| Basic gene information | Gene symbol | NPHS2 |
| Gene name | nephrosis 2, idiopathic, steroid-resistant (podocin) |
| Synonyms | PDCN|SRN1 |
| Cytomap | UCSC genome browser: 1q25.2 |
| Type of gene | protein-coding |
| RefGenes | NM_001297575.1,
NM_014625.3, |
| Description | podocin |
| Modification date | 20141207 |
| dbXrefs | MIM : 604766 |
| HGNC : HGNC |
| Ensembl : ENSG00000116218 |
| HPRD : 05303 |
| Vega : OTTHUMG00000035252 |
| Protein | UniProt:
go to UniProt's Cross Reference DB Table |
| Expression | CleanEX: HS_NPHS2 |
| BioGPS: 7827 |
| Pathway | NCI Pathway Interaction Database: NPHS2 |
| KEGG: NPHS2 |
| REACTOME: NPHS2 |
| Pathway Commons: NPHS2 |
| Context | iHOP: NPHS2 |
| ligand binding site mutation search in PubMed: NPHS2 |
| UCL Cancer Institute: NPHS2 |
| Assigned class in TissGDB* | A |
| Included tissue-specific gene expression resources | HPA,TiGER,GTEx |
| Specific-tissues in normal samples (assigned by TissGDB using HPA, TiGER, and GTEx) | Kidney |
| Cancer types related to the specific-tissues in cancer samples (assigned by TissGDB using TCGA) | KIRC,KIRP,KICH |
| Reference showing the relevant tissue of NPHS2 | Human Urine-Derived Renal Progenitors for Personalized Modeling of Genetic Kidney Disorders. Lazzeri E, Ronconi E, Angelotti ML, Peired A, Mazzinghi B, Becherucci F, Conti S, Sansavini G, Sisti A, Ravaglia F, Lombardi D, Provenzano A, Manonelles A, Cruzado JM, Giglio S, Roperto RM, Materassi M, Lasagni L, Romagnani P. J Am Soc Nephrol. 2015 Aug;26(8):1961-74. doi: 10.1681/ASN.2014010057. Epub 2015 Jan 7. (pmid:25568173)
go to article |
| Description by TissGene annotations | |
| BRCA (tumor) | BRCA (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
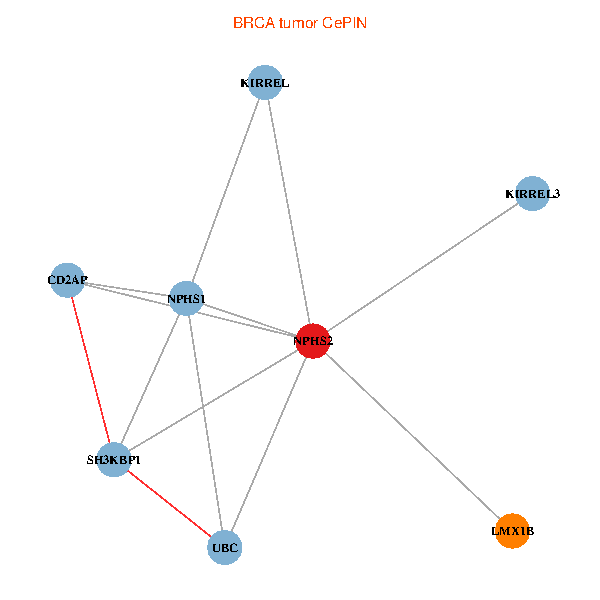 | 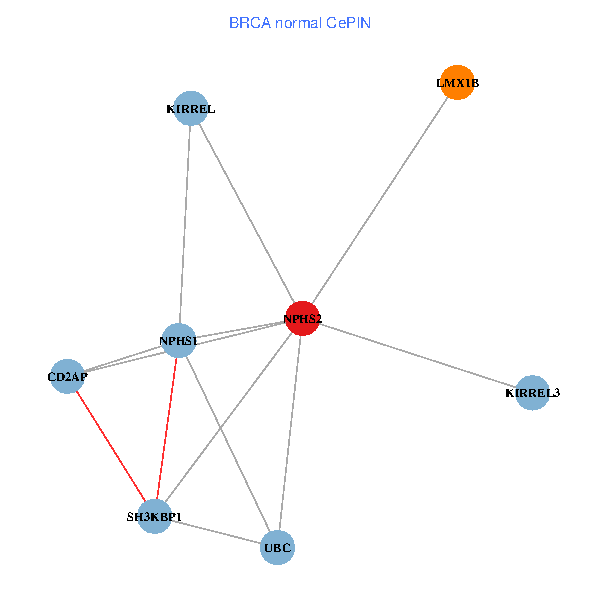 |
|
| COAD (tumor) | COAD (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
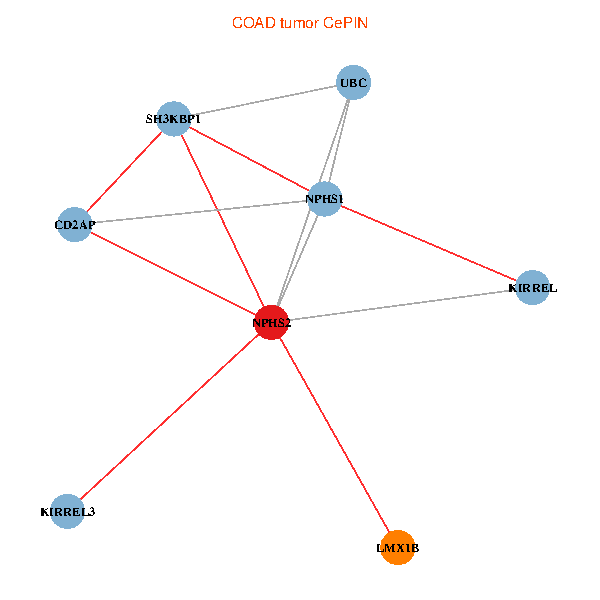 | 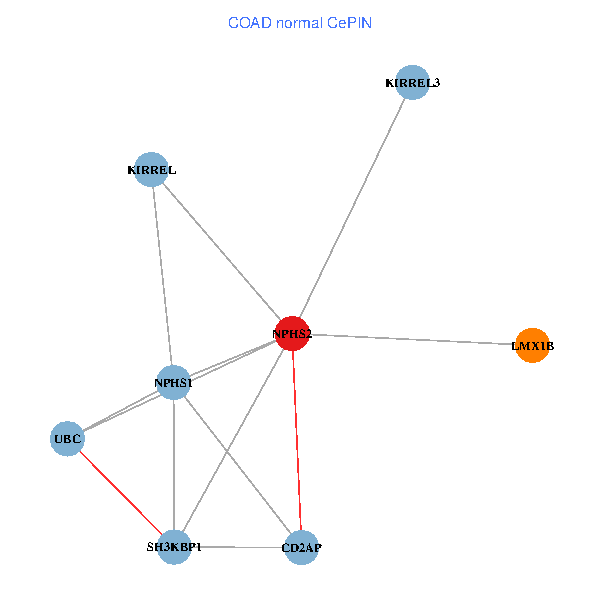 |
|
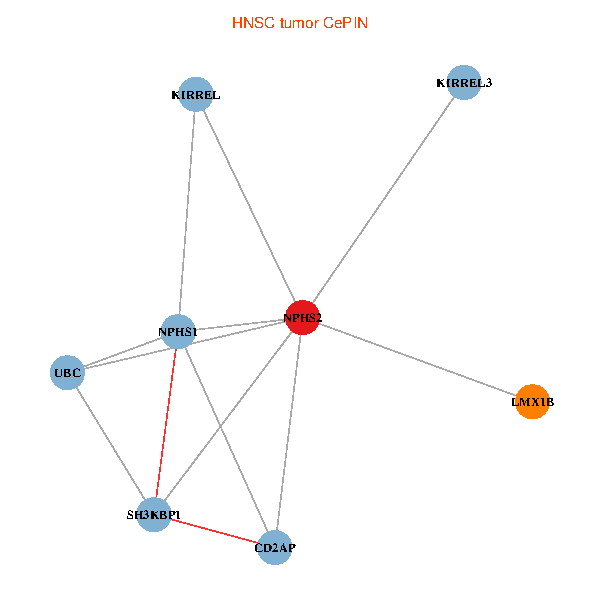
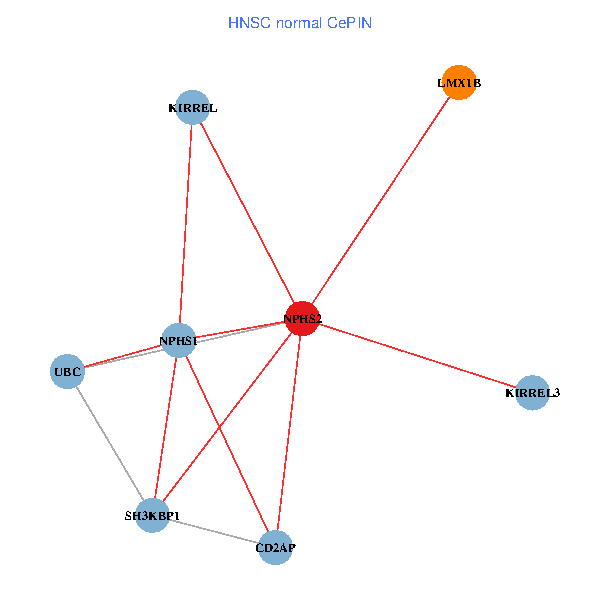
| HNSC (tumor) | HNSC (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
 |  |
|
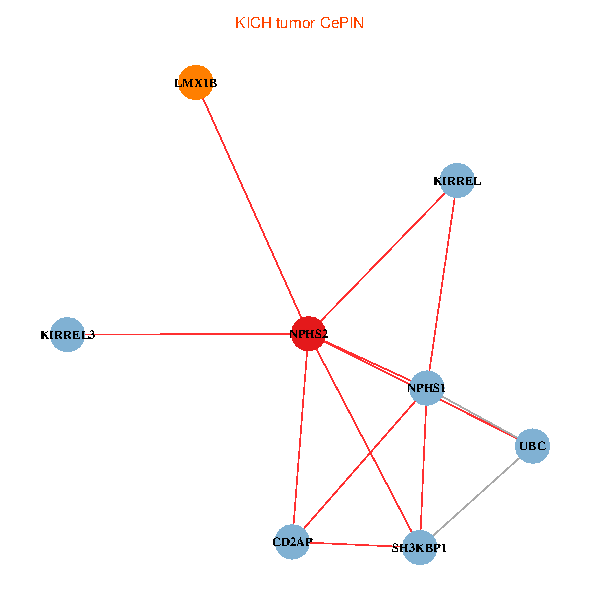
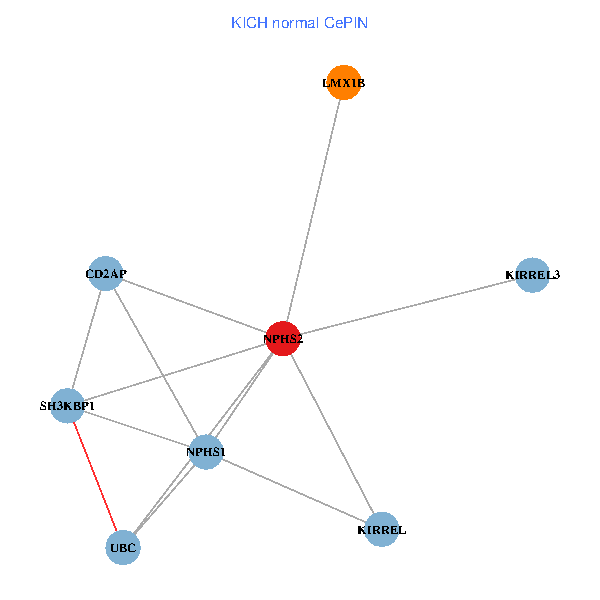
| KICH (tumor) | KICH (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
 |  |
|
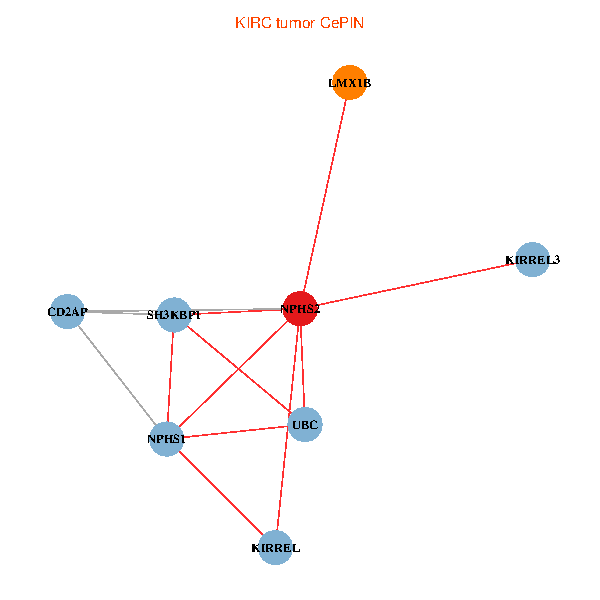
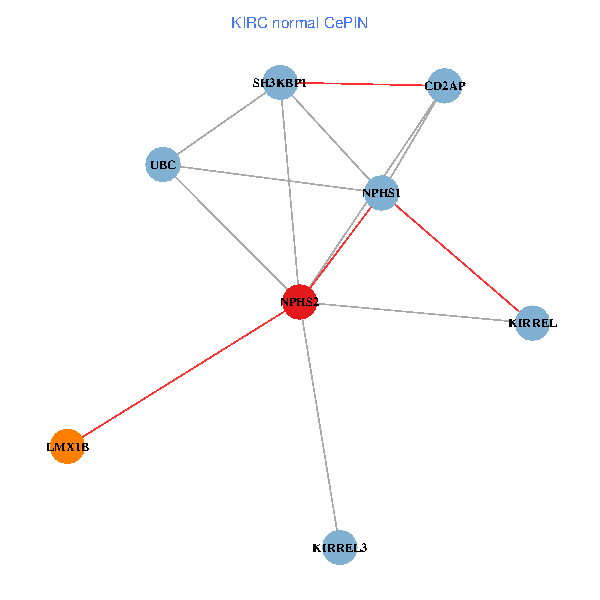
| KIRC (tumor) | KIRC (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
 |  |
|
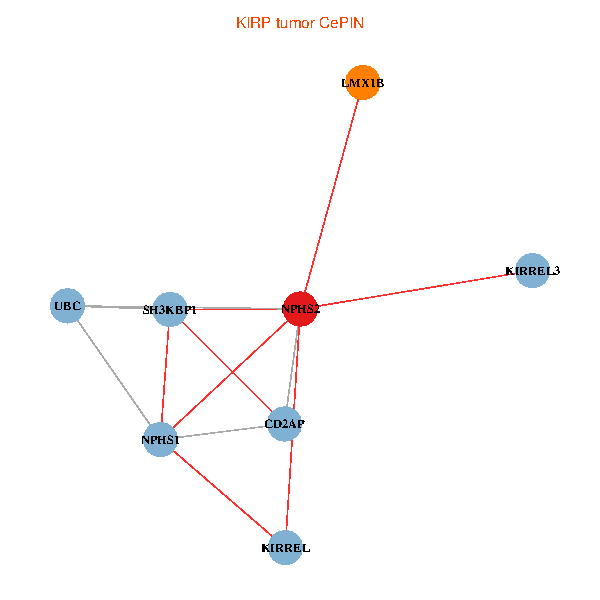
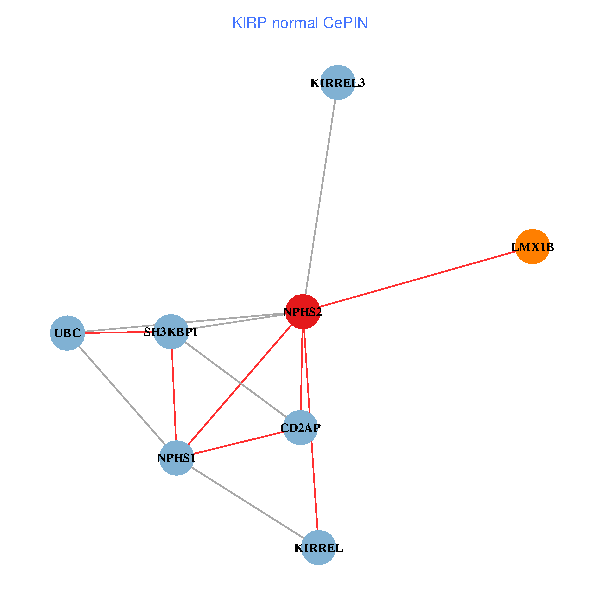
| KIRP (tumor) | KIRP (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
 |  |
|
| LIHC (tumor) | LIHC (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
 |  |
|
| LUAD (tumor) | LUAD (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
 |  |
|
| LUSC (tumor) | LUSC (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
 |  |
|
| PRAD (tumor) | PRAD (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
 |  |
|
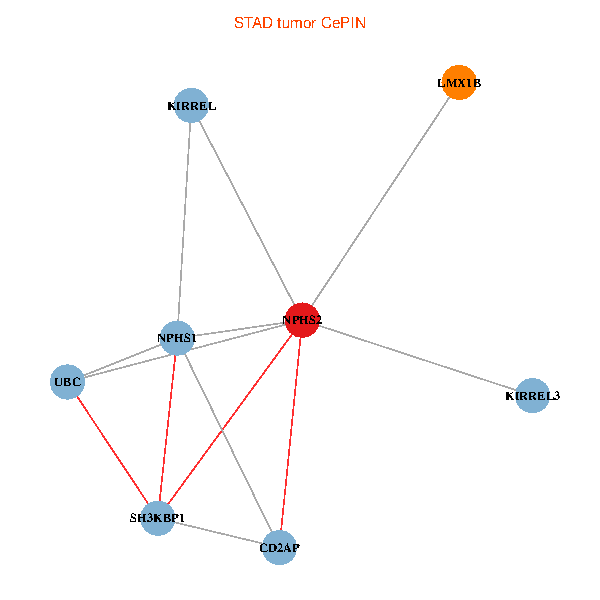
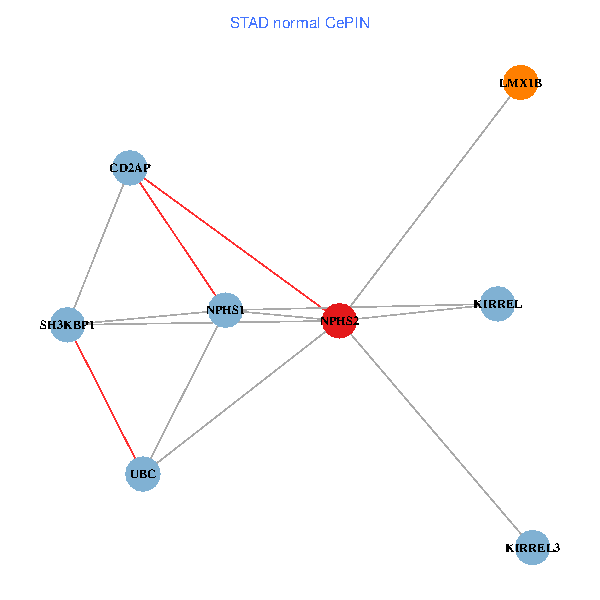
| STAD (tumor) | STAD (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
 |  |
|
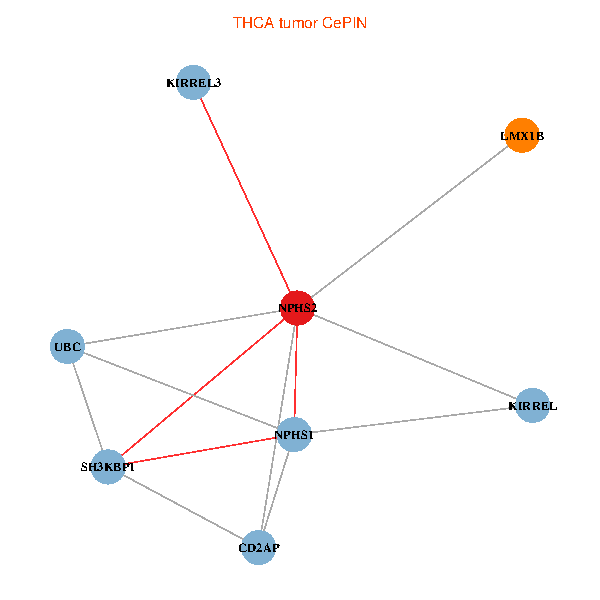
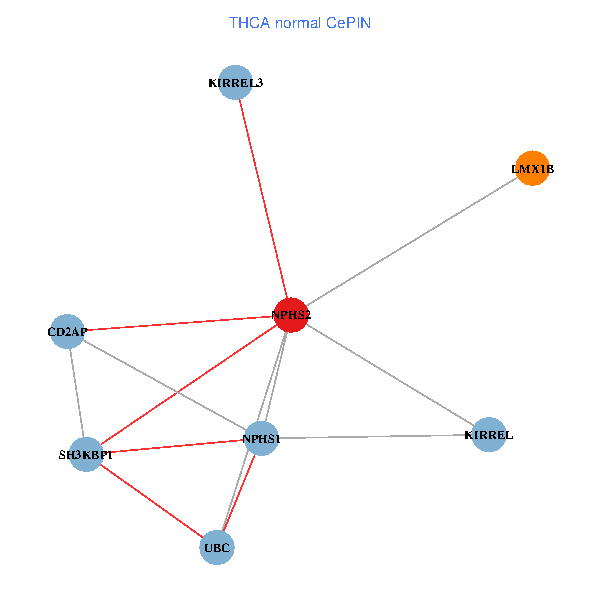
| THCA (tumor) | THCA (normal) |
| NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (tumor) | NPHS2, SH3KBP1, UBC, CD2AP, LMX1B, NPHS1, KIRREL, KIRREL3 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0027726 | Nephrotic Syndrome | 73 | BeFree,CTD_human,GAD,LHGDN,RGD |
| umls:C0403397 | Steroid-resistant nephrotic syndrome | 68 | BeFree |
| umls:C0017668 | Focal glomerulosclerosis | 41 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0333497 | Segmental glomerulosclerosis | 23 | BeFree |
| umls:C0022658 | Kidney Diseases | 17 | BeFree,GAD |
| umls:C0022661 | Kidney Failure, Chronic | 17 | BeFree,GAD,LHGDN |
| umls:C3496337 | Nephrotic Syndrome, Idiopathic | 9 | MGD,UNIPROT |
| umls:C0035078 | Kidney Failure | 8 | BeFree |
| umls:C2316810 | Chronic kidney disease stage 5 | 8 | BeFree |
| umls:C0268731 | Renal glomerular disease | 6 | BeFree |
| umls:C0011881 | Diabetic Nephropathy | 5 | BeFree,GAD,LHGDN |
| umls:C0017661 | IGA Glomerulonephritis | 5 | BeFree |
| umls:C0027720 | Nephrosis | 5 | BeFree,CTD_human,GAD |
| umls:C1704321 | Nephrotic Syndrome, Minimal Change | 5 | BeFree |
| umls:C1704320 | Glomerulonephritis, Minimal Change | 4 | BeFree |
| umls:C0001925 | Albuminuria | 3 | CTD_human,GAD |
| umls:C0027721 | Lipoid nephrosis | 3 | BeFree |
| umls:C0033687 | Proteinuria | 3 | CLINVAR,GAD,RGD |
| umls:C1709661 | Primary Focal Segmental Glomerulosclerosis | 3 | BeFree |
| umls:C0017665 | Membranous glomerulonephritis | 2 | BeFree,RGD |
| umls:C0020538 | Hypertensive disease | 2 | GAD |
| umls:C0027697 | Nephritis | 2 | BeFree,LHGDN |
| umls:C0178664 | Glomerulosclerosis (disorder) | 2 | BeFree |
| umls:C0265962 | Ichthyosis linearis circumflexa | 2 | BeFree |
| umls:C0268732 | Nephritic syndrome | 2 | BeFree |
| umls:C0268747 | Diffuse mesangial sclerosis (disorder) | 2 | BeFree |
| umls:C0403396 | Steroid-sensitive nephrotic syndrome | 2 | BeFree |
| umls:C0403447 | Chronic Kidney Insufficiency | 2 | BeFree |
| umls:C0730345 | Microalbuminuria | 2 | BeFree |
| umls:C1868672 | NEPHROTIC SYNDROME, STEROID-RESISTANT, AUTOSOMAL RECESSIVE | 2 | CLINVAR,CTD_human |
| umls:C0002726 | Amyloidosis | 1 | BeFree |
| umls:C0003850 | Arteriosclerosis | 1 | BeFree |
| umls:C0004153 | Atherosclerosis | 1 | BeFree |
| umls:C0004509 | Azoospermia | 1 | BeFree |
| umls:C0011847 | Diabetes | 1 | BeFree |
| umls:C0011849 | Diabetes Mellitus | 1 | BeFree |
| umls:C0011854 | Diabetes Mellitus, Insulin-Dependent | 1 | BeFree,GAD |
| umls:C0012546 | Diphtheria | 1 | BeFree |
| umls:C0015397 | Disorder of eye | 1 | GAD |
| umls:C0018799 | Heart Diseases | 1 | BeFree |
| umls:C0019693 | HIV Infections | 1 | BeFree |
| umls:C0027708 | Nephroblastoma | 1 | BeFree |
| umls:C0027719 | Nephrosclerosis | 1 | BeFree |
| umls:C0034150 | Purpura | 1 | LHGDN |
| umls:C0040336 | Tobacco Use Disorder | 1 | GAD |
| umls:C0078911 | AIDS-Associated Nephropathy | 1 | GAD |
| umls:C0151744 | Myocardial Ischemia | 1 | BeFree |
| umls:C0342793 | Malonic aciduria | 1 | BeFree |
| umls:C0403398 | Steroid-dependent nephrotic syndrome | 1 | BeFree |
| umls:C0403399 | Finnish congenital nephrotic syndrome | 1 | BeFree |
| umls:C0403440 | Thin basement membrane disease | 1 | BeFree |
| umls:C0445118 | Nephrotic range proteinuria | 1 | BeFree |
| umls:C1305904 | Familial hematuria | 1 | BeFree |
| umls:C1384583 | Congenital absence of germinal epithelium of testes | 1 | BeFree |
| umls:C1636149 | Macular dystrophy, corneal type 1 | 1 | BeFree |
| umls:C1962972 | Proteinuria Adverse Event | 1 | GAD |
| umls:C3825926 | Nephrotic syndrome in children | 1 | BeFree |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))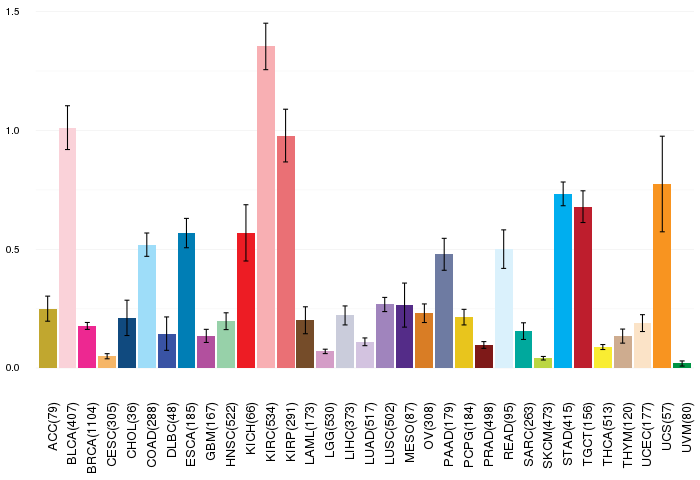
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))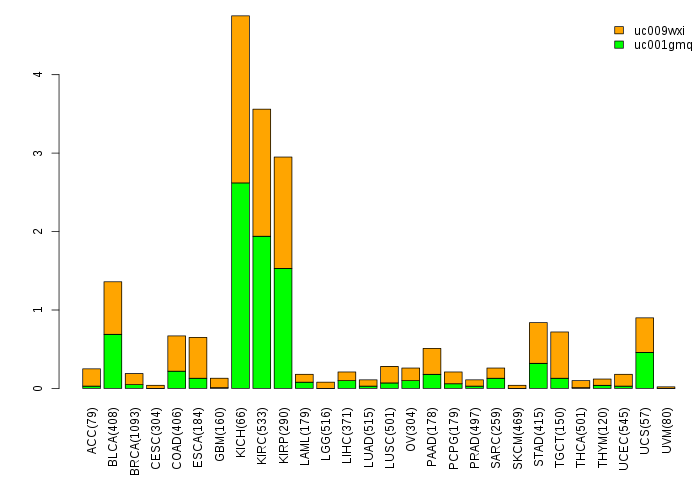
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data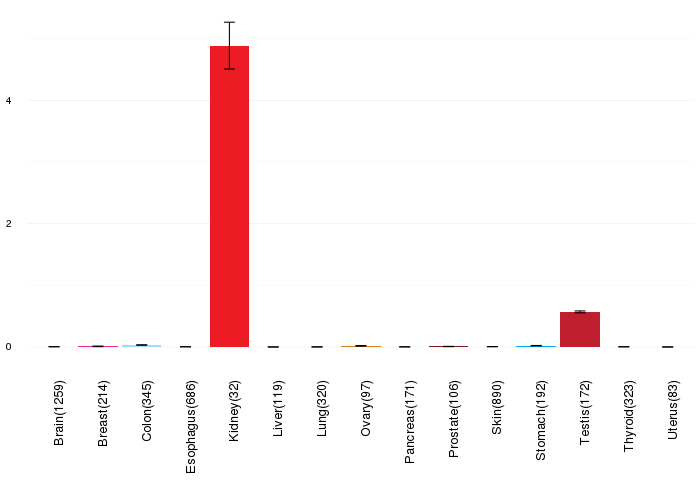
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))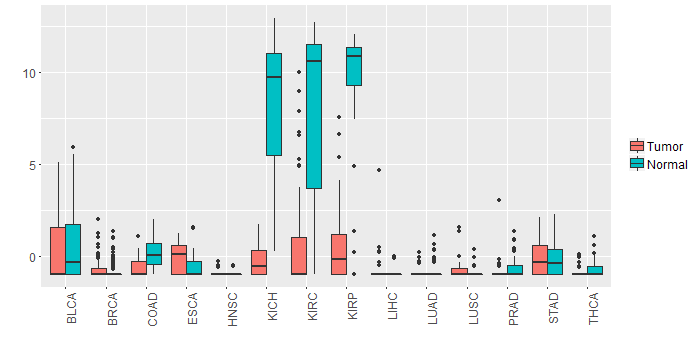
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.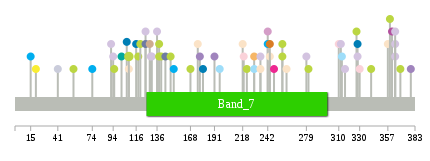

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 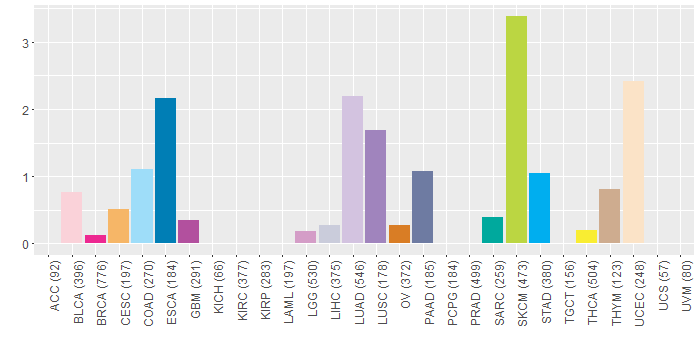
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)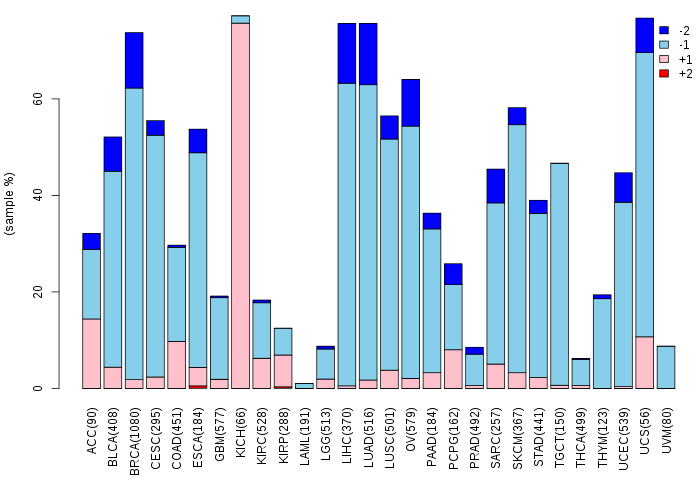
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 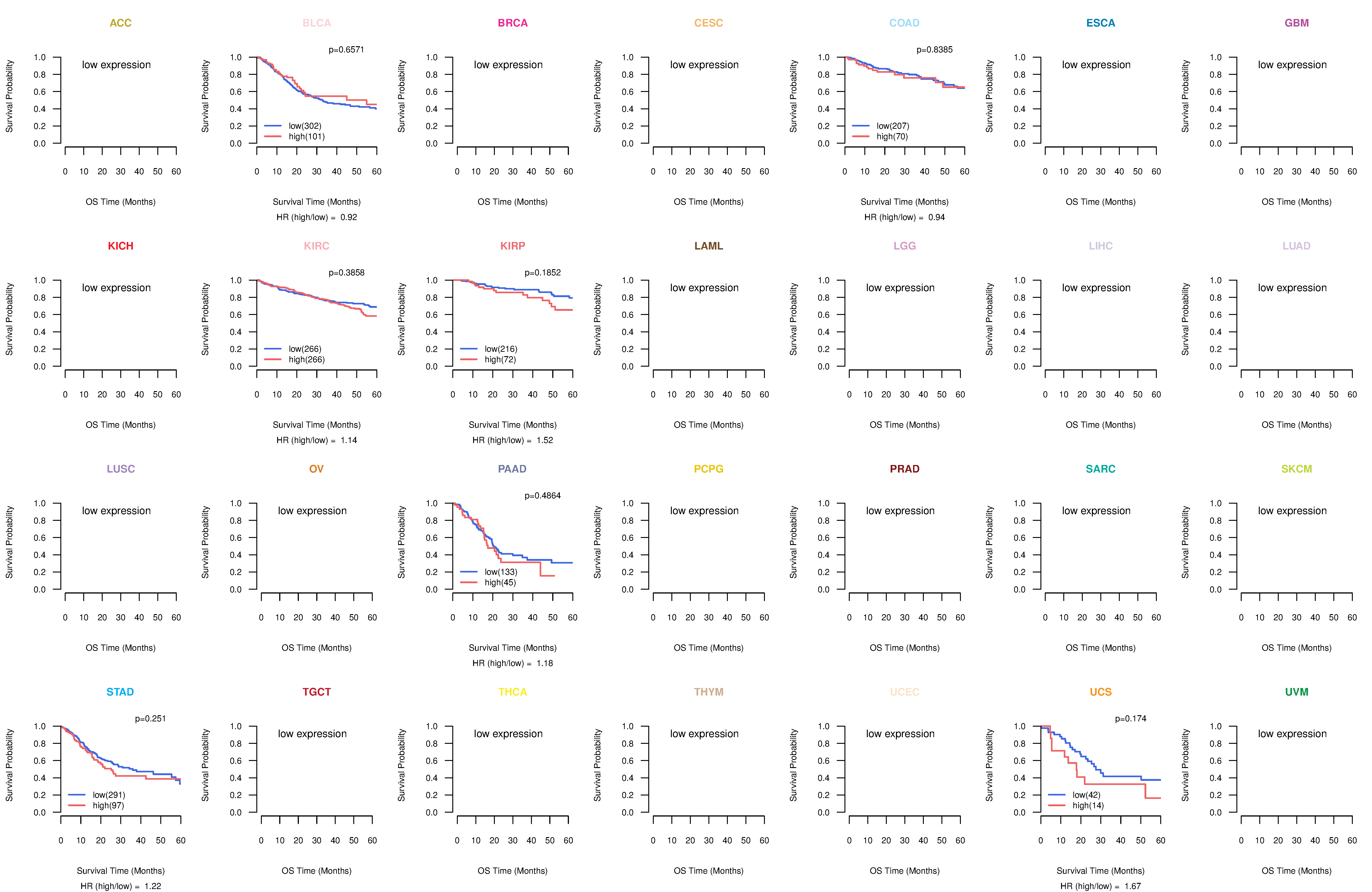
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 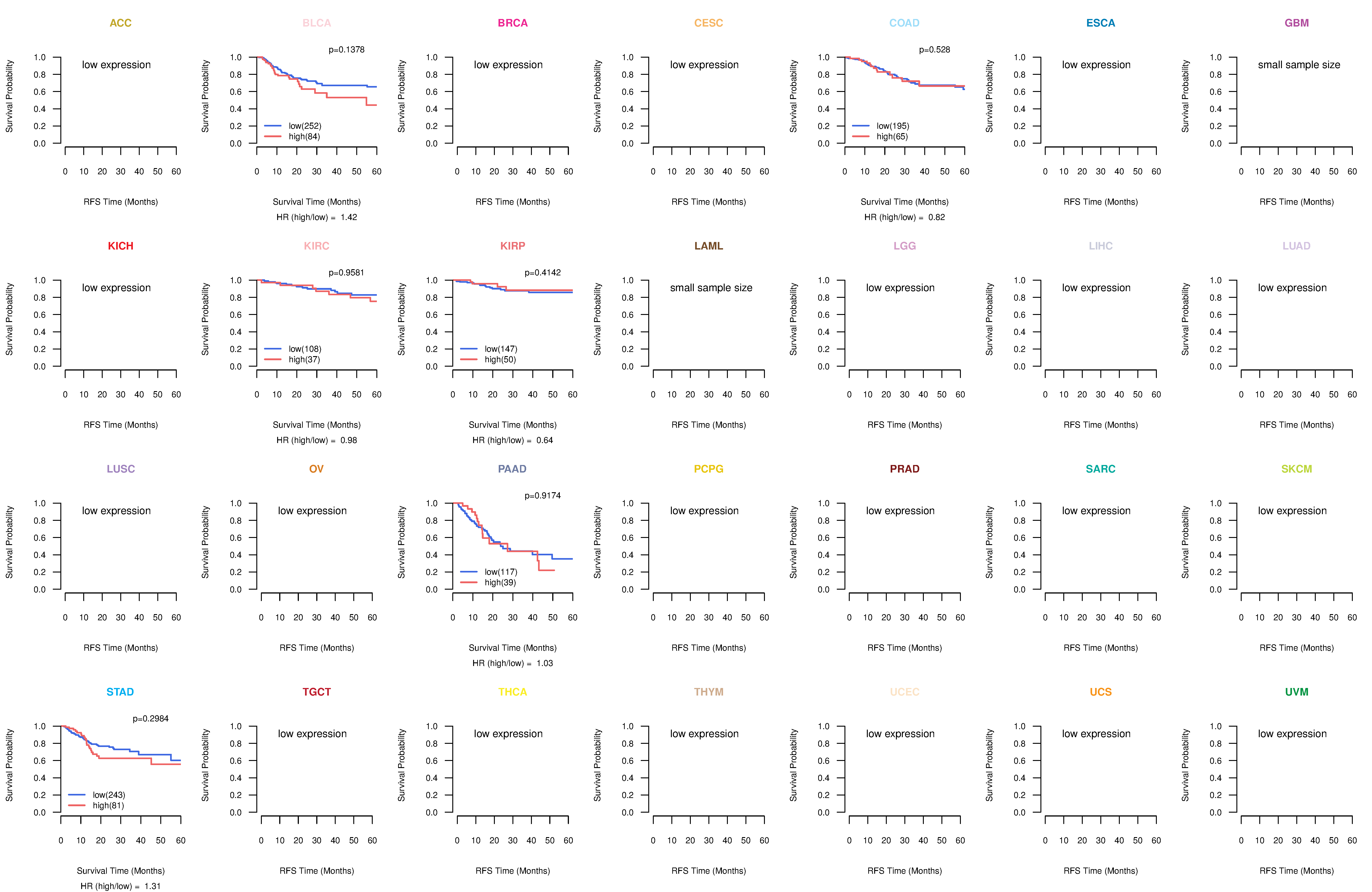
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 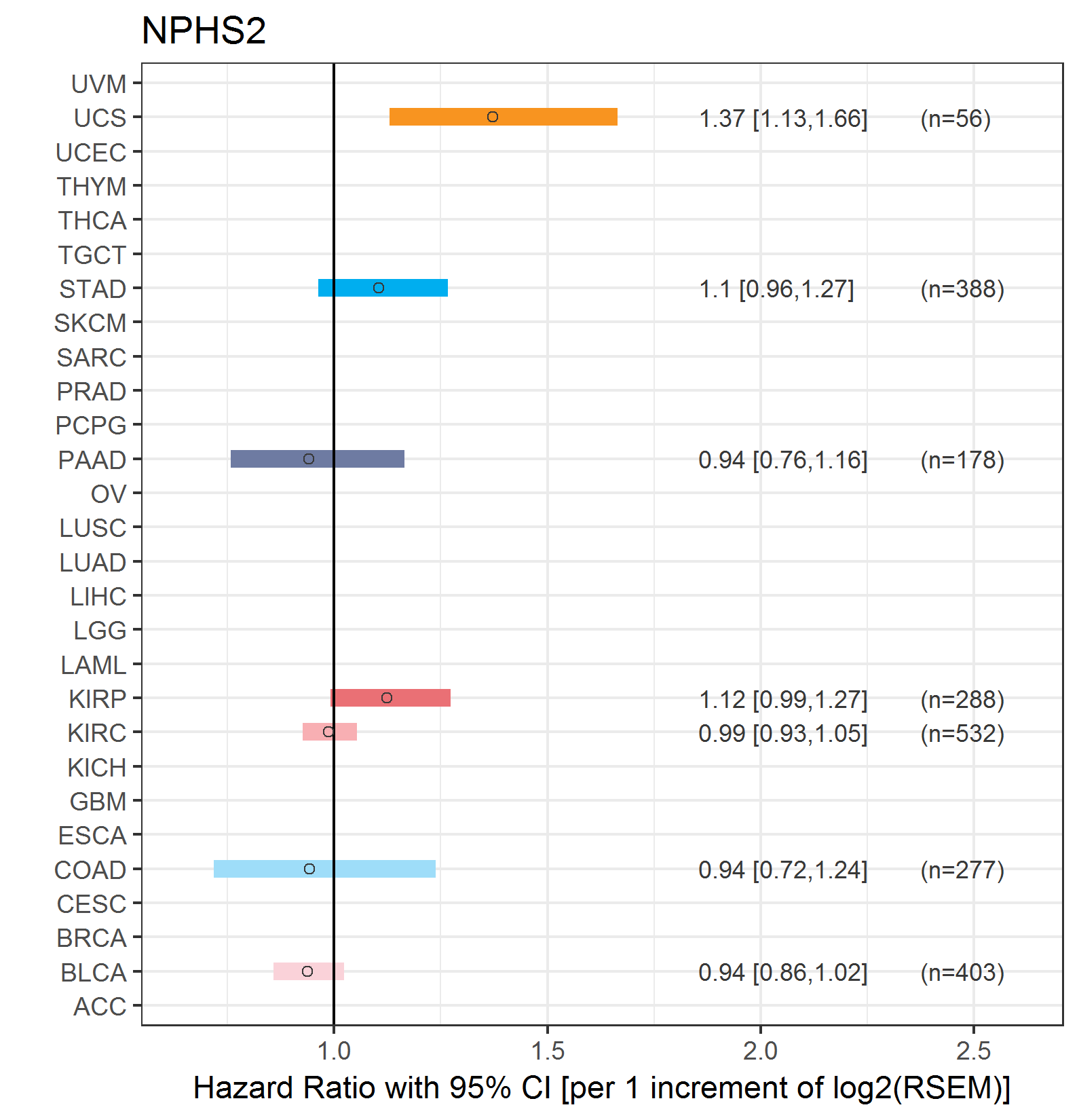
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 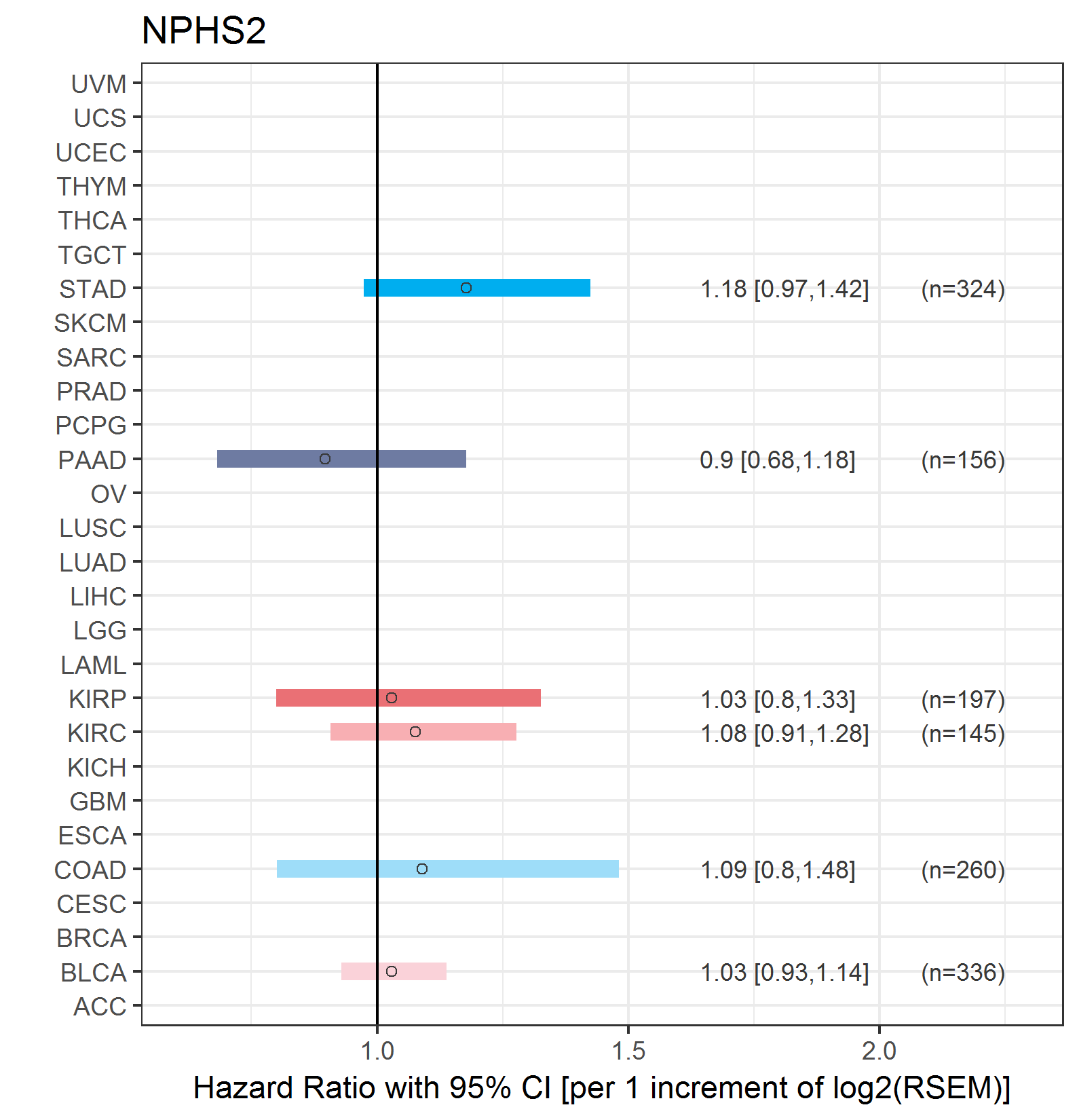
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























