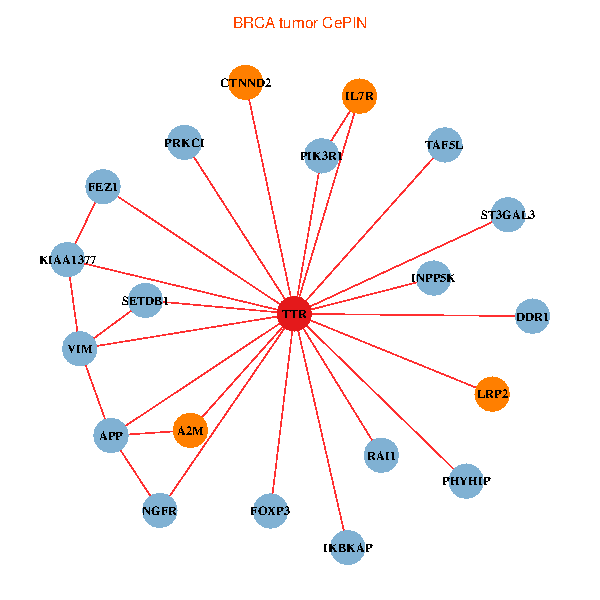
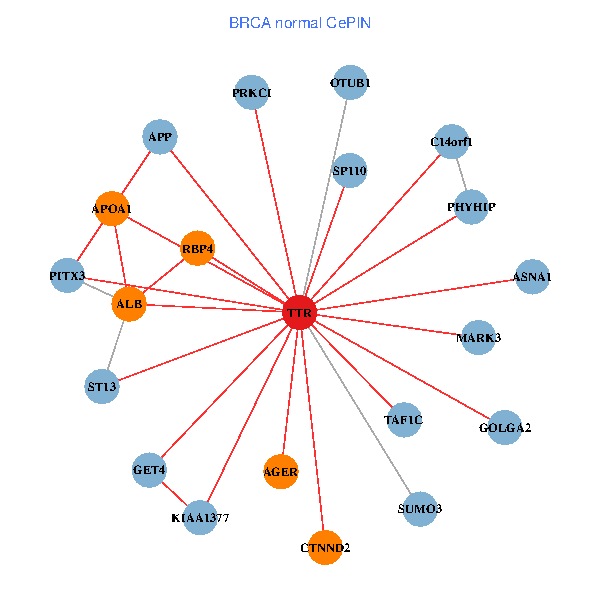
| BRCA (tumor) | BRCA (normal) |
| TTR, SETDB1, IKBKAP, APP, PIK3R1, FEZ1, NGFR, IL7R, RAI1, A2M, KIAA1377, PHYHIP, DDR1, PRKCI, VIM, TAF5L, FOXP3, LRP2, ST3GAL3, CTNND2, INPP5K (tumor) | TTR, TAF1C, GOLGA2, APP, PITX3, ALB, GET4, C14orf1, APOA1, KIAA1377, SUMO3, AGER, OTUB1, PHYHIP, ST13, PRKCI, MARK3, SP110, CTNND2, RBP4, ASNA1 (normal) |
 |  |
|
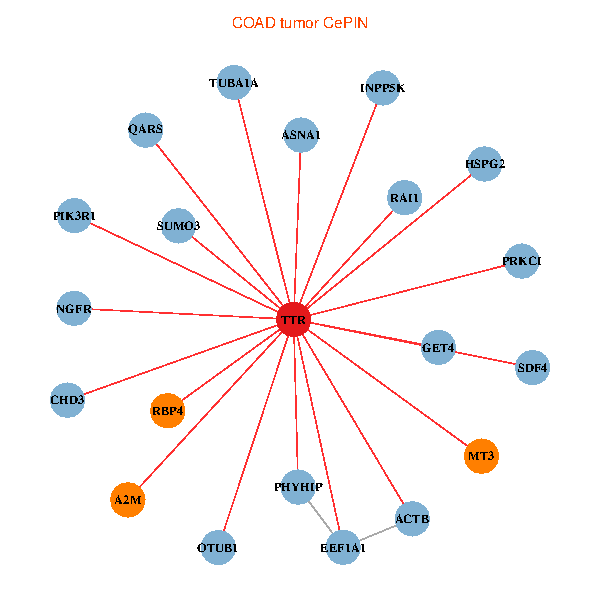
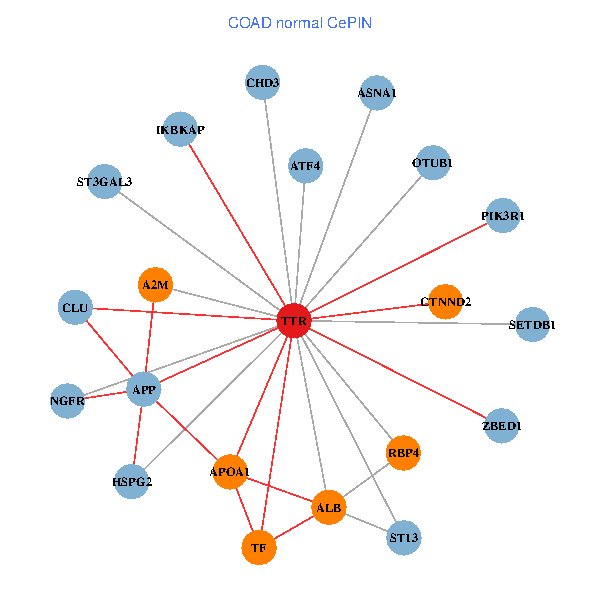
| COAD (tumor) | COAD (normal) |
| TTR, CHD3, ACTB, PIK3R1, NGFR, GET4, RAI1, A2M, EEF1A1, HSPG2, TUBA1A, SUMO3, OTUB1, PHYHIP, PRKCI, QARS, INPP5K, SDF4, RBP4, MT3, ASNA1 (tumor) | TTR, CHD3, SETDB1, ZBED1, IKBKAP, APP, PIK3R1, ALB, NGFR, A2M, HSPG2, APOA1, OTUB1, ST13, TF, ATF4, CLU, ST3GAL3, CTNND2, RBP4, ASNA1 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
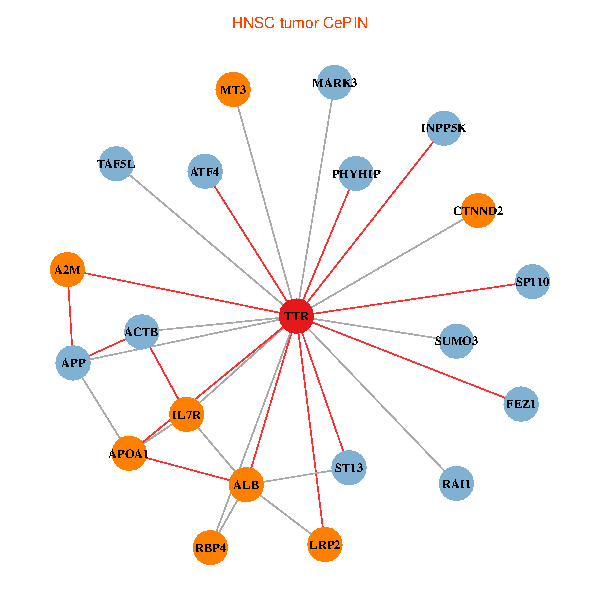
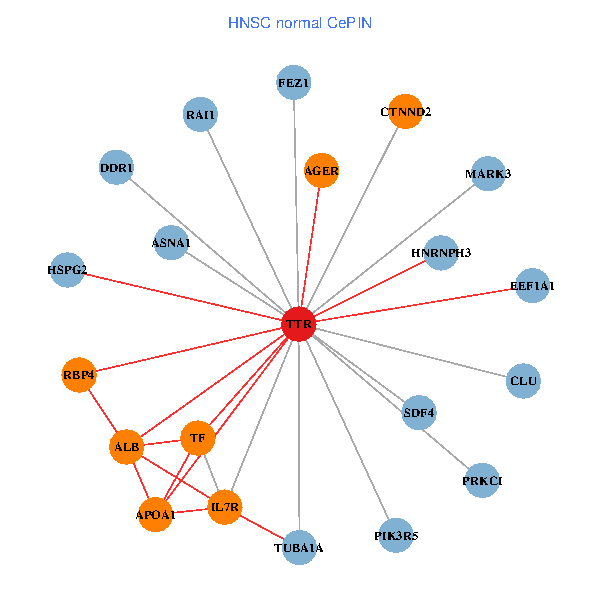
| TTR, APP, ACTB, ALB, FEZ1, IL7R, RAI1, A2M, APOA1, SUMO3, PHYHIP, ST13, MARK3, TAF5L, ATF4, LRP2, SP110, CTNND2, INPP5K, RBP4, MT3 (tumor) | TTR, ALB, FEZ1, IL7R, RAI1, EEF1A1, HSPG2, TUBA1A, APOA1, AGER, DDR1, PRKCI, MARK3, TF, HNRNPH3, PIK3R5, CLU, CTNND2, SDF4, RBP4, ASNA1 (normal) |
 |  |
|
| KICH (tumor) | KICH (normal) |
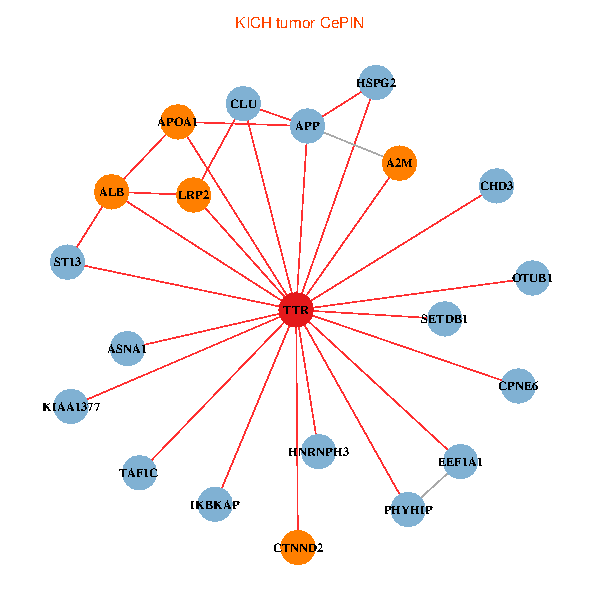
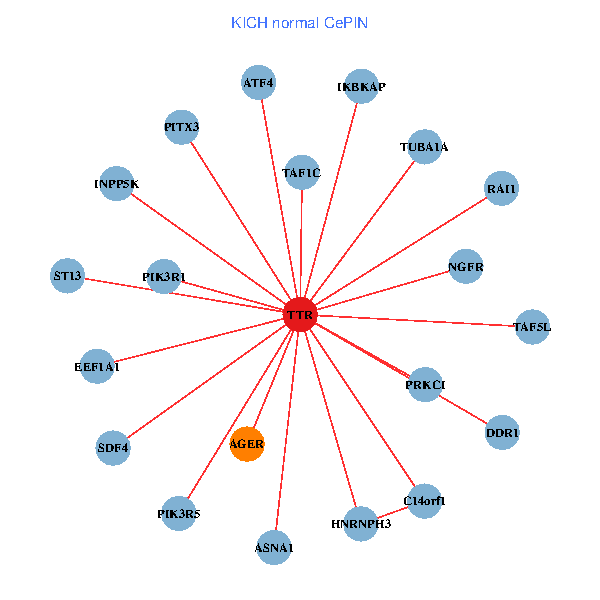
| TTR, TAF1C, CHD3, SETDB1, IKBKAP, APP, ALB, A2M, EEF1A1, HSPG2, APOA1, KIAA1377, OTUB1, PHYHIP, ST13, HNRNPH3, LRP2, CLU, CTNND2, ASNA1, CPNE6 (tumor) | TTR, TAF1C, IKBKAP, PIK3R1, PITX3, NGFR, C14orf1, RAI1, EEF1A1, TUBA1A, AGER, ST13, DDR1, PRKCI, TAF5L, ATF4, HNRNPH3, PIK3R5, INPP5K, SDF4, ASNA1 (normal) |
 |  |
|
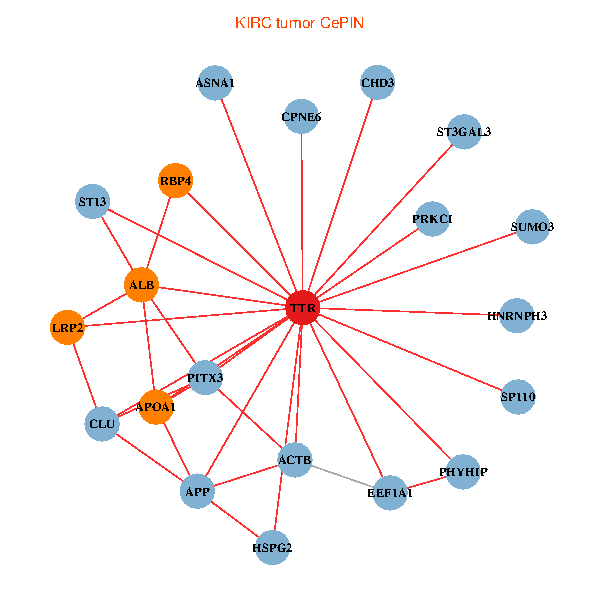
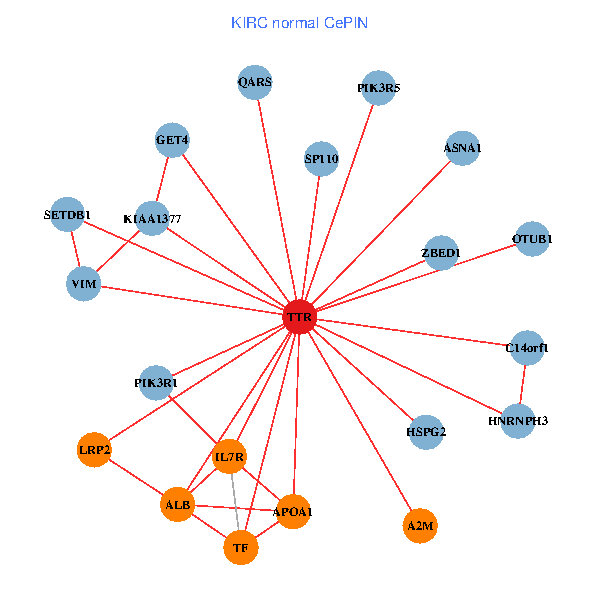
| KIRC (tumor) | KIRC (normal) |
| TTR, CHD3, APP, ACTB, PITX3, ALB, EEF1A1, HSPG2, APOA1, SUMO3, PHYHIP, ST13, PRKCI, HNRNPH3, LRP2, CLU, SP110, ST3GAL3, RBP4, ASNA1, CPNE6 (tumor) | TTR, SETDB1, ZBED1, PIK3R1, ALB, GET4, C14orf1, IL7R, A2M, HSPG2, APOA1, KIAA1377, OTUB1, TF, VIM, HNRNPH3, LRP2, PIK3R5, SP110, QARS, ASNA1 (normal) |
 |  |
|
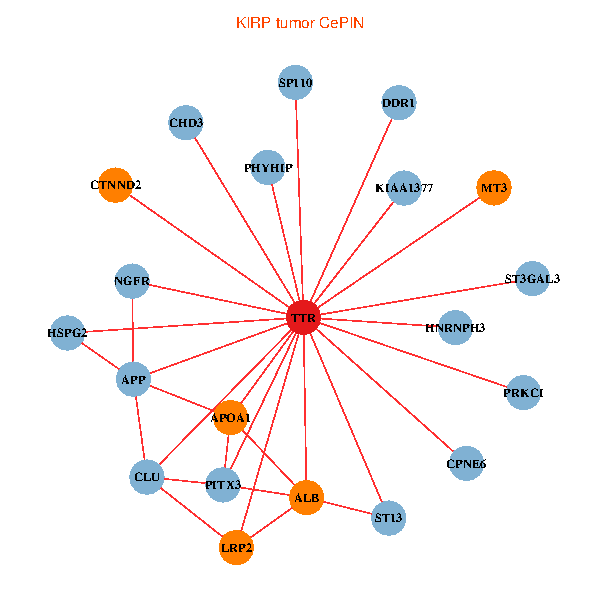
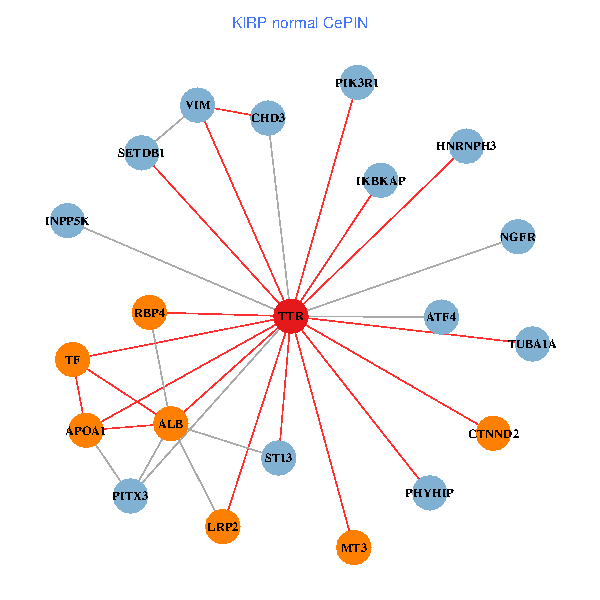
| KIRP (tumor) | KIRP (normal) |
| TTR, CHD3, APP, PITX3, ALB, NGFR, HSPG2, APOA1, KIAA1377, PHYHIP, ST13, DDR1, PRKCI, HNRNPH3, LRP2, CLU, SP110, ST3GAL3, CTNND2, MT3, CPNE6 (tumor) | TTR, CHD3, SETDB1, IKBKAP, PIK3R1, PITX3, ALB, NGFR, TUBA1A, APOA1, PHYHIP, ST13, TF, VIM, ATF4, HNRNPH3, LRP2, CTNND2, INPP5K, RBP4, MT3 (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| TTR, CHD3, IKBKAP, ALB, GET4, C14orf1, IL7R, HSPG2, TUBA1A, APOA1, KIAA1377, ST13, PRKCI, TF, VIM, PIK3R5, CLU, SP110, ST3GAL3, SDF4, RBP4 (tumor) | TTR, CHD3, GOLGA2, PIK3R1, PITX3, ALB, C14orf1, RAI1, TUBA1A, APOA1, KIAA1377, DDR1, PRKCI, TF, VIM, HNRNPH3, CLU, ST3GAL3, CTNND2, RBP4, CPNE6 (normal) |
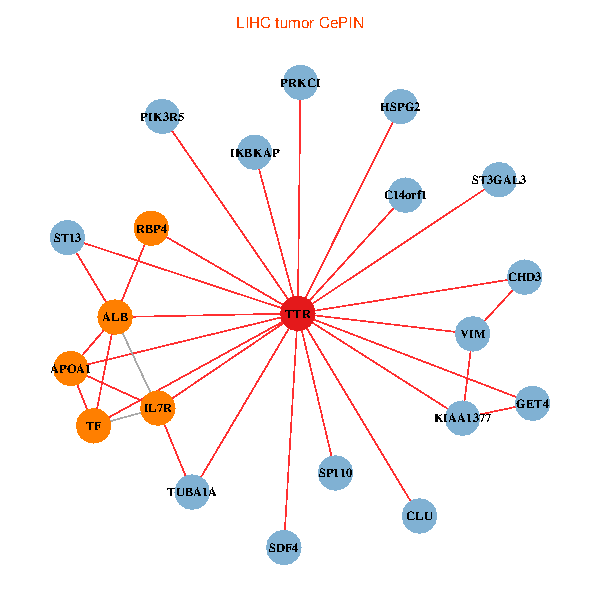 | 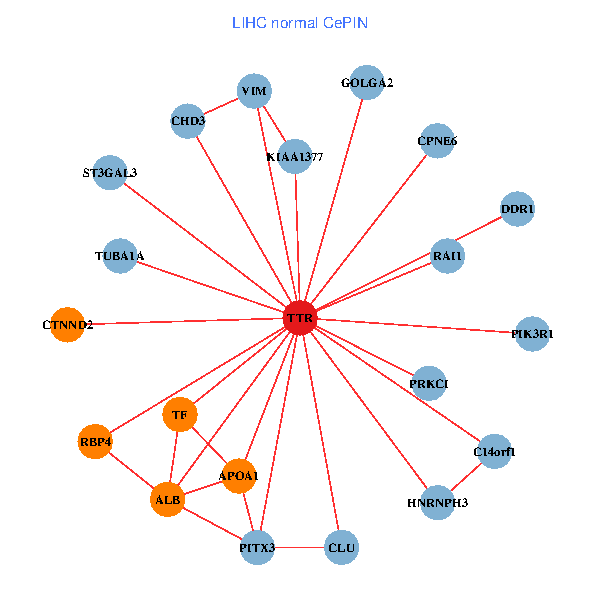 |
|
| LUAD (tumor) | LUAD (normal) |
| TTR, TAF1C, APP, PITX3, IL7R, TUBA1A, KIAA1377, SUMO3, OTUB1, PHYHIP, ST13, PRKCI, MARK3, TF, VIM, FOXP3, PIK3R5, CLU, ST3GAL3, INPP5K, MT3 (tumor) | TTR, TAF1C, ZBED1, APP, ACTB, ALB, GET4, APOA1, KIAA1377, SUMO3, OTUB1, DDR1, TF, VIM, TAF5L, ATF4, CLU, CTNND2, SDF4, RBP4, CPNE6 (normal) |
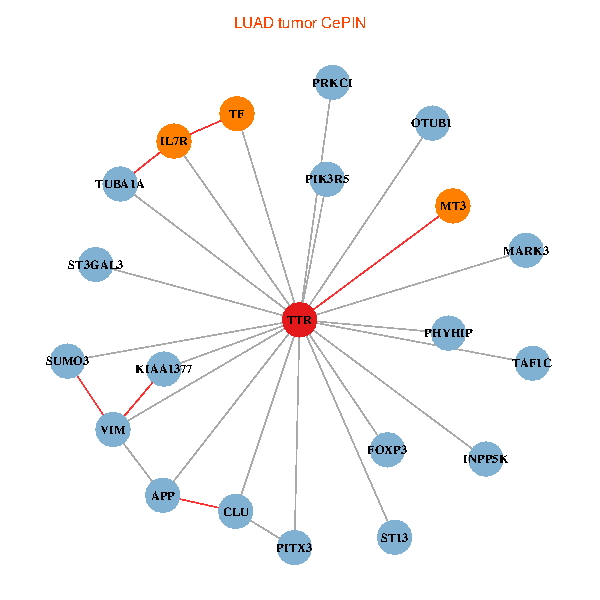 | 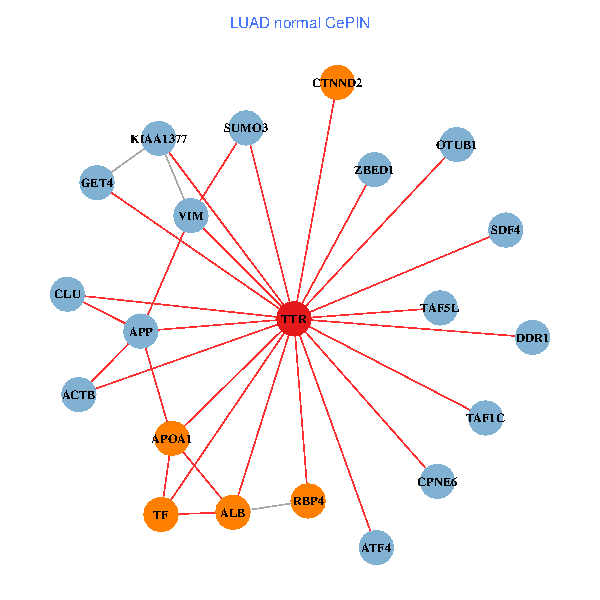 |
|
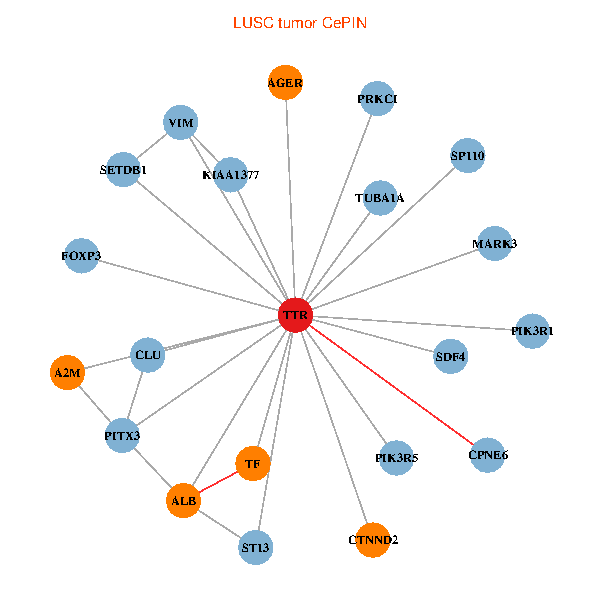
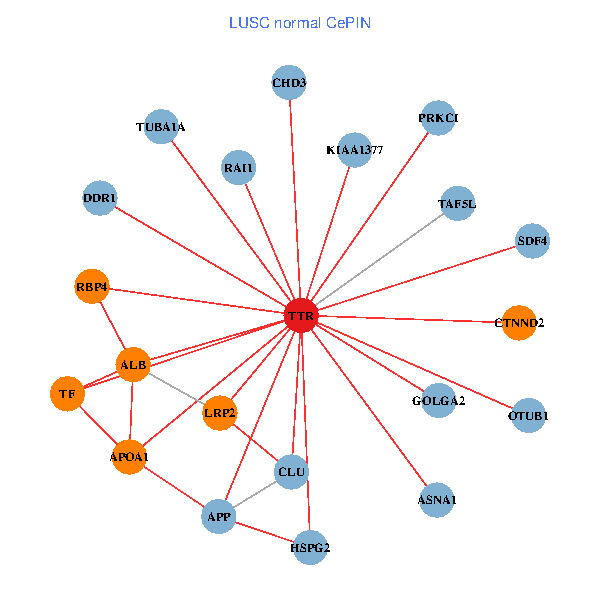
| LUSC (tumor) | LUSC (normal) |
| TTR, SETDB1, PIK3R1, PITX3, ALB, A2M, TUBA1A, KIAA1377, AGER, ST13, PRKCI, MARK3, TF, VIM, FOXP3, PIK3R5, CLU, SP110, CTNND2, SDF4, CPNE6 (tumor) | TTR, CHD3, GOLGA2, APP, ALB, RAI1, HSPG2, TUBA1A, APOA1, KIAA1377, OTUB1, DDR1, PRKCI, TF, TAF5L, LRP2, CLU, CTNND2, SDF4, RBP4, ASNA1 (normal) |
 |  |
|
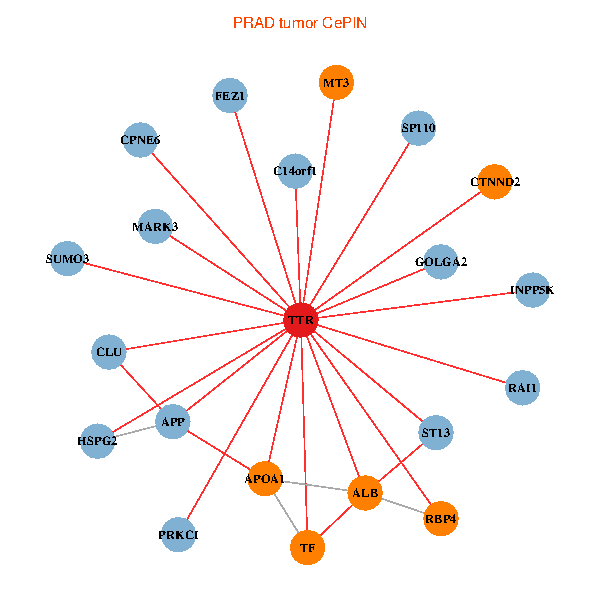
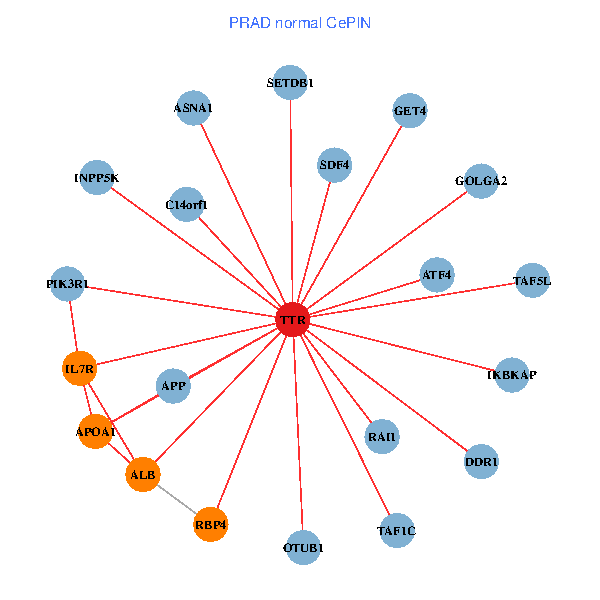
| PRAD (tumor) | PRAD (normal) |
| TTR, GOLGA2, APP, ALB, FEZ1, C14orf1, RAI1, HSPG2, APOA1, SUMO3, ST13, PRKCI, MARK3, TF, CLU, SP110, CTNND2, INPP5K, RBP4, MT3, CPNE6 (tumor) | TTR, TAF1C, SETDB1, GOLGA2, IKBKAP, APP, PIK3R1, ALB, GET4, C14orf1, IL7R, RAI1, APOA1, OTUB1, DDR1, TAF5L, ATF4, INPP5K, SDF4, RBP4, ASNA1 (normal) |
 |  |
|
| STAD (tumor) | STAD (normal) |
| TTR, CHD3, GOLGA2, ZBED1, IKBKAP, ALB, NGFR, IL7R, RAI1, APOA1, KIAA1377, SUMO3, PHYHIP, MARK3, TF, PIK3R5, SP110, CTNND2, SDF4, RBP4, ASNA1 (tumor) | TTR, SETDB1, GOLGA2, ACTB, ALB, FEZ1, GET4, C14orf1, RAI1, A2M, EEF1A1, APOA1, KIAA1377, AGER, TF, LRP2, CLU, SP110, CTNND2, RBP4, ASNA1 (normal) |
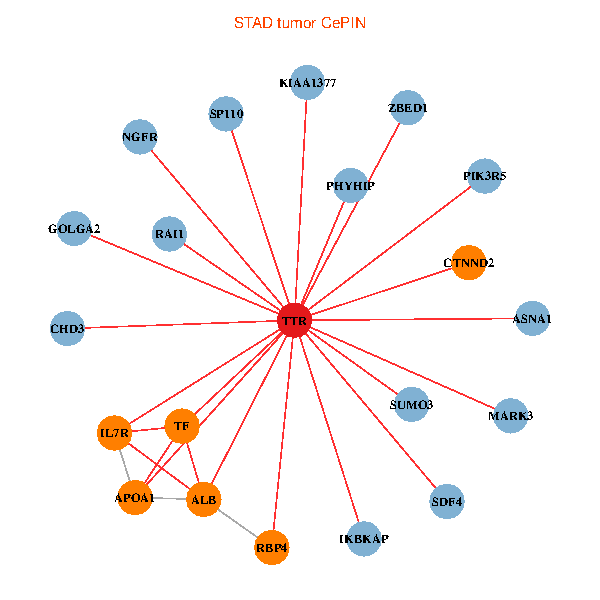 | 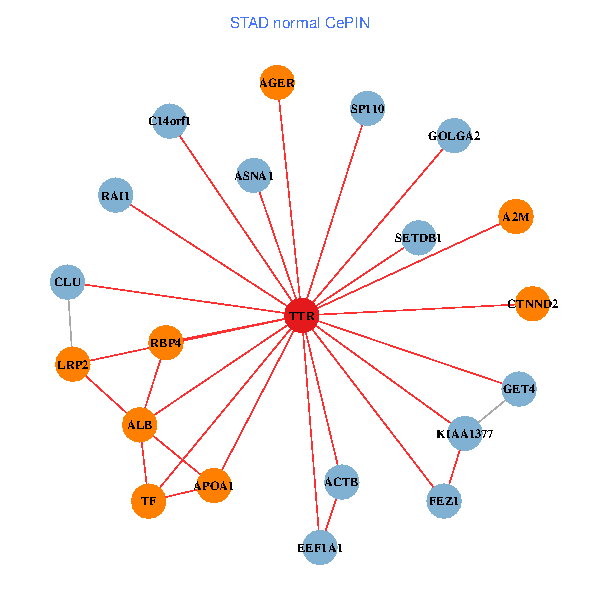 |
|
| THCA (tumor) | THCA (normal) |
| TTR, TAF1C, CHD3, GOLGA2, PIK3R1, PITX3, FEZ1, NGFR, C14orf1, RAI1, EEF1A1, HSPG2, APOA1, KIAA1377, SUMO3, AGER, PHYHIP, DDR1, SP110, SDF4, RBP4 (tumor) | TTR, TAF1C, GOLGA2, IKBKAP, ALB, GET4, C14orf1, RAI1, APOA1, KIAA1377, OTUB1, PRKCI, TF, VIM, TAF5L, HNRNPH3, ST3GAL3, CTNND2, INPP5K, SDF4, CPNE6 (normal) |
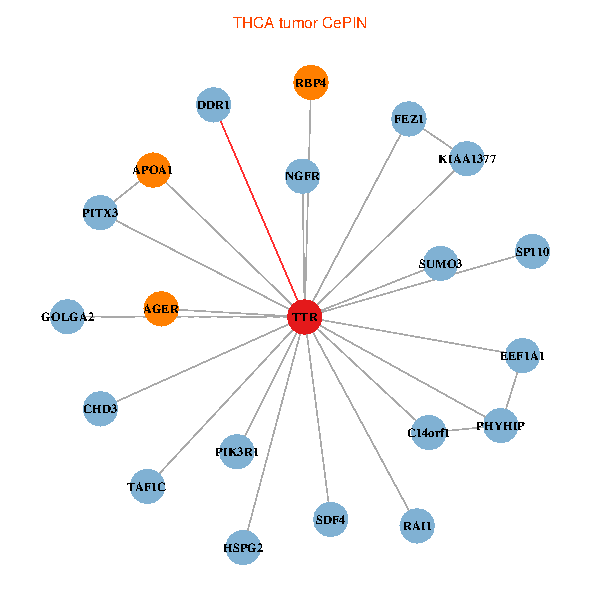 | 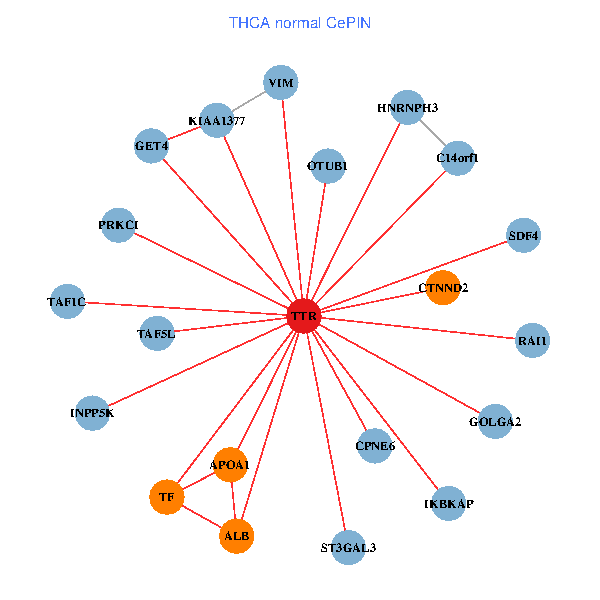 |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0002726 | Amyloidosis | 257 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0152025 | Polyneuropathy | 173 | BeFree,LHGDN |
| umls:C0206245 | Amyloid Neuropathies, Familial | 160 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0032580 | Adenomatous Polyposis Coli | 110 | BeFree |
| umls:C0268407 | Senile cardiac amyloidosis | 81 | BeFree,GAD |
| umls:C2751492 | AMYLOIDOSIS, HEREDITARY, TRANSTHYRETIN-RELATED | 67 | BeFree,CLINVAR,CTD_human,ORPHANET,UNIPROT |
| umls:C2936349 | Plaque, Amyloid | 66 | BeFree |
| umls:C0878544 | Cardiomyopathies | 43 | BeFree |
| umls:C0342623 | Senile systemic amyloidosis | 41 | BeFree |
| umls:C0268380 | Systemic amyloidosis | 38 | BeFree |
| umls:C0281479 | Primary Systemic Amyloidosis | 38 | BeFree |
| umls:C0740340 | Amyloidosis, Familial | 34 | BeFree,CTD_human |
| umls:C0206247 | Amyloid Neuropathies | 32 | BeFree,GAD |
| umls:C0206246 | Amyloidosis, Hereditary | 25 | BeFree |
| umls:C0442874 | Neuropathy | 22 | BeFree |
| umls:C0031117 | Peripheral Neuropathy | 17 | BeFree |
| umls:C0007286 | Carpal Tunnel Syndrome | 16 | BeFree,CTD_human,LHGDN,UNIPROT |
| umls:C0268384 | Familial Amyloid Neuropathy, Portuguese Type | 16 | BeFree |
| umls:C0342613 | Danish type familial amyloid cardiomyopathy | 12 | BeFree |
| umls:C0524851 | Neurodegenerative Disorders | 11 | BeFree |
| umls:C0018802 | Congestive heart failure | 10 | BeFree |
| umls:C0259749 | Autonomic neuropathy | 10 | BeFree |
| umls:C0002395 | Alzheimer's Disease | 9 | BeFree,GAD,LHGDN |
| umls:C0020551 | Hyperthyroxinemia | 9 | BeFree |
| umls:C2239176 | Liver carcinoma | 8 | BeFree |
| umls:C0018801 | Heart failure | 7 | BeFree |
| umls:C0027819 | Neuroblastoma | 7 | BeFree |
| umls:C0700095 | Central neuroblastoma | 7 | BeFree |
| umls:C0221014 | Reactive systemic amyloidosis | 6 | BeFree |
| umls:C0271682 | Mixed sensory-motor polyneuropathy | 6 | BeFree |
| umls:C0007196 | Restrictive cardiomyopathy | 5 | BeFree |
| umls:C1842937 | AURAL ATRESIA, CONGENITAL | 5 | BeFree |
| umls:C0024121 | Lung Neoplasms | 4 | CTD_human,LHGDN |
| umls:C0085220 | Cerebral Amyloid Angiopathy | 4 | BeFree |
| umls:C0333463 | Senile Plaques | 4 | BeFree |
| umls:C0339562 | Amyloid of vitreous | 4 | BeFree |
| umls:C0002871 | Anemia | 3 | BeFree |
| umls:C0007134 | Renal Cell Carcinoma | 3 | BeFree |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 3 | BeFree |
| umls:C0014474 | Ependymoma | 3 | BeFree |
| umls:C0017638 | Glioma | 3 | BeFree |
| umls:C0018799 | Heart Diseases | 3 | BeFree,GAD |
| umls:C0022658 | Kidney Diseases | 3 | BeFree |
| umls:C0023903 | Liver neoplasms | 3 | BeFree |
| umls:C0027765 | nervous system disorder | 3 | BeFree |
| umls:C0268381 | Primary amyloidosis | 3 | BeFree |
| umls:C0268406 | Age-related amyloidosis | 3 | BeFree |
| umls:C0271683 | Polyneuropathy, Motor | 3 | BeFree |
| umls:C0598589 | Inherited neuropathies | 3 | BeFree |
| umls:C0002736 | Amyotrophic Lateral Sclerosis | 2 | BeFree |
| umls:C0003850 | Arteriosclerosis | 2 | BeFree |
| umls:C0011265 | Presenile dementia | 2 | BeFree |
| umls:C0013080 | Down Syndrome | 2 | BeFree |
| umls:C0017601 | Glaucoma | 2 | BeFree,LHGDN |
| umls:C0021400 | Influenza | 2 | BeFree |
| umls:C0022661 | Kidney Failure, Chronic | 2 | BeFree |
| umls:C0027627 | Neoplasm Metastasis | 2 | BeFree,LHGDN |
| umls:C0042373 | Vascular Diseases | 2 | BeFree |
| umls:C0162429 | Malnutrition | 2 | BeFree |
| umls:C0235025 | Peripheral motor neuropathy | 2 | BeFree |
| umls:C0338656 | Impaired cognition | 2 | BeFree |
| umls:C0497327 | Dementia | 2 | BeFree |
| umls:C0555278 | Cerebral metastasis | 2 | BeFree |
| umls:C0677932 | Progressive Neoplastic Disease | 2 | BeFree |
| umls:C0679407 | Gastrointestinal dysfunction | 2 | BeFree |
| umls:C0686377 | CNS metastases | 2 | BeFree |
| umls:C0751448 | Polyneuropathy, Familial | 2 | BeFree |
| umls:C1559271 | Non-Malignant Ascites Adverse Event | 2 | BeFree |
| umls:C1719315 | Hereditary cardiac amyloidosis | 2 | BeFree |
| umls:C2316810 | Chronic kidney disease stage 5 | 2 | BeFree |
| umls:C2931784 | Amyloid angiopathy | 2 | BeFree |
| umls:C2939462 | Immunoglobulin deposition disease | 2 | BeFree |
| umls:C3539781 | Progressive cGVHD | 2 | BeFree |
| umls:C0004153 | Atherosclerosis | 1 | BeFree |
| umls:C0005956 | Bone Marrow Diseases | 1 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 1 | BeFree |
| umls:C0007095 | Carcinoid Tumor | 1 | BeFree |
| umls:C0007133 | Carcinoma, Papillary | 1 | BeFree |
| umls:C0007194 | Hypertrophic Cardiomyopathy | 1 | BeFree |
| umls:C0007682 | CNS disorder | 1 | BeFree |
| umls:C0007786 | Brain Ischemia | 1 | RGD |
| umls:C0007789 | Cerebral Palsy | 1 | BeFree |
| umls:C0008350 | Cholelithiasis | 1 | BeFree |
| umls:C0008925 | Cleft Palate | 1 | GAD |
| umls:C0009319 | Colitis | 1 | BeFree |
| umls:C0009421 | Comatose | 1 | BeFree |
| umls:C0010266 | Cranial nerve diseases | 1 | BeFree |
| umls:C0011570 | Mental Depression | 1 | BeFree |
| umls:C0011581 | Depressive disorder | 1 | BeFree |
| umls:C0011609 | Drug Eruptions | 1 | CTD_human |
| umls:C0011854 | Diabetes Mellitus, Insulin-Dependent | 1 | BeFree |
| umls:C0013404 | Dyspnea | 1 | BeFree |
| umls:C0015397 | Disorder of eye | 1 | BeFree |
| umls:C0016085 | Filariasis | 1 | BeFree |
| umls:C0018790 | Cardiac Arrest | 1 | BeFree |
| umls:C0019158 | Hepatitis | 1 | BeFree |
| umls:C0019159 | Hepatitis A | 1 | BeFree |
| umls:C0020538 | Hypertensive disease | 1 | LHGDN |
| umls:C0020676 | Hypothyroidism | 1 | GAD |
| umls:C0022116 | Ischemia | 1 | BeFree |
| umls:C0022806 | Kwashiorkor | 1 | BeFree |
| umls:C0023467 | Leukemia, Myelocytic, Acute | 1 | BeFree |
| umls:C0024408 | Machado-Joseph Disease | 1 | BeFree |
| umls:C0025149 | Medulloblastoma | 1 | BeFree |
| umls:C0025286 | Meningioma | 1 | BeFree |
| umls:C0025362 | Mental Retardation | 1 | BeFree |
| umls:C0026470 | Monoclonal Gammopathy of Undetermined Significance | 1 | BeFree |
| umls:C0027796 | Neuralgia | 1 | BeFree |
| umls:C0027809 | Neurilemmoma | 1 | BeFree |
| umls:C0029408 | Degenerative polyarthritis | 1 | BeFree |
| umls:C0030353 | Papilledema | 1 | BeFree |
| umls:C0030354 | Papilloma | 1 | BeFree |
| umls:C0030421 | Paraganglioma | 1 | BeFree |
| umls:C0031511 | Pheochromocytoma | 1 | BeFree |
| umls:C0033075 | Presbyopia | 1 | BeFree |
| umls:C0036341 | Schizophrenia | 1 | BeFree,GAD |
| umls:C0036572 | Seizures | 1 | BeFree |
| umls:C0037019 | Shy-Drager Syndrome | 1 | BeFree |
| umls:C0037061 | Siderosis | 1 | BeFree |
| umls:C0037928 | Spinal Cord Diseases | 1 | BeFree |
| umls:C0038525 | Subarachnoid Hemorrhage | 1 | BeFree |
| umls:C0040156 | Thyrotoxicosis | 1 | BeFree |
| umls:C0085400 | Neurofibrillary degeneration (morphologic abnormality) | 1 | BeFree |
| umls:C0149721 | Left Ventricular Hypertrophy | 1 | BeFree |
| umls:C0149893 | Secondary glaucoma | 1 | BeFree |
| umls:C0152096 | Complete trisomy 18 syndrome | 1 | BeFree |
| umls:C0155552 | Hearing Loss, Mixed Conductive-Sensorineural | 1 | BeFree |
| umls:C0155765 | Disease of capillaries | 1 | BeFree |
| umls:C0205770 | Choroid Plexus Papilloma | 1 | BeFree |
| umls:C0206624 | Hepatoblastoma | 1 | BeFree |
| umls:C0206698 | Cholangiocarcinoma | 1 | LHGDN |
| umls:C0234366 | Ataxic | 1 | BeFree |
| umls:C0238190 | Inclusion Body Myositis (disorder) | 1 | BeFree |
| umls:C0239233 | Early satiety | 1 | BeFree |
| umls:C0242363 | Islet Cell Tumor | 1 | BeFree |
| umls:C0267839 | Hepatic amyloidosis | 1 | BeFree |
| umls:C0268382 | Amyloid nephropathy | 1 | BeFree |
| umls:C0268392 | Localized amyloidosis | 1 | BeFree |
| umls:C0268397 | Amyloidosis, Primary Cutaneous | 1 | BeFree |
| umls:C0268579 | Propionic acidemia | 1 | BeFree |
| umls:C0270921 | Axonal neuropathy | 1 | BeFree |
| umls:C0272247 | Biclonal gammopathy | 1 | BeFree |
| umls:C0278996 | Cancer of Head and Neck | 1 | BeFree |
| umls:C0281361 | Adenocarcinoma of pancreas | 1 | BeFree |
| umls:C0334299 | Carcinoid tumor no ICD-O subtype | 1 | BeFree |
| umls:C0338484 | Familial Hemiplegic Migraine | 1 | BeFree |
| umls:C0342199 | Iodine deficiency syndrome | 1 | BeFree |
| umls:C0342276 | Maturity onset diabetes mellitus in young | 1 | BeFree |
| umls:C0342618 | Amyloid myopathy | 1 | BeFree |
| umls:C0393571 | Multiple System Atrophy | 1 | BeFree |
| umls:C0474853 | Plasma cell tumor, malignant | 1 | BeFree |
| umls:C0547030 | Visual disturbance | 1 | BeFree |
| umls:C0555198 | Malignant Glioma | 1 | BeFree |
| umls:C0559758 | Multisystem disorder | 1 | BeFree |
| umls:C0595936 | Aqueous Humor Disorders | 1 | BeFree |
| umls:C0596452 | disabling disease | 1 | BeFree |
| umls:C0678222 | Breast Carcinoma | 1 | BeFree |
| umls:C0730345 | Microalbuminuria | 1 | BeFree |
| umls:C0741933 | cardiac symptom | 1 | BeFree |
| umls:C0751409 | Upper Extremity Paresis | 1 | BeFree |
| umls:C0751498 | Sigmoid Cancer | 1 | BeFree |
| umls:C0751658 | Ulnar Nerve Entrapment Syndrome | 1 | BeFree |
| umls:C0751713 | Inclusion Body Myopathy, Sporadic | 1 | BeFree |
| umls:C0751870 | Heredodegenerative Disorders, Nervous System | 1 | BeFree |
| umls:C0860207 | Drug-Induced Liver Injury | 1 | CTD_human |
| umls:C0936273 | Familial Amyloid Polyneuropathy, Type IV | 1 | BeFree |
| umls:C0947622 | Cholecystolithiasis | 1 | BeFree |
| umls:C1112256 | Peripheral sensorimotor neuropathy | 1 | GAD |
| umls:C1136084 | Plasma cell dyscrasia | 1 | BeFree |
| umls:C1136085 | Monoclonal Gammapathies | 1 | BeFree |
| umls:C1270972 | Mild cognitive disorder | 1 | BeFree |
| umls:C1285162 | Degenerative disorder | 1 | BeFree |
| umls:C1328479 | Pancreatic Endocrine Carcinoma | 1 | BeFree |
| umls:C1335302 | Pancreatic Ductal Adenocarcinoma | 1 | BeFree |
| umls:C1861735 | Dementia, familial Danish | 1 | BeFree |
| umls:C2718001 | Protein Misfolding Disorders | 1 | BeFree |
| umls:C2750824 | Dystransthyretinemic Euthyroidal Hyperthyroxinemia | 1 | CLINVAR,CTD_human,UNIPROT |
| umls:C3536715 | AA amyloidosis | 1 | BeFree |
| umls:C3665346 | Unspecified visual loss | 1 | BeFree |
| umls:C3151470 | AMYLOIDOSIS, LEPTOMENINGEAL, TRANSTHYRETIN-RELATED | 0 | CLINVAR |
| umls:C3151471 | AMYLOID CARDIOMYOPATHY, TRANSTHYRETIN-RELATED | 0 | CLINVAR |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))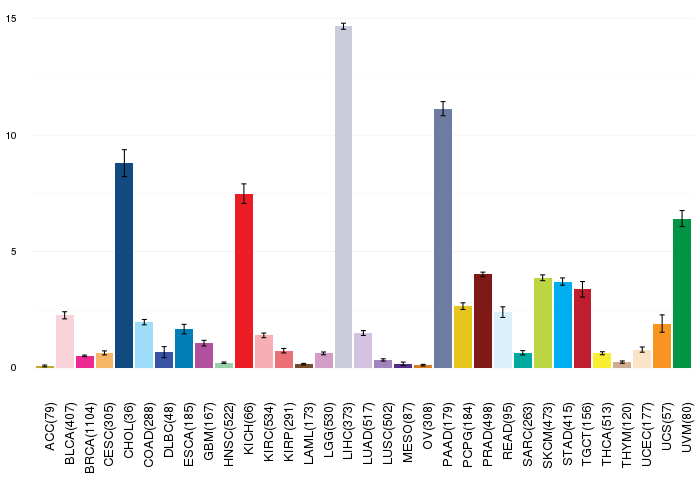
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))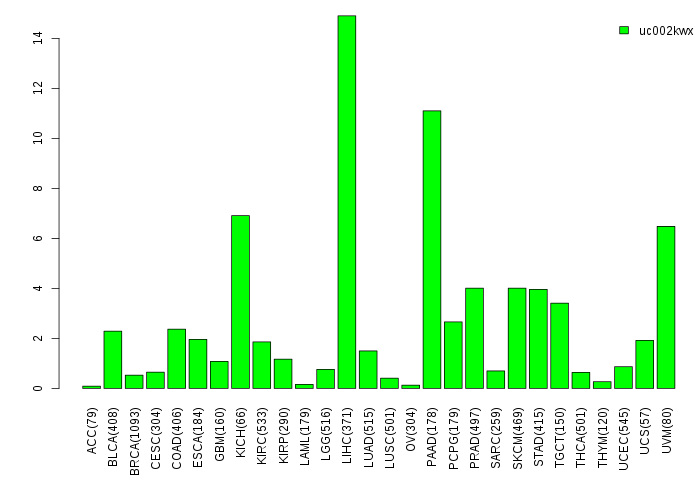
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data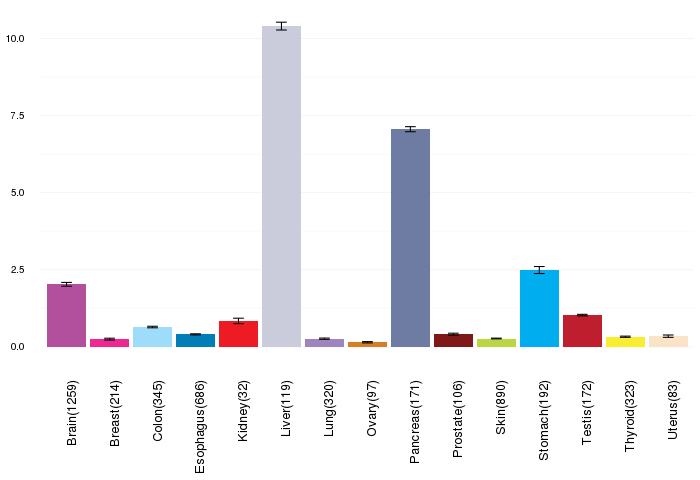
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))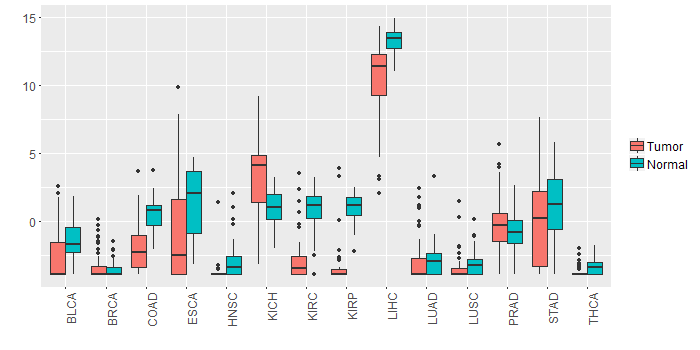
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.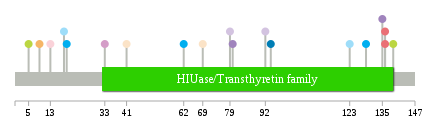

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 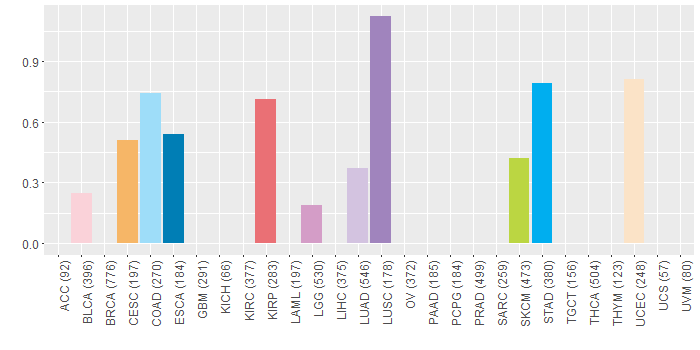
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)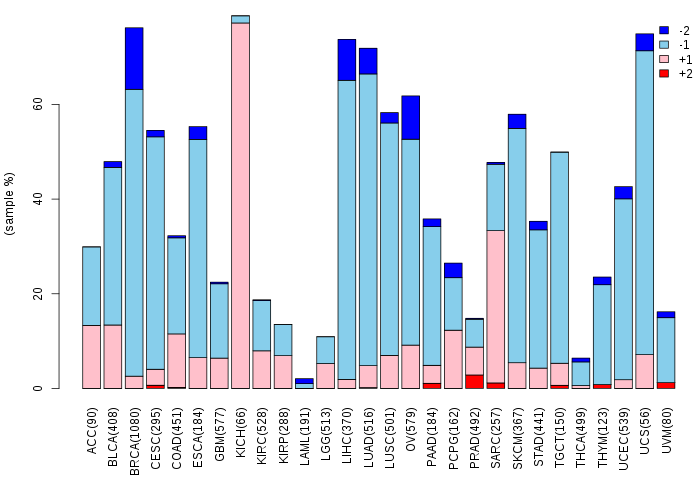
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 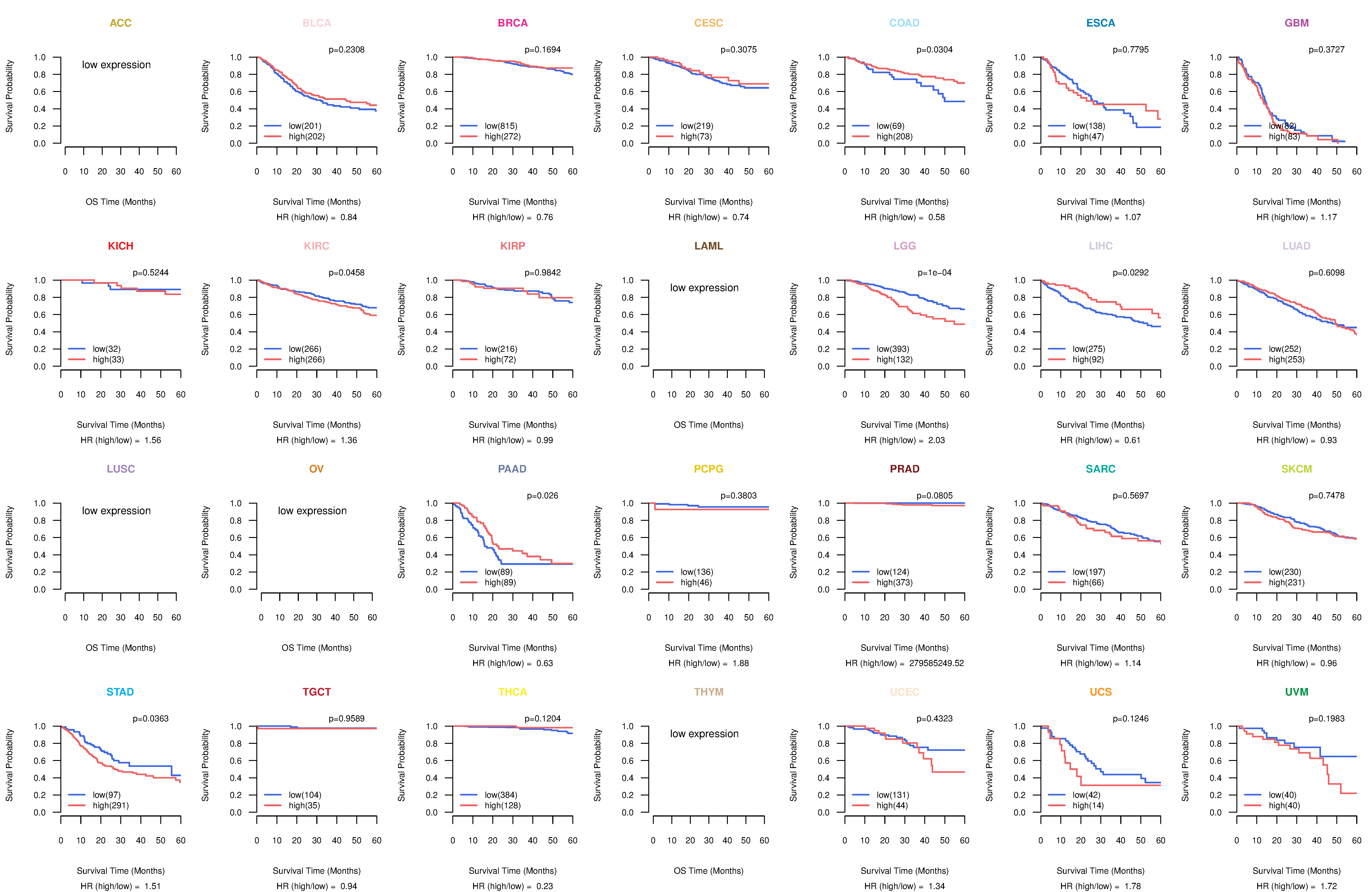
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 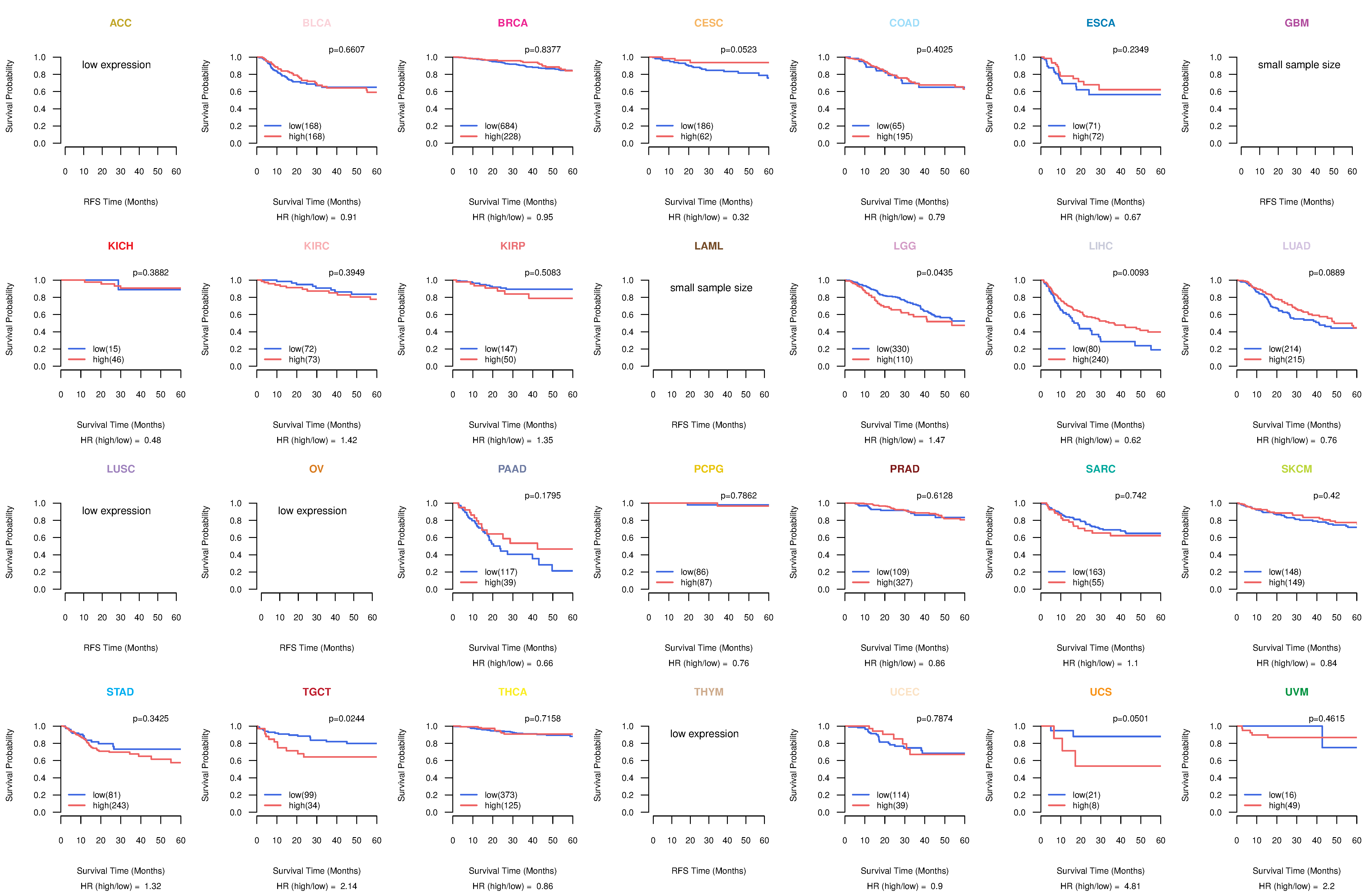
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 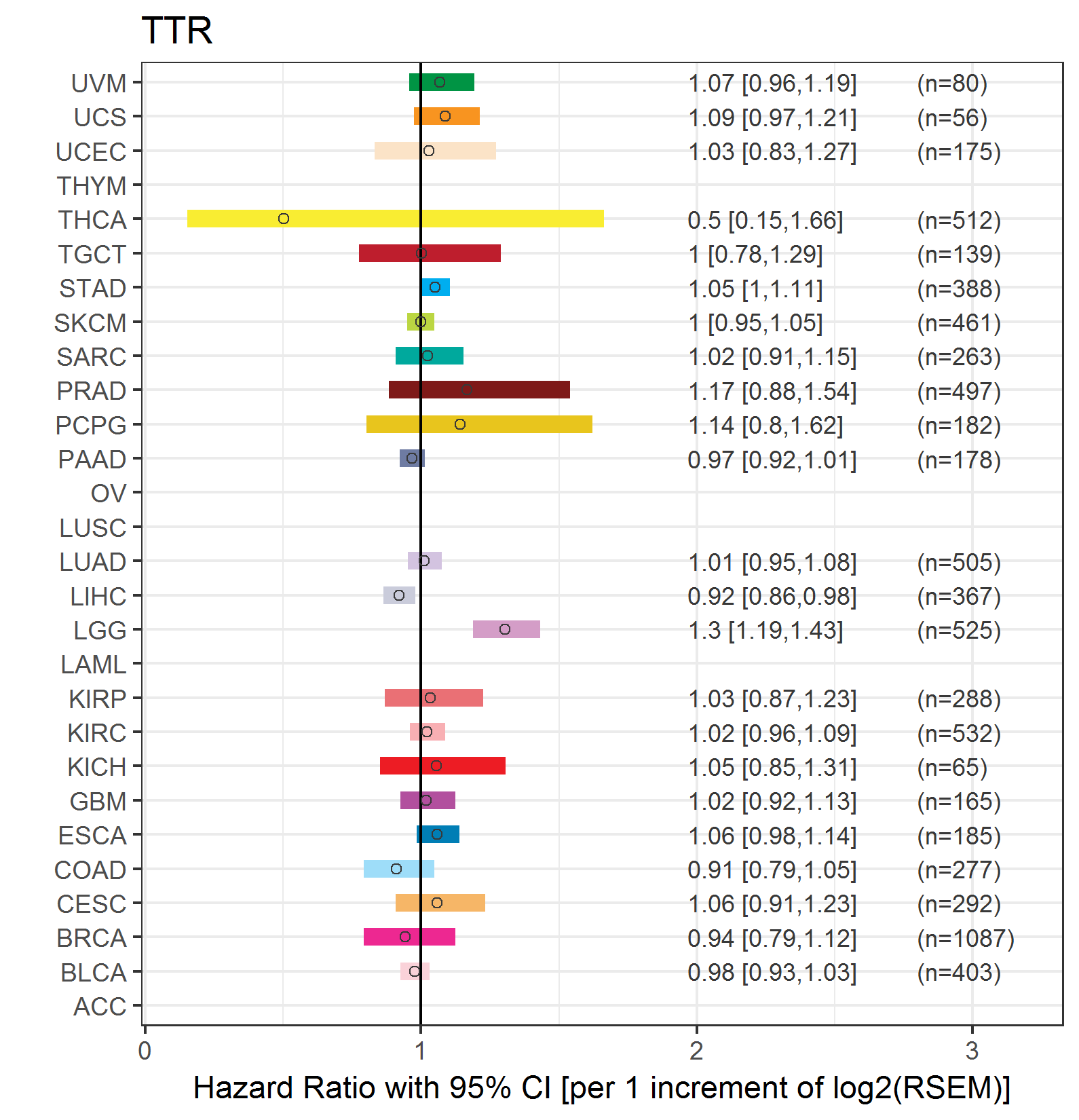
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 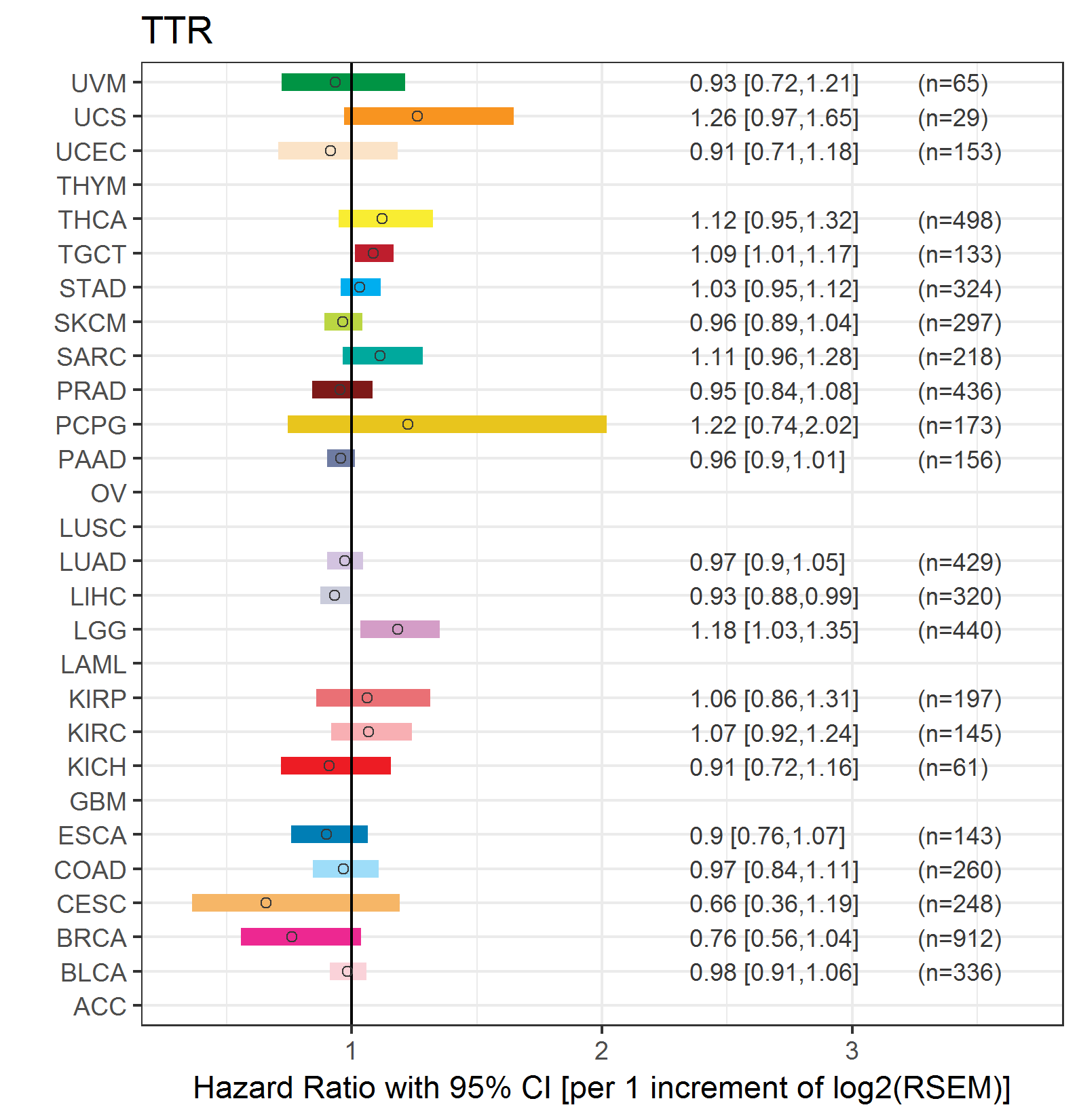
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























