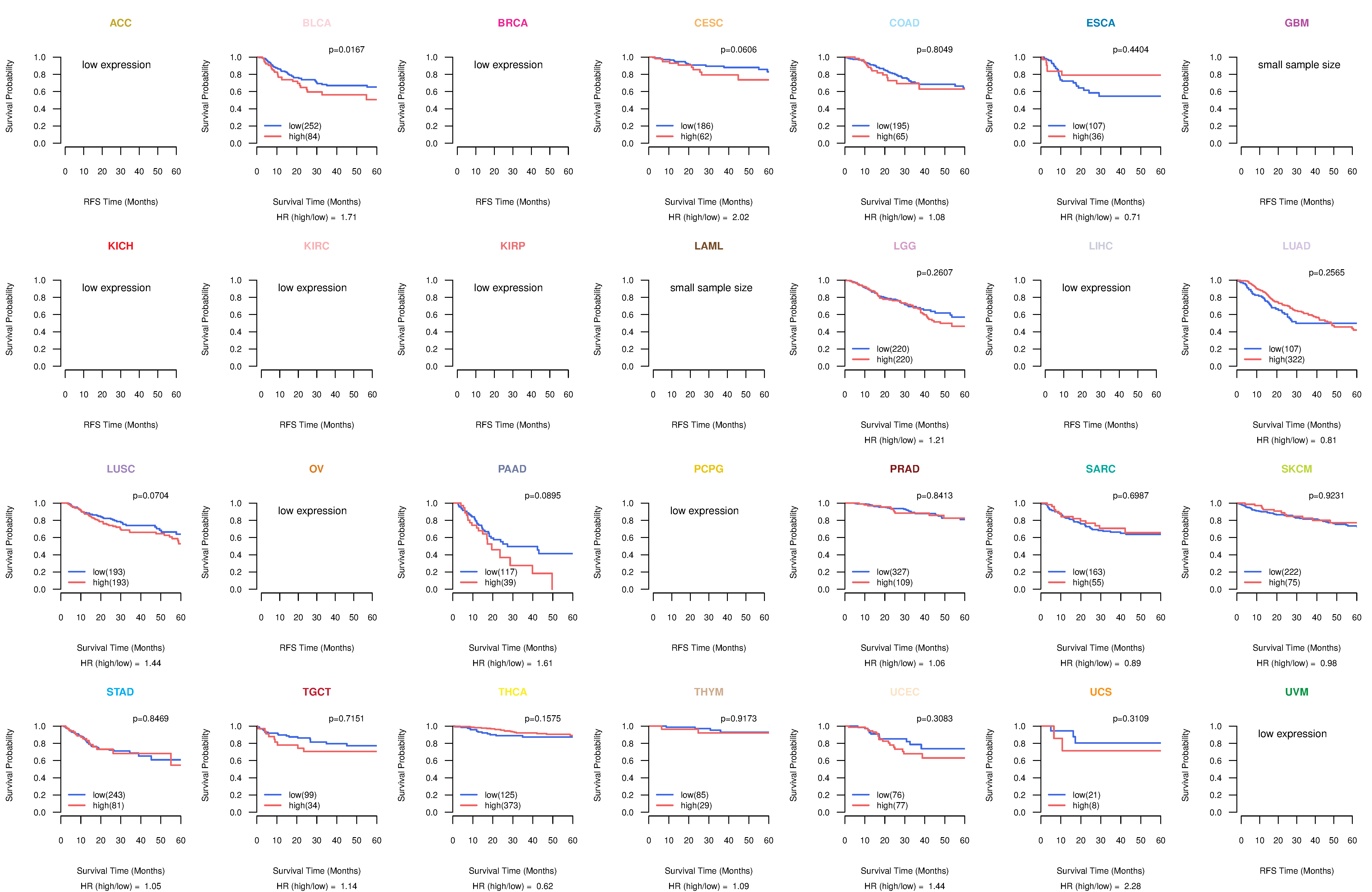
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 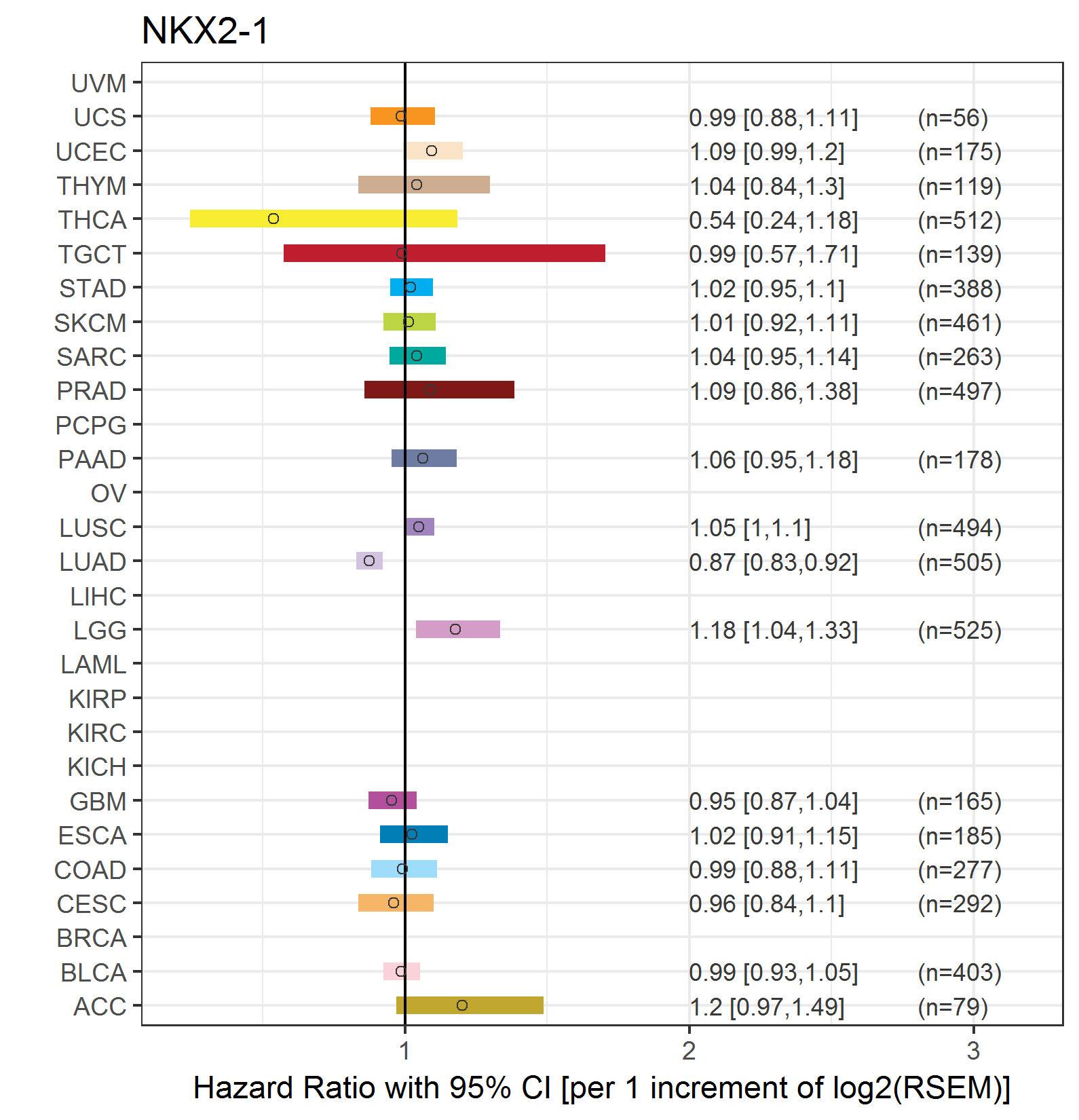
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 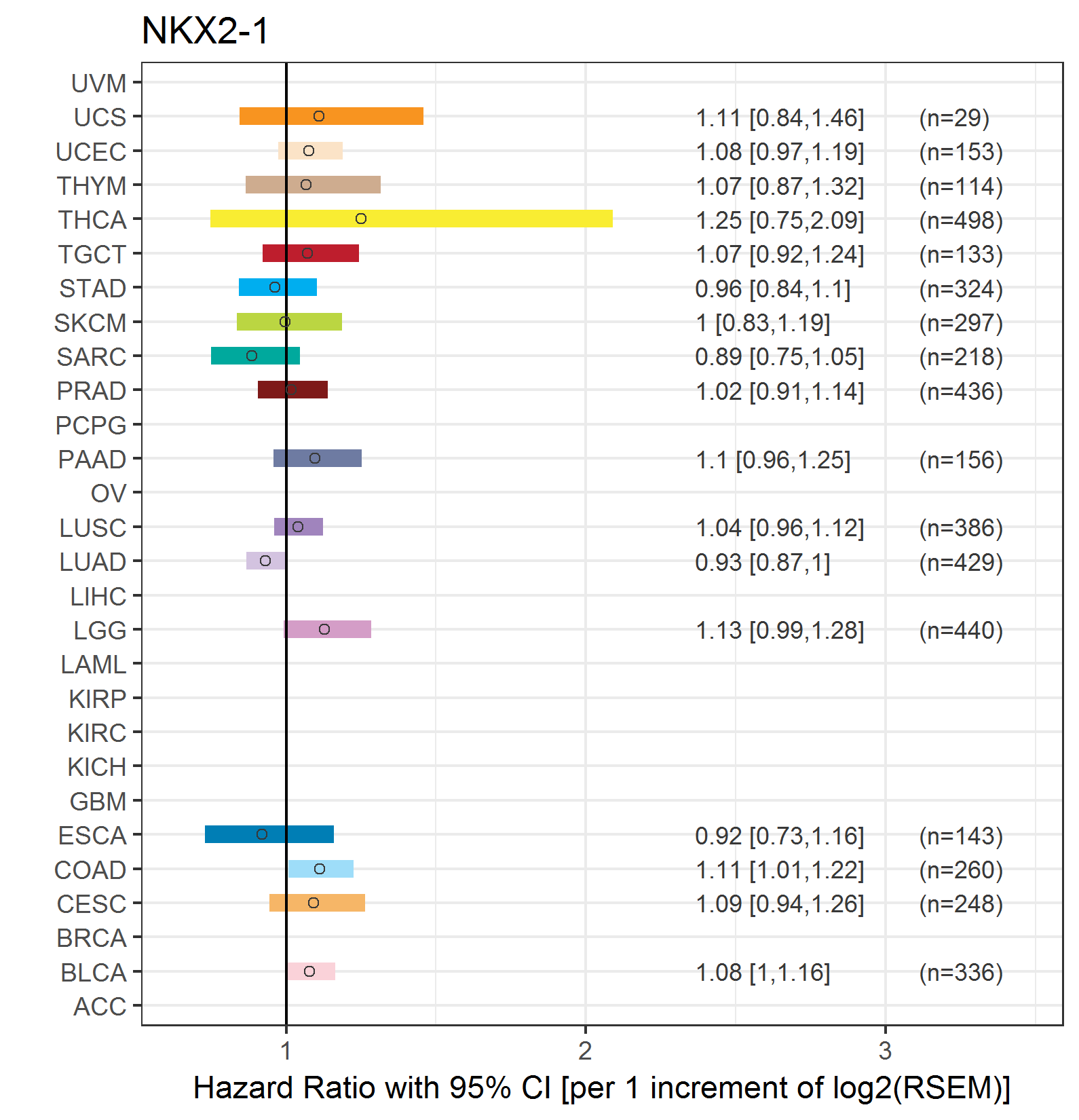
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























