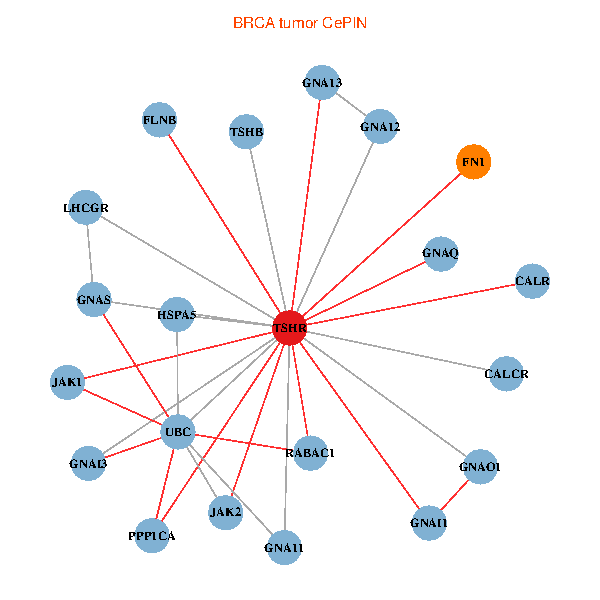
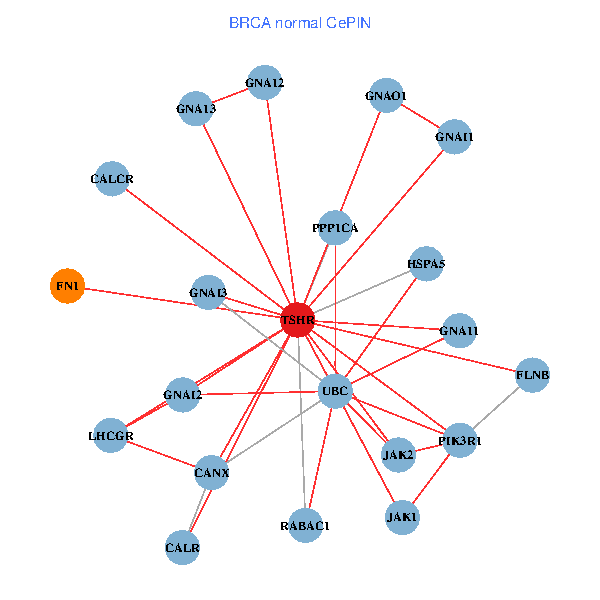
| BRCA (tumor) | BRCA (normal) |
| TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, JAK2, GNAQ, RABAC1, CALR, HSPA5, CALCR, GNA12, GNAS, GNAO1, GNAI1, GNA11, GNAI3, GNA13, TSHB, LHCGR (tumor) | TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, JAK2, RABAC1, PIK3R1, CALR, HSPA5, CANX, GNAI2, CALCR, GNA12, GNAO1, GNAI1, GNA11, GNAI3, GNA13, LHCGR (normal) |
 |  |
|
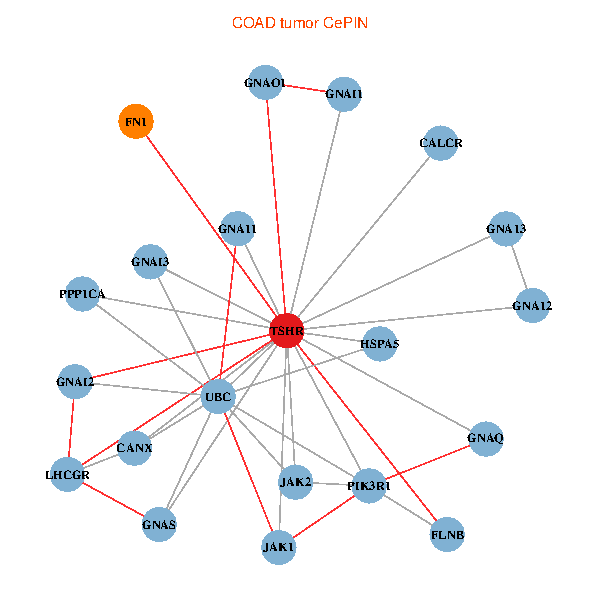
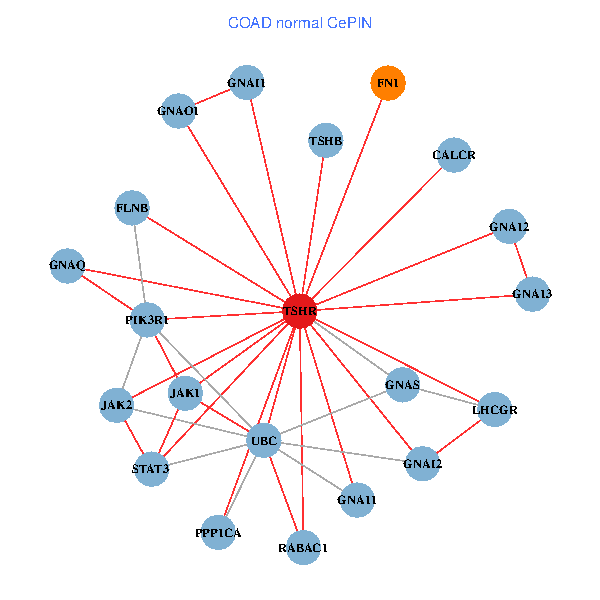
| COAD (tumor) | COAD (normal) |
| TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, JAK2, GNAQ, PIK3R1, HSPA5, CANX, GNAI2, CALCR, GNA12, GNAS, GNAO1, GNAI1, GNA11, GNAI3, GNA13, LHCGR (tumor) | TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, PIK3R1, GNAI2, CALCR, GNA12, GNAS, GNAO1, GNAI1, GNA11, GNA13, TSHB, LHCGR (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
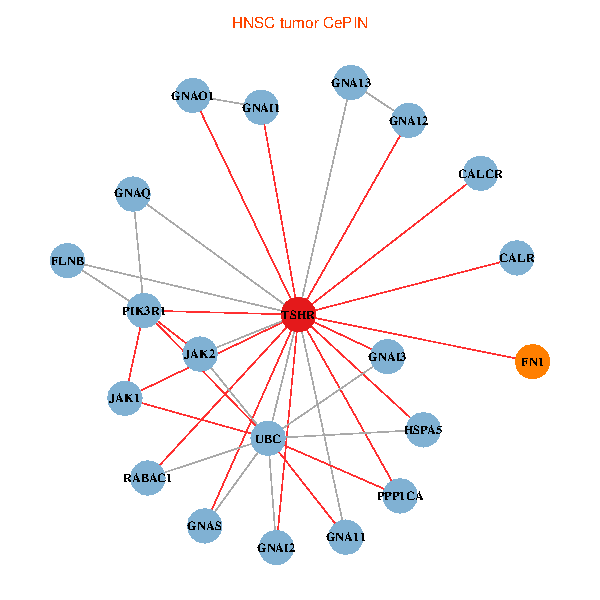
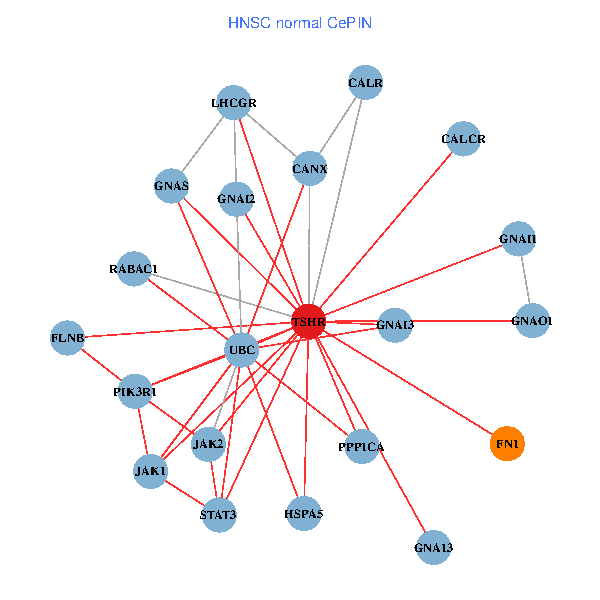
| TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, JAK2, GNAQ, RABAC1, PIK3R1, CALR, HSPA5, GNAI2, CALCR, GNA12, GNAS, GNAO1, GNAI1, GNA11, GNAI3, GNA13 (tumor) | TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, STAT3, JAK2, RABAC1, PIK3R1, CALR, HSPA5, CANX, GNAI2, CALCR, GNAS, GNAO1, GNAI1, GNAI3, GNA13, LHCGR (normal) |
 |  |
|
| KICH (tumor) | KICH (normal) |
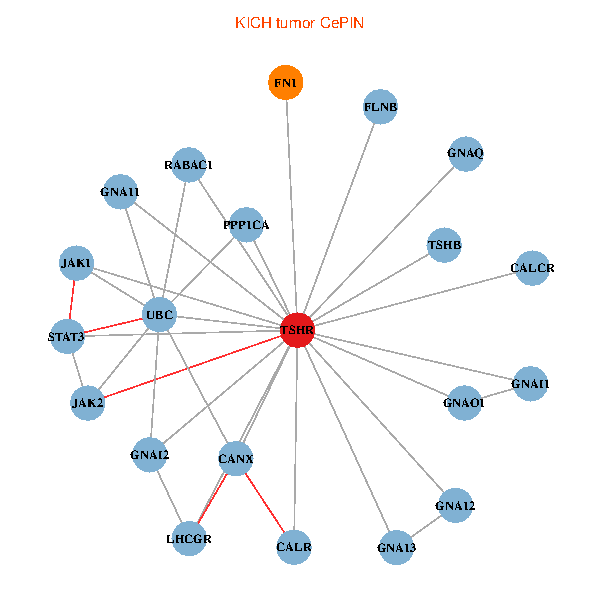
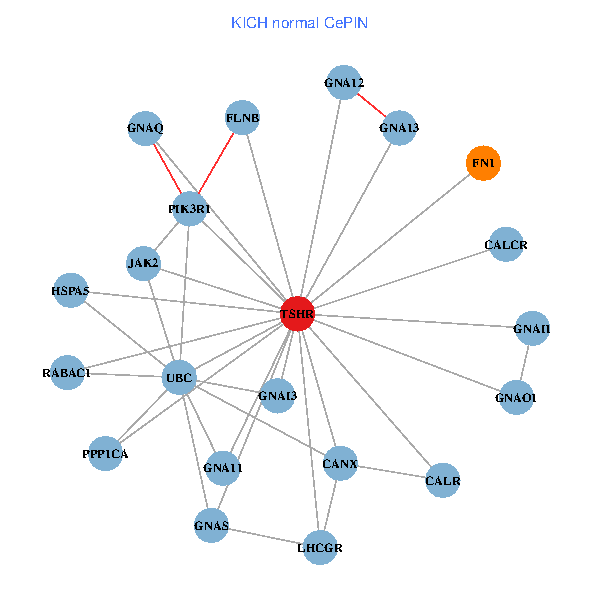
| TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, CALR, CANX, GNAI2, CALCR, GNA12, GNAO1, GNAI1, GNA11, GNA13, TSHB, LHCGR (tumor) | TSHR, PPP1CA, UBC, FLNB, FN1, JAK2, GNAQ, RABAC1, PIK3R1, CALR, HSPA5, CANX, CALCR, GNA12, GNAS, GNAO1, GNAI1, GNA11, GNAI3, GNA13, LHCGR (normal) |
 |  |
|
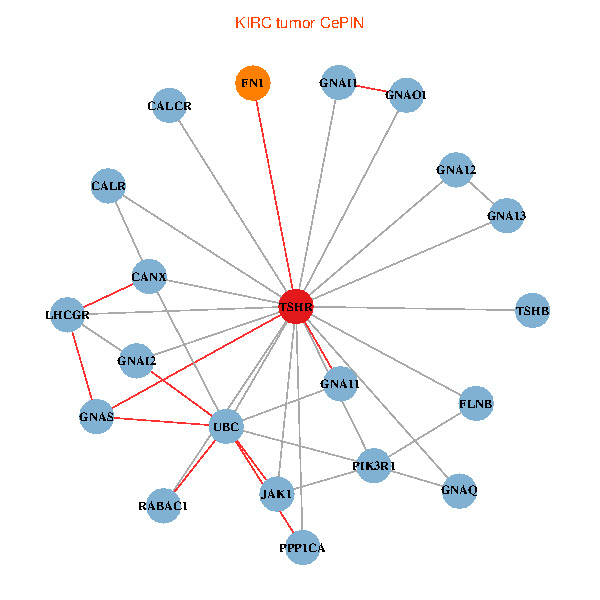
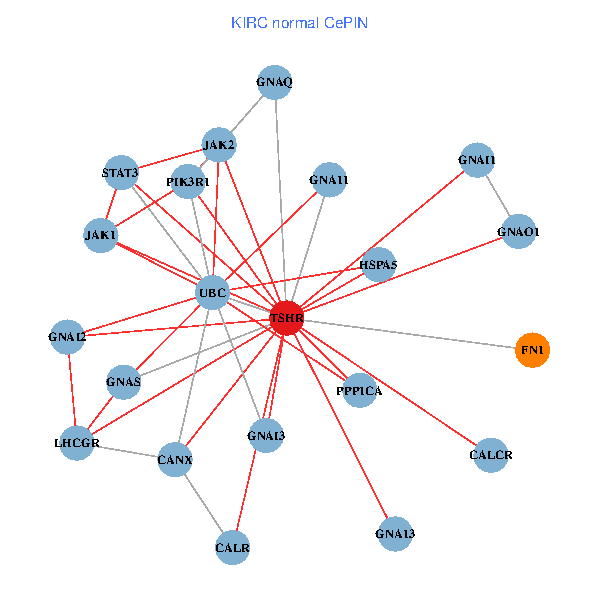
| KIRC (tumor) | KIRC (normal) |
| TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, GNAQ, RABAC1, PIK3R1, CALR, CANX, GNAI2, CALCR, GNA12, GNAS, GNAO1, GNAI1, GNA11, GNA13, TSHB, LHCGR (tumor) | TSHR, PPP1CA, UBC, JAK1, FN1, STAT3, JAK2, GNAQ, PIK3R1, CALR, HSPA5, CANX, GNAI2, CALCR, GNAS, GNAO1, GNAI1, GNA11, GNAI3, GNA13, LHCGR (normal) |
 |  |
|
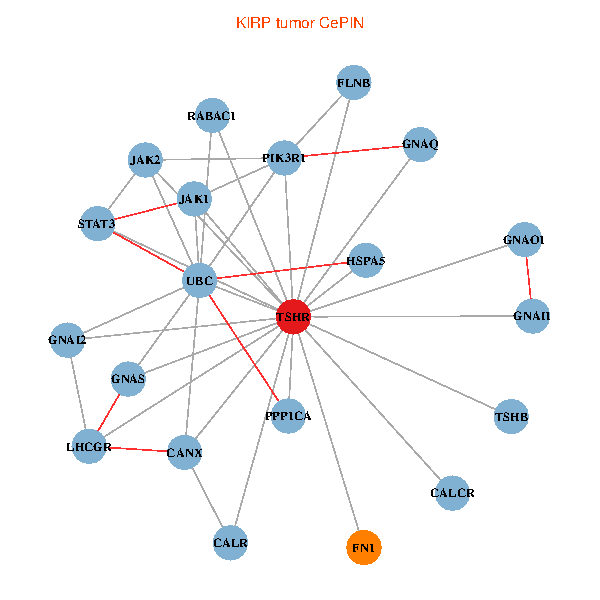
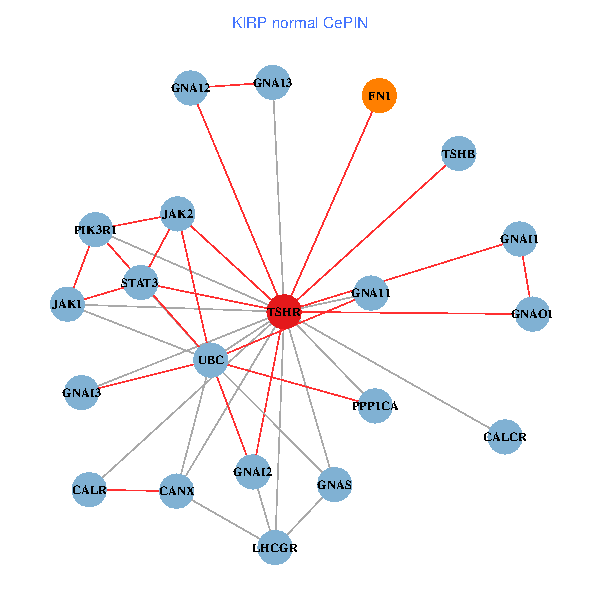
| KIRP (tumor) | KIRP (normal) |
| TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, PIK3R1, CALR, HSPA5, CANX, GNAI2, CALCR, GNAS, GNAO1, GNAI1, TSHB, LHCGR (tumor) | TSHR, PPP1CA, UBC, JAK1, FN1, STAT3, JAK2, PIK3R1, CALR, CANX, GNAI2, CALCR, GNA12, GNAS, GNAO1, GNAI1, GNA11, GNAI3, GNA13, TSHB, LHCGR (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, CALR, HSPA5, CANX, CALCR, GNA12, GNAS, GNAO1, GNAI1, GNA11, GNAI3, TSHB (tumor) | TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, PIK3R1, GNAI2, CALCR, GNA12, GNAS, GNAO1, GNA11, GNAI3, GNA13, TSHB, LHCGR (normal) |
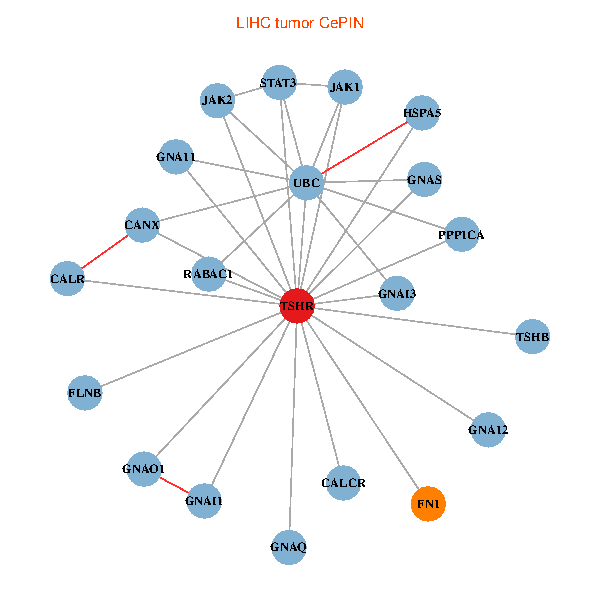 | 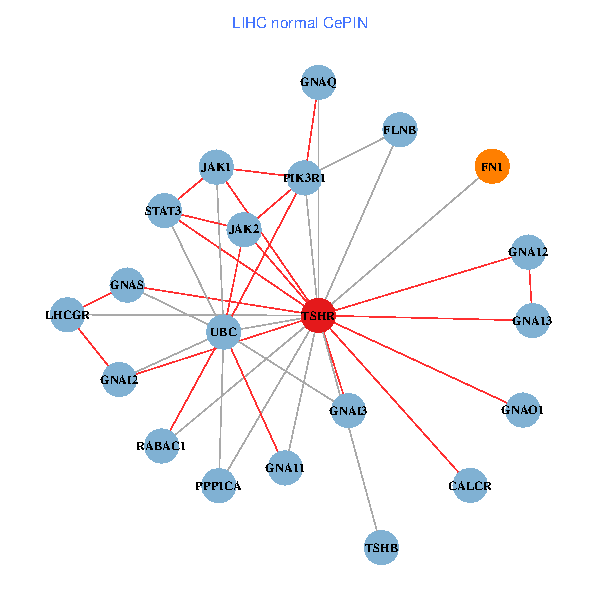 |
|
| LUAD (tumor) | LUAD (normal) |
| TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, STAT3, GNAQ, RABAC1, PIK3R1, CALR, HSPA5, GNAI2, GNA12, GNAS, GNAO1, GNAI1, GNAI3, GNA13, TSHB, LHCGR (tumor) | TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, PIK3R1, CALR, HSPA5, GNAI2, GNAS, GNAO1, GNA11, GNAI3, GNA13, TSHB, LHCGR (normal) |
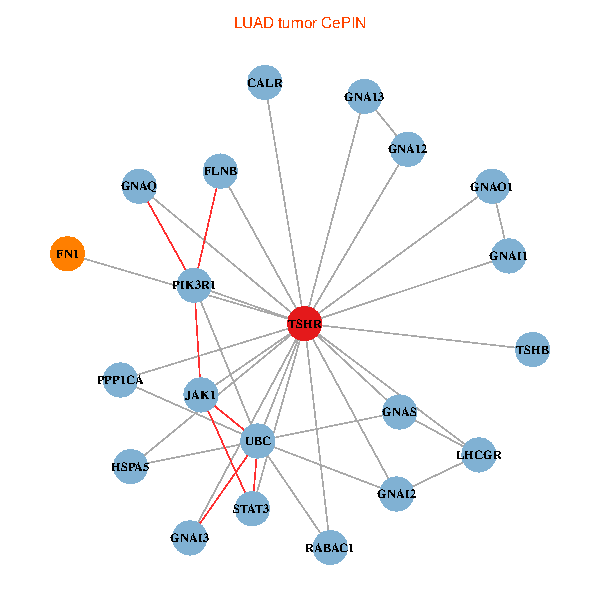 | 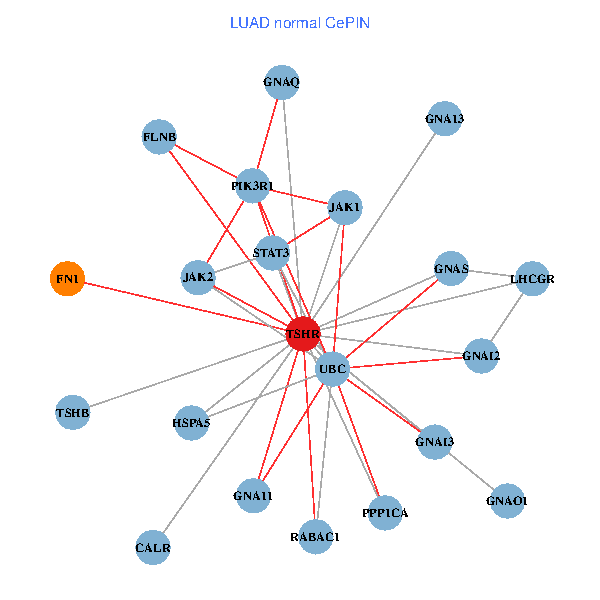 |
|
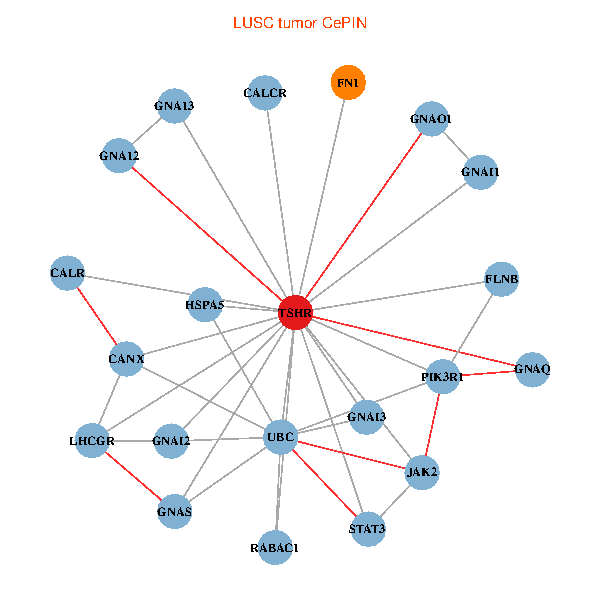
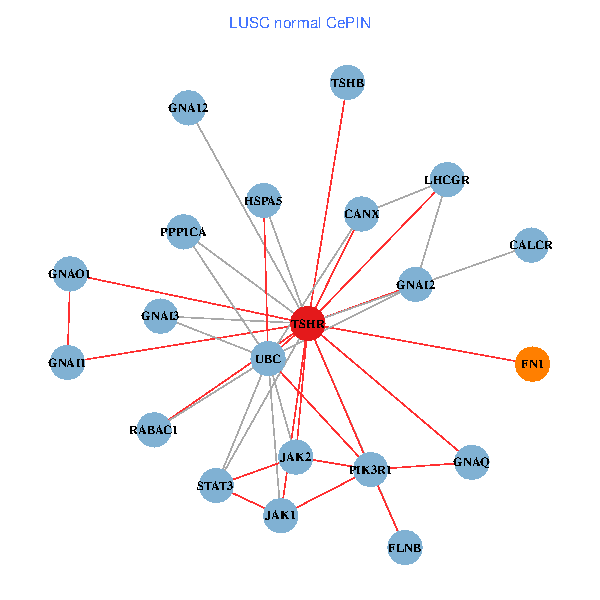
| LUSC (tumor) | LUSC (normal) |
| TSHR, UBC, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, PIK3R1, CALR, HSPA5, CANX, GNAI2, CALCR, GNA12, GNAS, GNAO1, GNAI1, GNAI3, GNA13, LHCGR (tumor) | TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, PIK3R1, HSPA5, CANX, GNAI2, CALCR, GNA12, GNAO1, GNAI1, GNAI3, TSHB, LHCGR (normal) |
 |  |
|
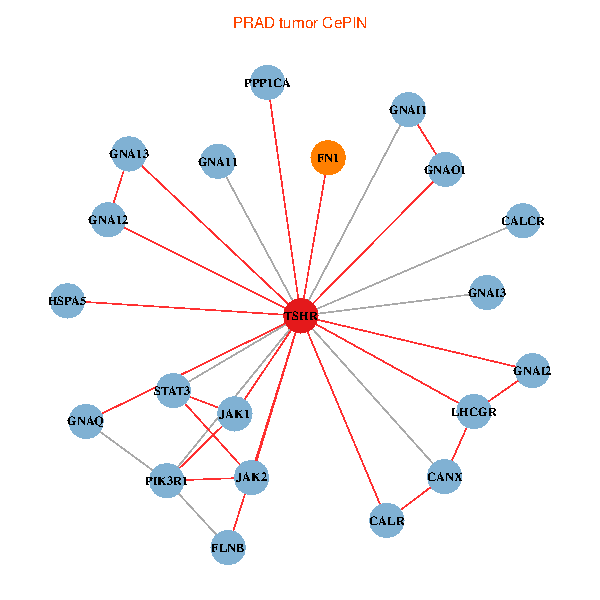
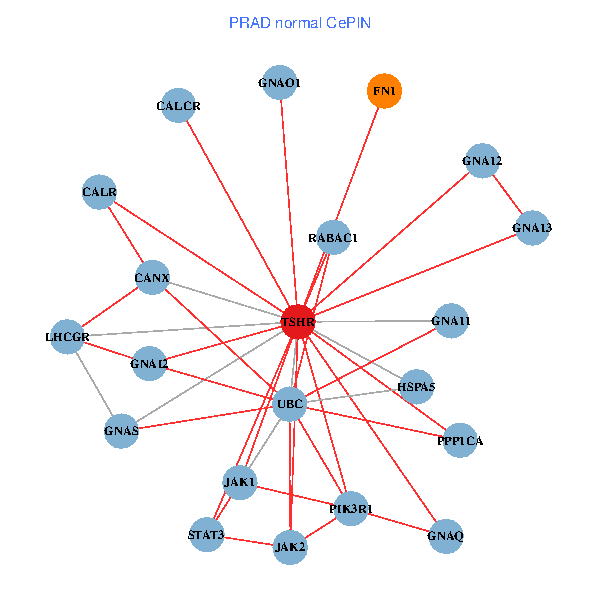
| PRAD (tumor) | PRAD (normal) |
| TSHR, PPP1CA, JAK1, FLNB, FN1, STAT3, JAK2, GNAQ, PIK3R1, CALR, HSPA5, CANX, GNAI2, CALCR, GNA12, GNAO1, GNAI1, GNA11, GNAI3, GNA13, LHCGR (tumor) | TSHR, PPP1CA, UBC, JAK1, FN1, STAT3, JAK2, GNAQ, RABAC1, PIK3R1, CALR, HSPA5, CANX, GNAI2, CALCR, GNA12, GNAS, GNAO1, GNA11, GNA13, LHCGR (normal) |
 |  |
|
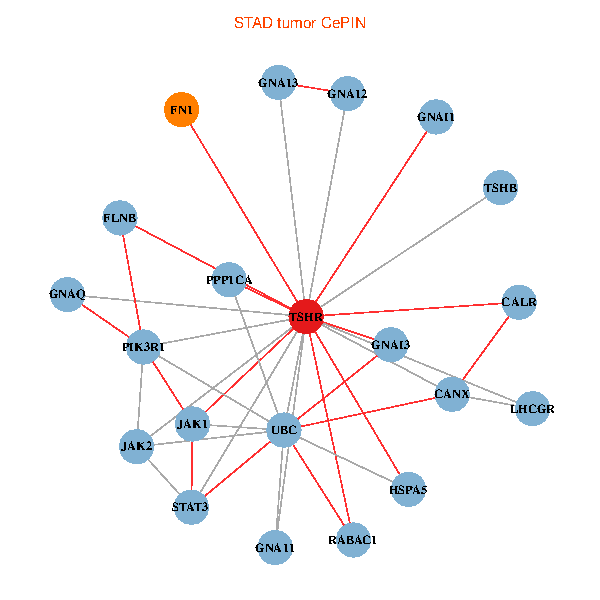
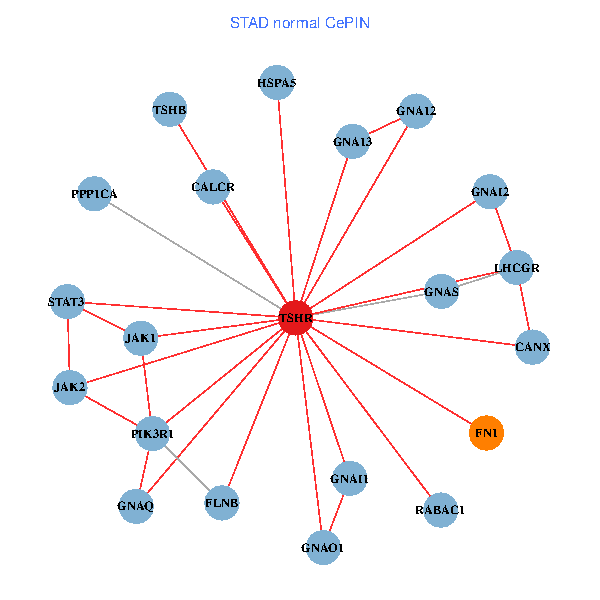
| STAD (tumor) | STAD (normal) |
| TSHR, PPP1CA, UBC, JAK1, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, PIK3R1, CALR, HSPA5, CANX, GNA12, GNAI1, GNA11, GNAI3, GNA13, TSHB, LHCGR (tumor) | TSHR, PPP1CA, JAK1, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, PIK3R1, HSPA5, CANX, GNAI2, CALCR, GNA12, GNAS, GNAO1, GNAI1, GNA13, TSHB, LHCGR (normal) |
 |  |
|
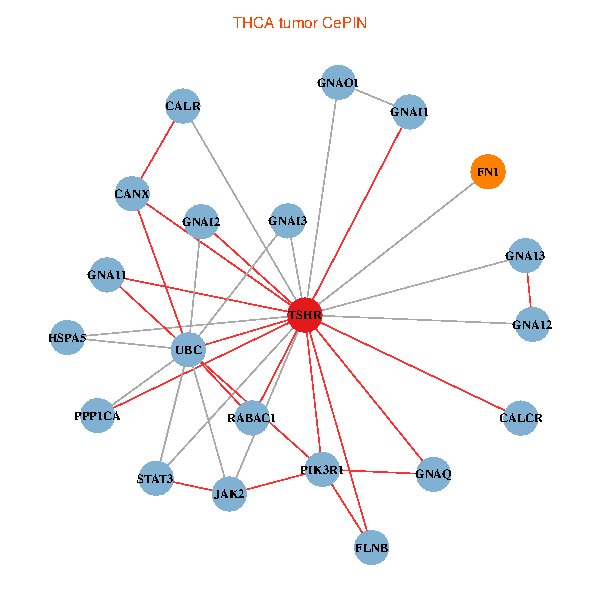
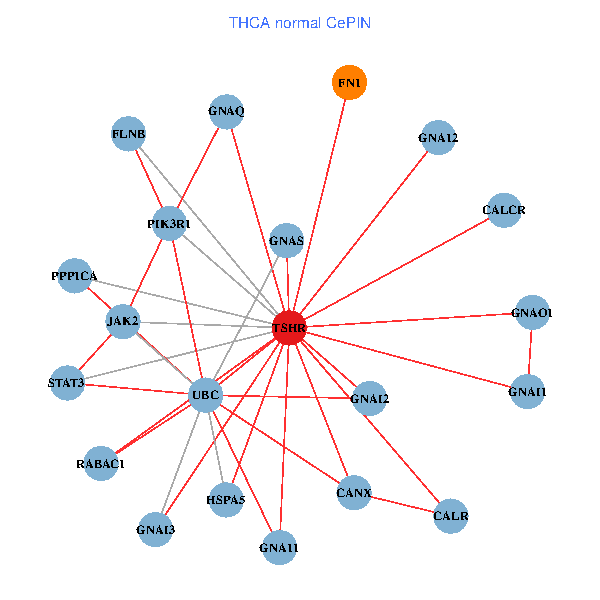
| THCA (tumor) | THCA (normal) |
| TSHR, PPP1CA, UBC, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, PIK3R1, CALR, HSPA5, CANX, GNAI2, CALCR, GNA12, GNAO1, GNAI1, GNA11, GNAI3, GNA13 (tumor) | TSHR, PPP1CA, UBC, FLNB, FN1, STAT3, JAK2, GNAQ, RABAC1, PIK3R1, CALR, HSPA5, CANX, GNAI2, CALCR, GNA12, GNAS, GNAO1, GNAI1, GNA11, GNAI3 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0018213 | Graves Disease | 125 | BeFree,CTD_human,GAD,GWASCAT,LHGDN |
| umls:C0020550 | Hyperthyroidism | 57 | BeFree,CTD_human,LHGDN |
| umls:C0010308 | Congenital Hypothyroidism | 30 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0342200 | Endemic Cretinism | 30 | BeFree |
| umls:C0178468 | Autoimmune thyroid disease | 28 | BeFree,GAD |
| umls:C1836706 | Hyperthyroidism, Nonautoimmune | 28 | BeFree,CLINVAR,CTD_human,ORPHANET,UNIPROT |
| umls:C0549473 | Thyroid carcinoma | 26 | BeFree,GAD |
| umls:C0339143 | Thyroid associated opthalmopathies | 25 | BeFree,GAD |
| umls:C0040137 | Thyroid Nodule | 22 | BeFree,GAD |
| umls:C0040136 | Thyroid Neoplasm | 21 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0001430 | Adenoma | 19 | BeFree,CTD_human |
| umls:C0020676 | Hypothyroidism | 19 | BeFree,GAD,LHGDN |
| umls:C0007115 | Malignant neoplasm of thyroid | 17 | BeFree |
| umls:C3493776 | HYPOTHYROIDISM, CONGENITAL, NONGOITROUS, 1 | 14 | BeFree,CLINVAR,MGD,ORPHANET,UNIPROT |
| umls:C0018021 | Goiter | 13 | BeFree |
| umls:C0040128 | Thyroid Diseases | 13 | BeFree,GAD |
| umls:C0040156 | Thyrotoxicosis | 13 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0151468 | Thyroid Gland Follicular Adenoma | 12 | BeFree |
| umls:C0004364 | Autoimmune Diseases | 10 | BeFree |
| umls:C0238463 | Papillary thyroid carcinoma | 10 | BeFree |
| umls:C1563716 | Thyroid Dysgenesis | 9 | BeFree,LHGDN |
| umls:C0302840 | Toxic thyroid adenoma | 8 | BeFree |
| umls:C1262098 | Congenital hyperthyroidism | 8 | BeFree |
| umls:C1337013 | Differentiated Thyroid Gland Carcinoma | 8 | BeFree |
| umls:C0015397 | Disorder of eye | 7 | BeFree,GAD,LHGDN |
| umls:C0700502 | Acquired hypothyroidism | 7 | BeFree |
| umls:C0027145 | Myxedema | 6 | BeFree |
| umls:C0029182 | orbit (eye disorders) | 6 | BeFree |
| umls:C0154143 | Toxic multinodular goiter | 6 | BeFree |
| umls:C0271790 | Subclinical hypothyroidism | 6 | BeFree |
| umls:C0342208 | Multinodular goiter | 6 | BeFree |
| umls:C2940786 | Thyroid Hormone Resistance Syndrome | 6 | BeFree |
| umls:C0028259 | Nodule | 5 | BeFree |
| umls:C0040021 | Thromboangiitis Obliterans | 5 | BeFree |
| umls:C0040147 | Thyroiditis | 5 | BeFree |
| umls:C0206682 | Follicular thyroid carcinoma | 5 | BeFree |
| umls:C0677607 | Hashimoto Disease | 5 | BeFree,GAD |
| umls:C0238462 | Medullary carcinoma of thyroid | 4 | BeFree |
| umls:C0869147 | Neonatal hyperthyroidism | 4 | BeFree |
| umls:C0002793 | Anaplasia | 3 | BeFree |
| umls:C0007133 | Carcinoma, Papillary | 3 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 3 | BeFree |
| umls:C0032002 | Pituitary Diseases | 3 | BeFree |
| umls:C0151516 | Thyroid Hypoplasia | 3 | BeFree,ORPHANET |
| umls:C0151526 | Premature Birth | 3 | GAD |
| umls:C0238461 | Anaplastic thyroid carcinoma | 3 | BeFree |
| umls:C1112776 | Thyroid hyperplasia | 3 | BeFree |
| umls:C0013080 | Down Syndrome | 2 | BeFree |
| umls:C0018023 | Nodular Goiter | 2 | GAD,LHGDN |
| umls:C0021368 | Inflammation | 2 | GAD |
| umls:C0151514 | Atrophic condition of skin | 2 | BeFree |
| umls:C0205647 | Follicular adenoma | 2 | BeFree |
| umls:C0205696 | Anaplastic carcinoma | 2 | BeFree |
| umls:C0206693 | Medullary carcinoma | 2 | BeFree |
| umls:C0271791 | Severe hypothyroidism | 2 | BeFree |
| umls:C0596263 | Carcinogenesis | 2 | BeFree |
| umls:C0749420 | Thyroid Agenesis | 2 | BeFree |
| umls:C0920350 | Autoimmune thyroiditis | 2 | BeFree,CTD_human,GAD |
| umls:C1998045 | Subclinical hyperthyroidism | 2 | BeFree |
| umls:C2940785 | HYPOTHYROIDISM, CONGENITAL, NONGOITROUS, 3 | 2 | BeFree |
| umls:C3714618 | Primary Hyperthyroidism | 2 | BeFree |
| umls:C0000768 | Congenital Abnormality | 1 | BeFree |
| umls:C0000809 | Abortion, Habitual | 1 | GAD |
| umls:C0001624 | Adrenal Gland Neoplasms | 1 | CTD_human |
| umls:C0007867 | Cervix Diseases | 1 | BeFree |
| umls:C0009782 | Connective Tissue Diseases | 1 | GAD |
| umls:C0014130 | Endocrine System Diseases | 1 | GAD |
| umls:C0015544 | Failure to Thrive | 1 | BeFree |
| umls:C0015929 | Fetal Diseases | 1 | GAD |
| umls:C0021361 | Female infertility | 1 | GAD |
| umls:C0024121 | Lung Neoplasms | 1 | CTD_human |
| umls:C0024232 | Lymphatic Metastasis | 1 | CTD_human |
| umls:C0024667 | Animal Mammary Neoplasms | 1 | GAD |
| umls:C0026267 | Mitral Valve Prolapse Syndrome | 1 | BeFree |
| umls:C0026764 | Multiple Myeloma | 1 | BeFree |
| umls:C0026769 | Multiple Sclerosis | 1 | GAD |
| umls:C0026857 | Musculoskeletal Diseases | 1 | GAD |
| umls:C0027819 | Neuroblastoma | 1 | BeFree |
| umls:C0030297 | Pancreatic Neoplasm | 1 | GAD |
| umls:C0032964 | Pregnancy Complications, Hematologic | 1 | GAD |
| umls:C0033844 | Pseudotumor | 1 | BeFree |
| umls:C0037274 | Dermatologic disorders | 1 | GAD |
| umls:C0038013 | Ankylosing spondylitis | 1 | GAD |
| umls:C0038379 | Strabismus | 1 | BeFree |
| umls:C0040115 | Thymus Hyperplasia | 1 | BeFree |
| umls:C0040336 | Tobacco Use Disorder | 1 | GAD |
| umls:C0154094 | Carcinoma in situ of eye | 1 | GAD |
| umls:C0158983 | Neonatal thyrotoxicosis | 1 | BeFree |
| umls:C0205642 | Adenocarcinoma, Oxyphilic | 1 | BeFree |
| umls:C0271335 | Orbital congestion | 1 | BeFree |
| umls:C0342114 | Diffuse goiter | 1 | BeFree |
| umls:C0342127 | Toxic nodular goiter | 1 | BeFree |
| umls:C0410158 | Muscle damage | 1 | BeFree |
| umls:C0600086 | Toxic goiter | 1 | BeFree |
| umls:C0686619 | Secondary malignant neoplasm of lymph node | 1 | BeFree |
| umls:C0700095 | Central neuroblastoma | 1 | BeFree |
| umls:C0749424 | Thyroid Hurthle Cell Carcinoma | 1 | BeFree |
| umls:C0920420 | cancer recurrence | 1 | BeFree |
| umls:C1262091 | Lymphocytic infiltration | 1 | BeFree |
| umls:C1266050 | Poorly Differentiated Thyroid Carcinoma | 1 | BeFree |
| umls:C1458155 | Mammary Neoplasms | 1 | GAD |
| umls:C1510502 | Oxyphilic Adenoma | 1 | BeFree |
| umls:C1863959 | Hyperthyroidism, Familial Gestational | 1 | CLINVAR,CTD_human,ORPHANET,UNIPROT |
| umls:C1955741 | Glucocorticoid deficiency | 1 | BeFree |
| umls:C2024965 | Caries of smooth surface of tooth | 1 | GWASCAT |
| umls:C3714514 | Infection | 1 | GAD |
| umls:C1863960 | Thyroid Adenoma, Hyperfunctioning | 0 | CLINVAR |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))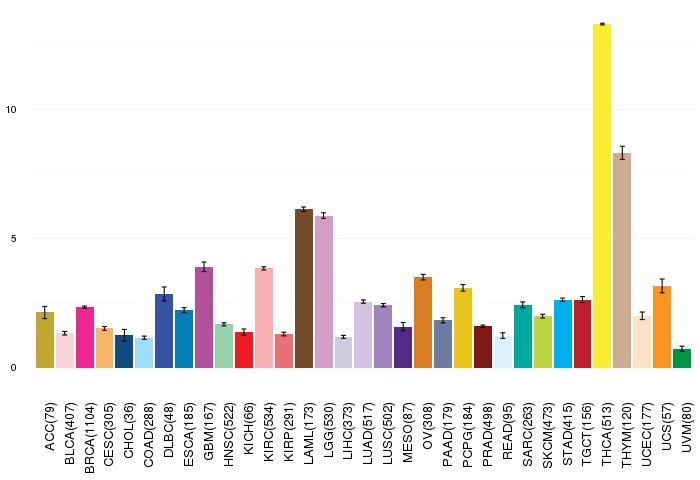
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))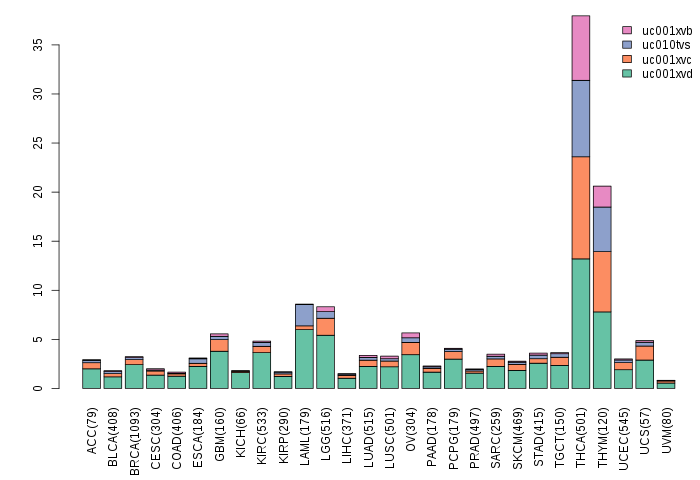
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data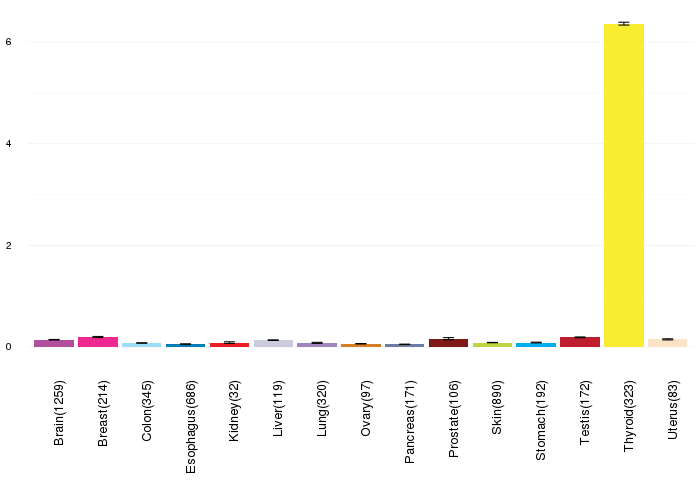
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))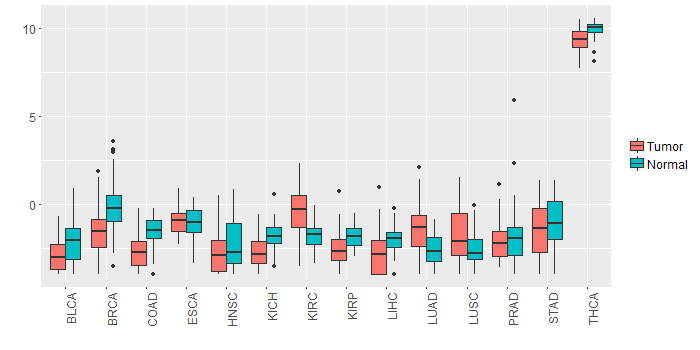
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.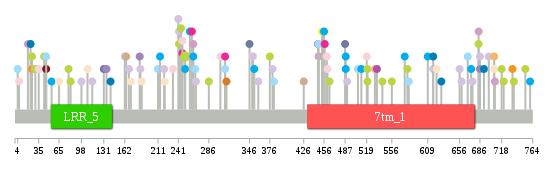

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 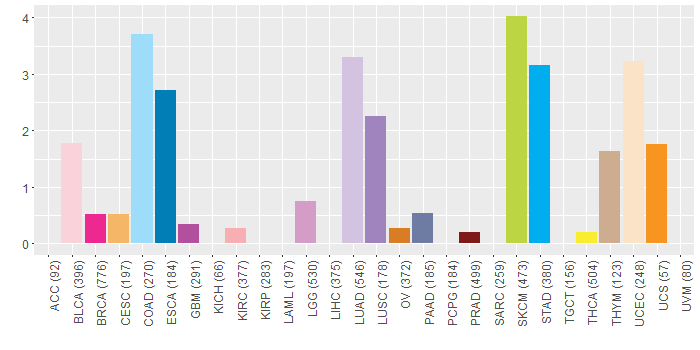
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)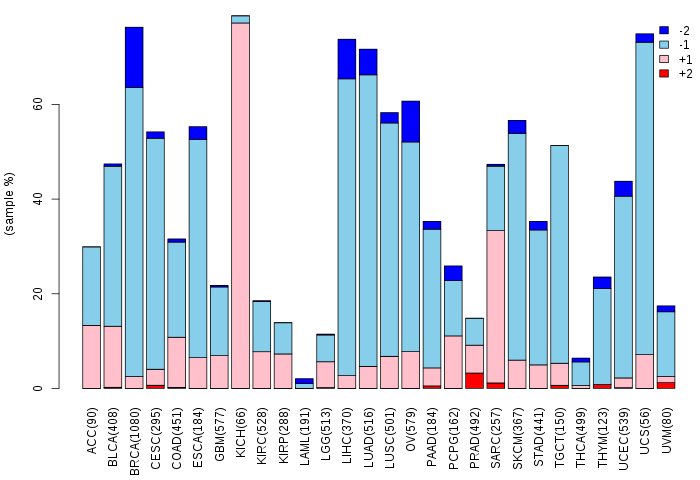
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 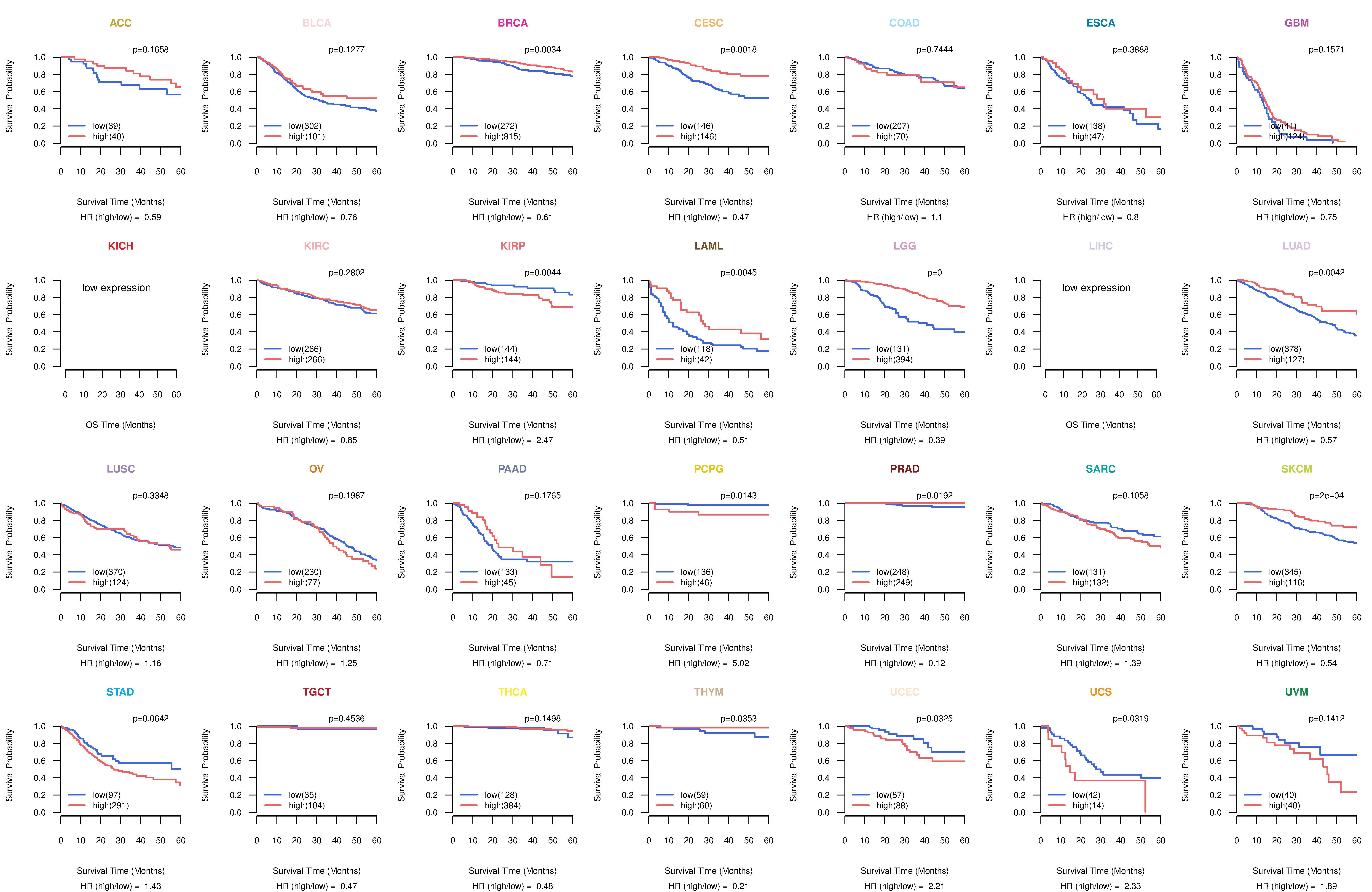
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 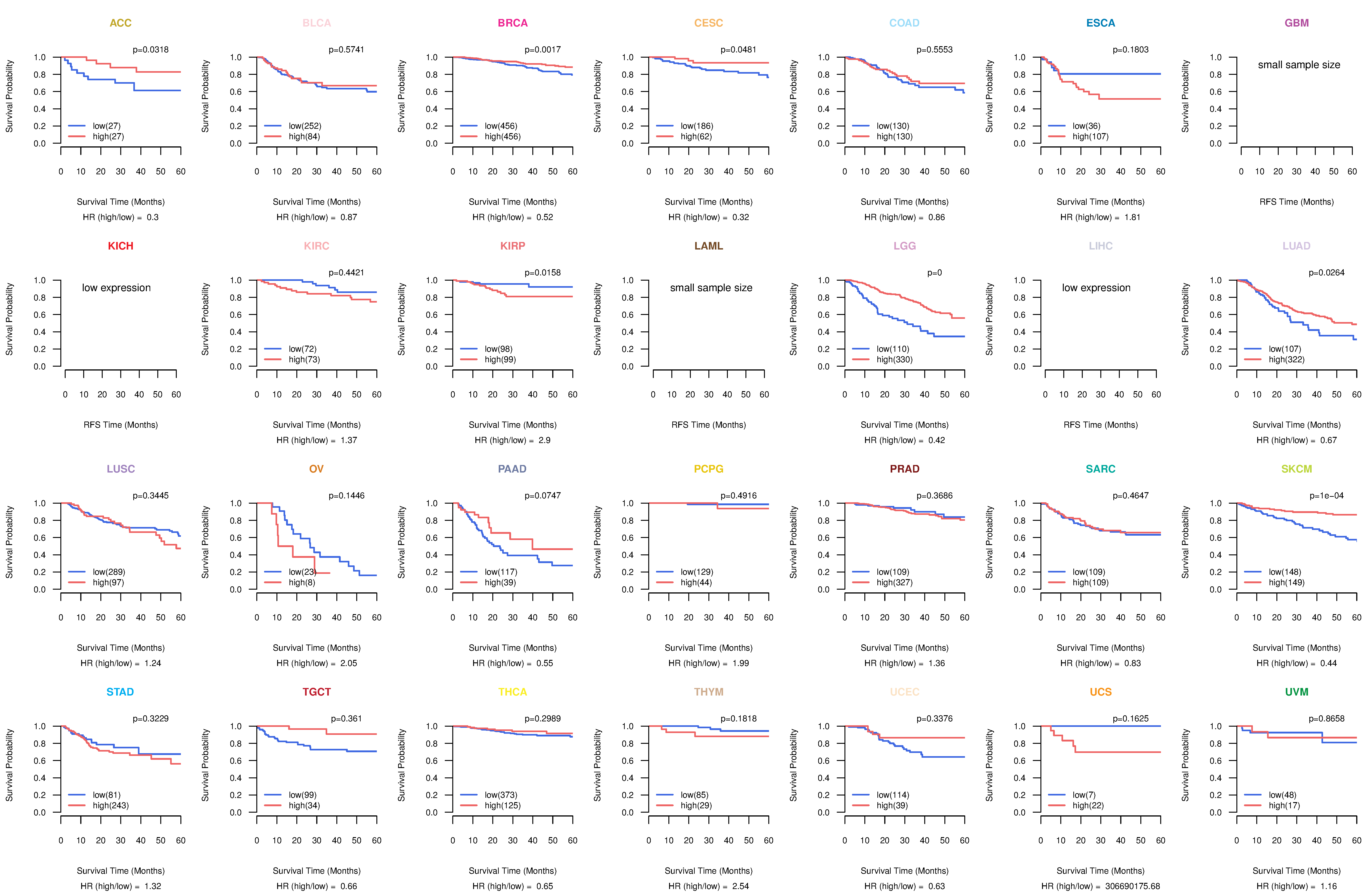
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 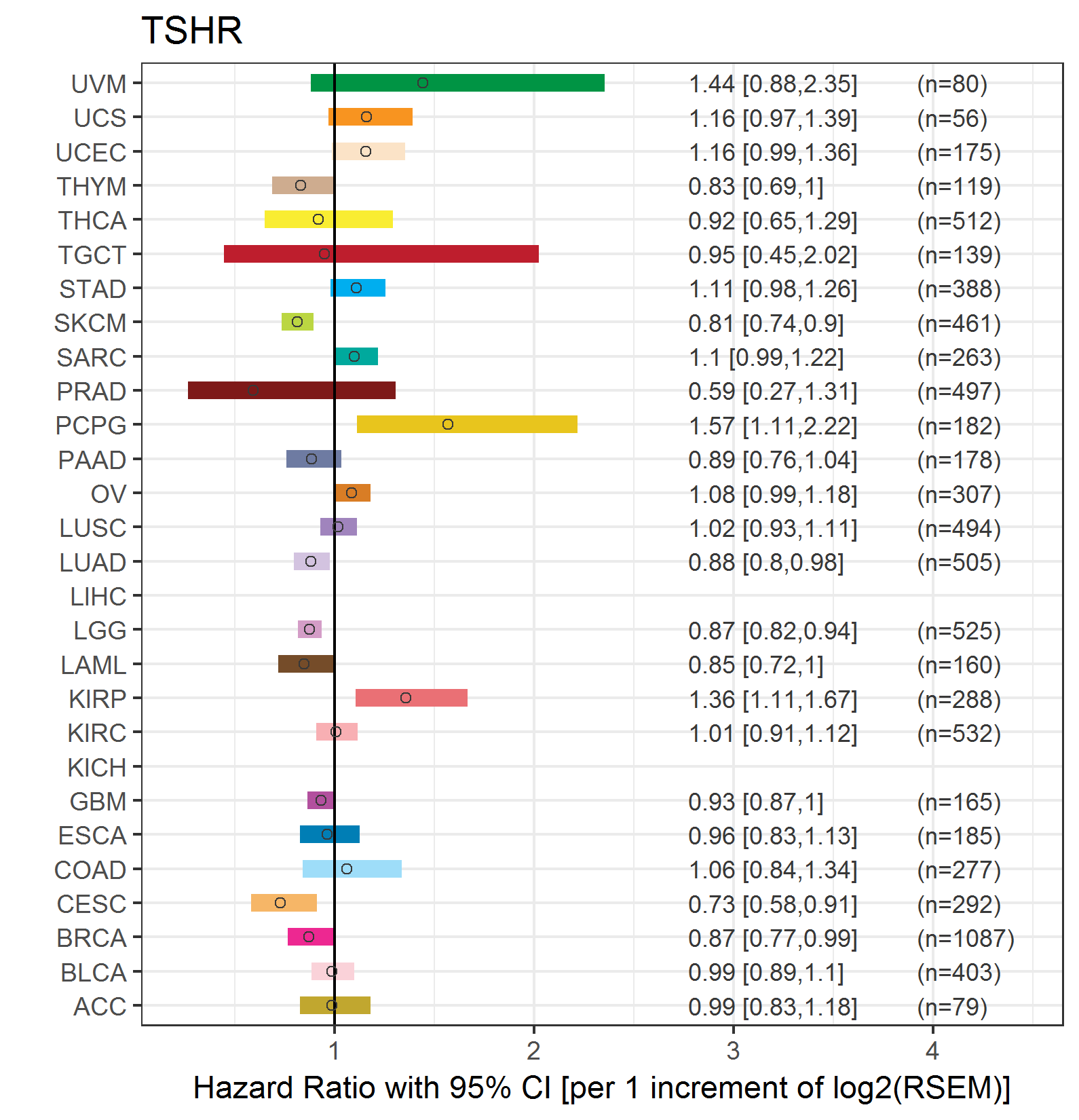
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 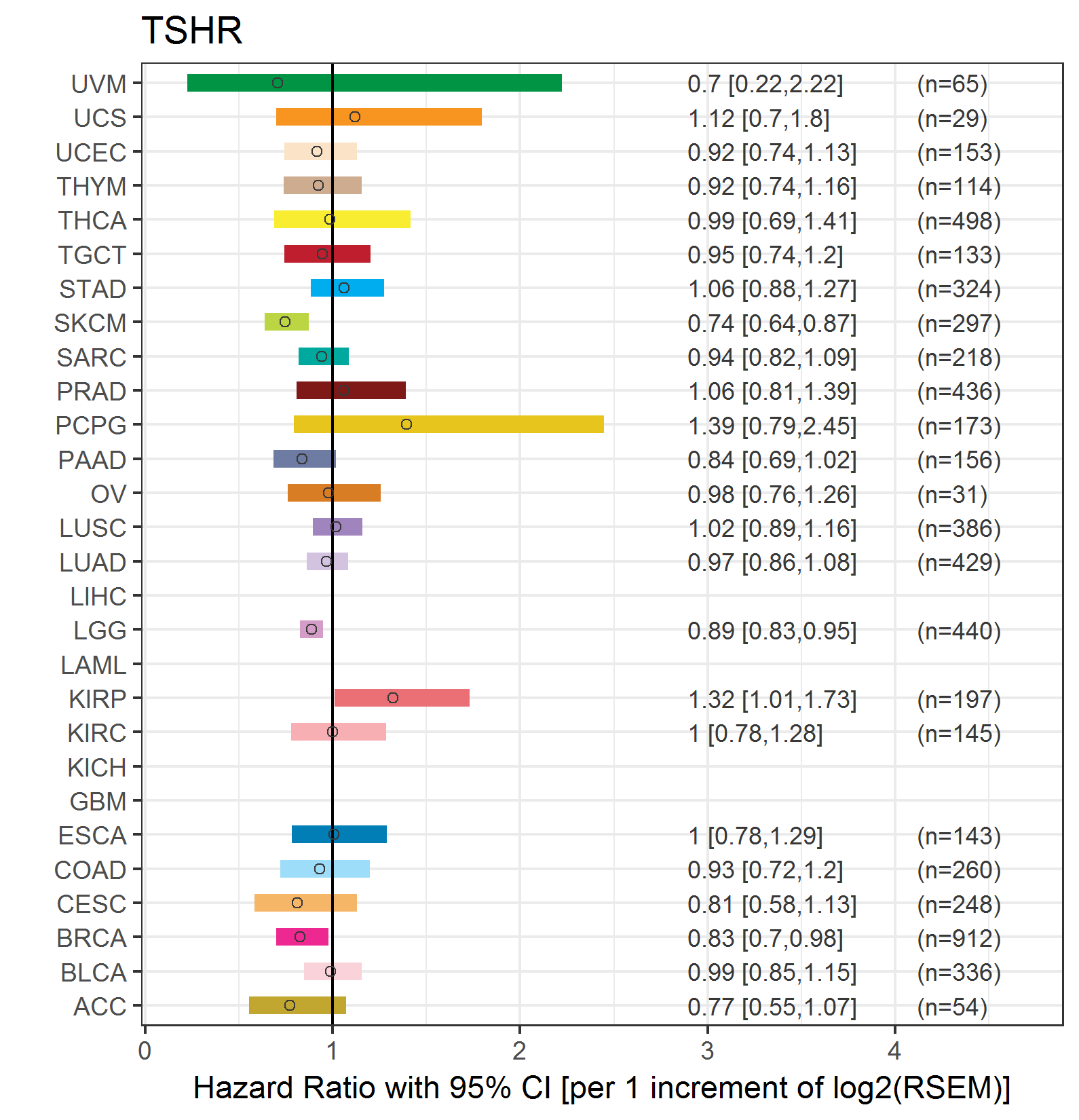
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























