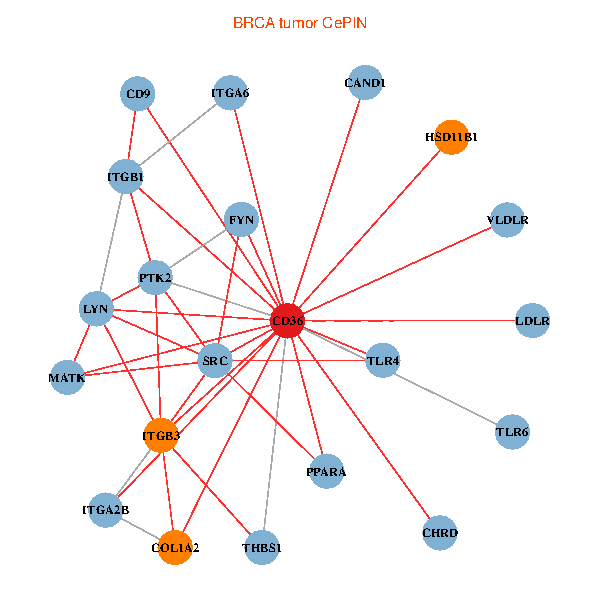
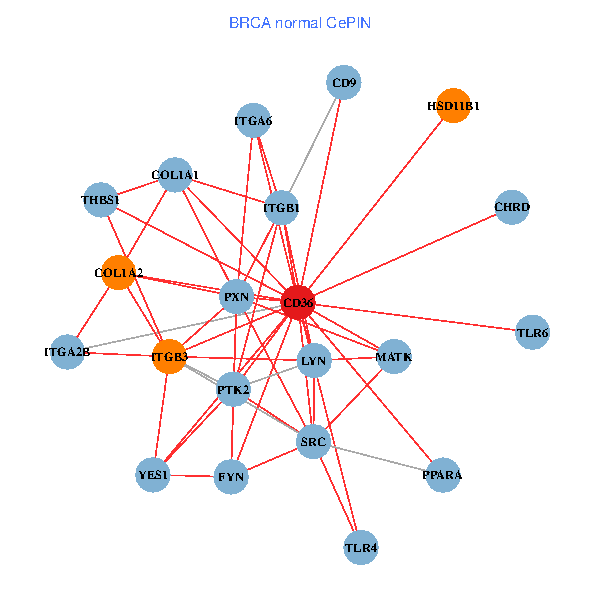
| BRCA (tumor) | BRCA (normal) |
| CD36, CAND1, SRC, LYN, FYN, PTK2, THBS1, ITGB1, ITGB3, COL1A2, PPARA, CHRD, TLR4, CD9, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, LDLR, ITGA6 (tumor) | CD36, SRC, LYN, YES1, FYN, PTK2, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, CHRD, TLR4, CD9, HSD11B1, TLR6, MATK, ITGA2B, ITGA6 (normal) |
 |  |
|
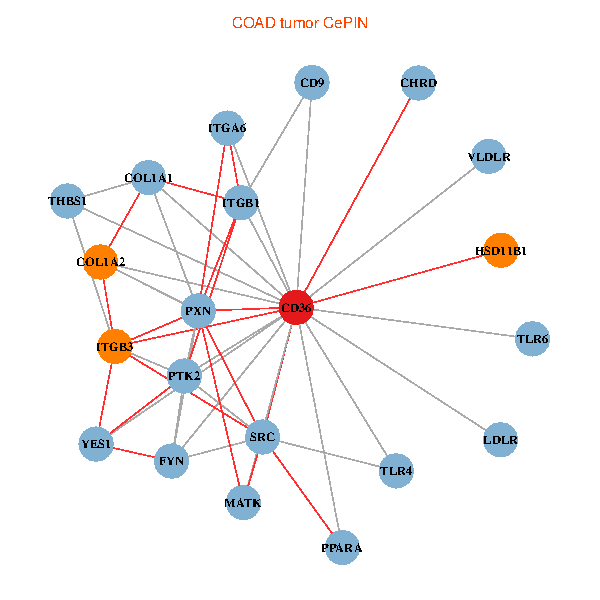
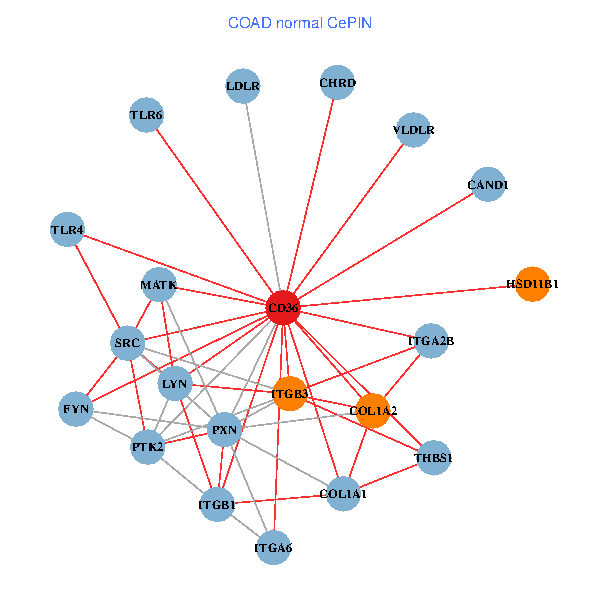
| COAD (tumor) | COAD (normal) |
| CD36, SRC, YES1, FYN, PTK2, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, CHRD, TLR4, CD9, HSD11B1, TLR6, VLDLR, MATK, LDLR, ITGA6 (tumor) | CD36, CAND1, SRC, LYN, FYN, PTK2, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, CHRD, TLR4, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, LDLR, ITGA6 (normal) |
 |  |
|
| HNSC (tumor) | HNSC (normal) |
| CD36, CAND1, SRC, LYN, YES1, FYN, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, CHRD, TLR4, CD9, HSD11B1, VLDLR, MATK, ITGA2B, LDLR (tumor) | CD36, CAND1, SRC, LYN, YES1, FYN, THBS1, COL1A1, ITGB3, COL1A2, PXN, PPARA, CHRD, TLR4, CD9, HSD11B1, VLDLR, MATK, ITGA2B, LDLR, ITGA6 (normal) |
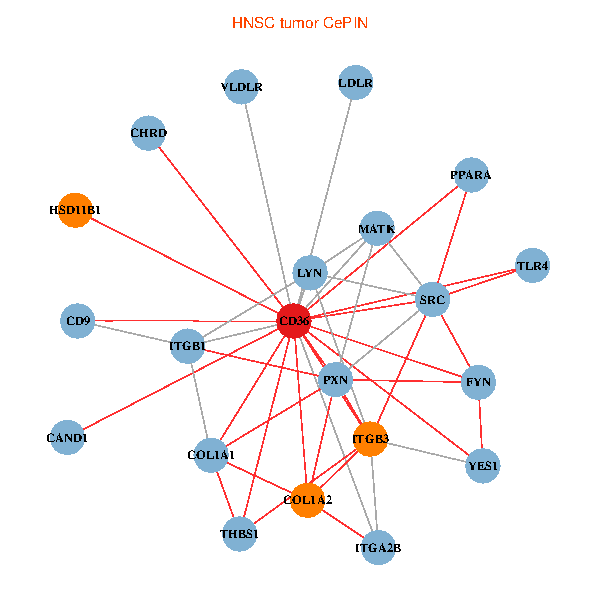 | 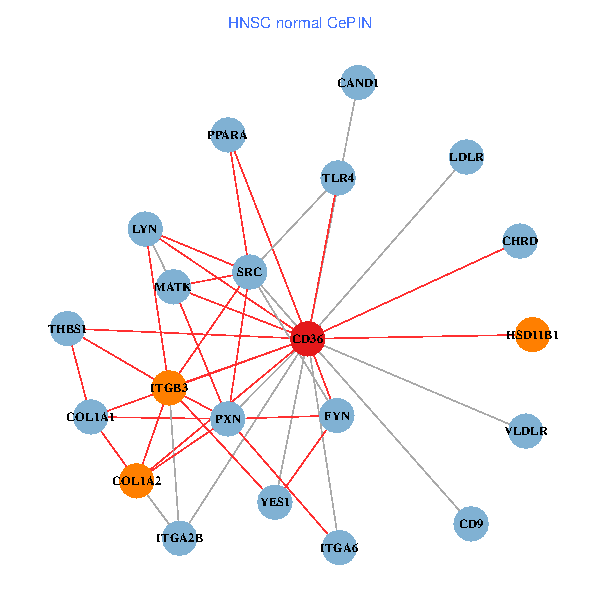 |
|
| KICH (tumor) | KICH (normal) |
| CD36, SRC, LYN, YES1, FYN, PTK2, THBS1, COL1A1, ITGB3, COL1A2, PXN, CHRD, TLR4, CD9, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, LDLR, ITGA6 (tumor) | CD36, CAND1, SRC, LYN, YES1, FYN, THBS1, ITGB1, COL1A1, ITGB3, PXN, PPARA, CHRD, TLR4, CD9, HSD11B1, TLR6, MATK, ITGA2B, LDLR, ITGA6 (normal) |
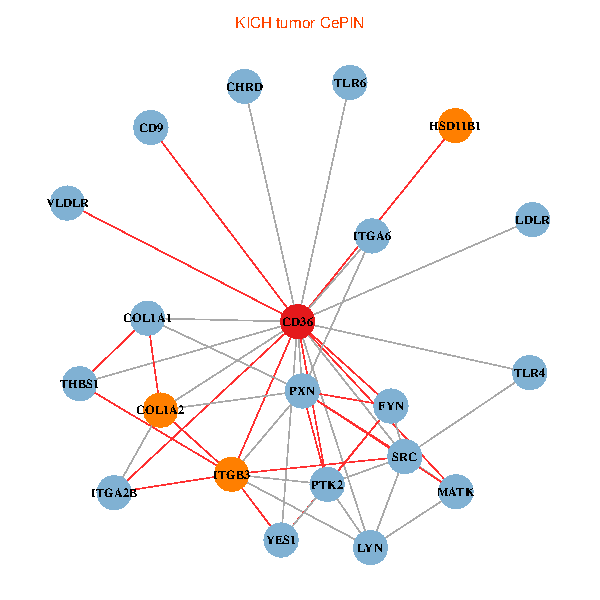 |  |
|
| KIRC (tumor) | KIRC (normal) |
| CD36, CAND1, SRC, LYN, YES1, FYN, PTK2, THBS1, ITGB1, ITGB3, PXN, PPARA, CHRD, TLR4, CD9, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, ITGA6 (tumor) | CD36, SRC, LYN, YES1, FYN, PTK2, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PPARA, CHRD, TLR4, CD9, HSD11B1, VLDLR, MATK, ITGA2B, LDLR, ITGA6 (normal) |
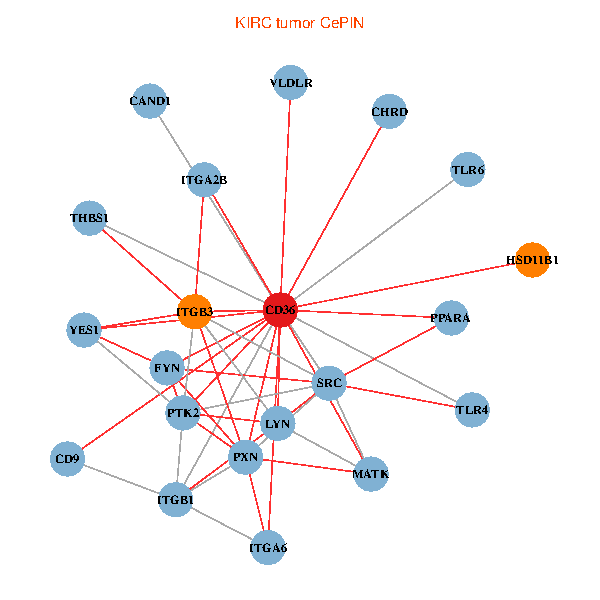 | 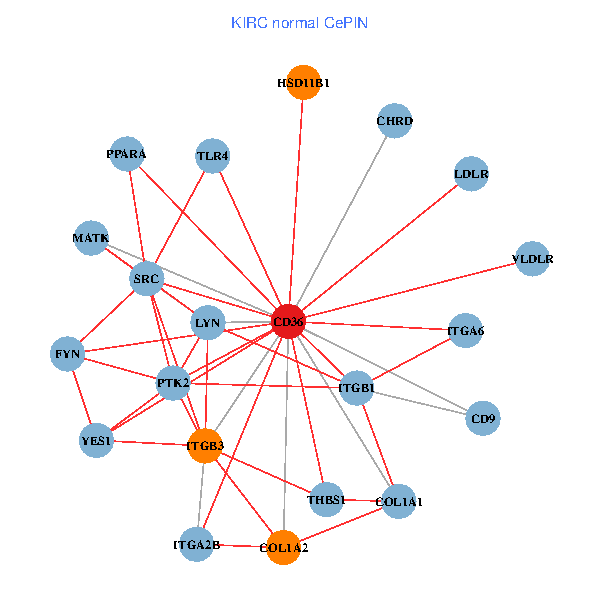 |
|
| KIRP (tumor) | KIRP (normal) |
| CD36, CAND1, SRC, LYN, YES1, FYN, PTK2, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, CHRD, CD9, HSD11B1, VLDLR, MATK, ITGA2B, LDLR, ITGA6 (tumor) | CD36, CAND1, SRC, LYN, YES1, FYN, PTK2, THBS1, ITGB1, COL1A1, COL1A2, CHRD, TLR4, CD9, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, LDLR, ITGA6 (normal) |
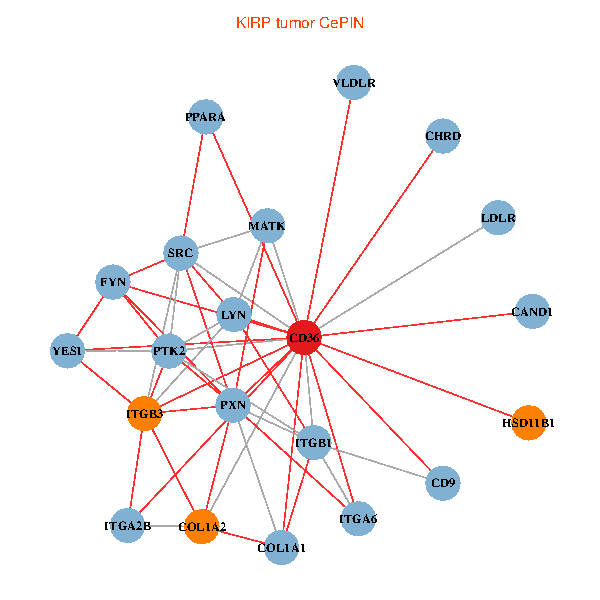 | 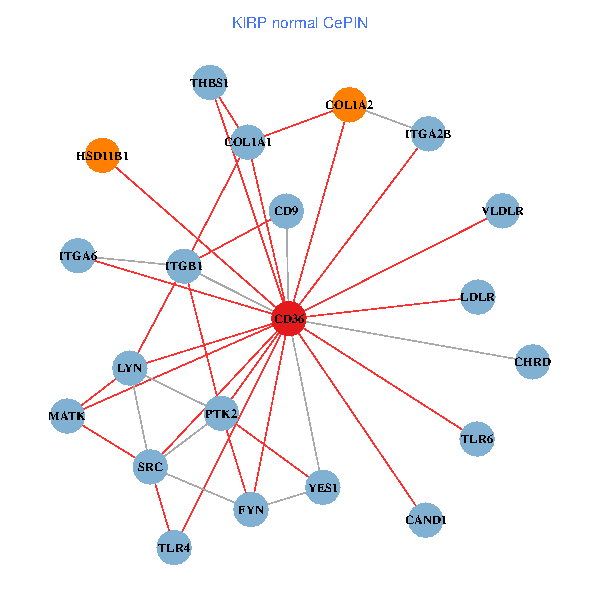 |
|
| LIHC (tumor) | LIHC (normal) |
| CD36, CAND1, LYN, YES1, PTK2, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, TLR4, CD9, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, LDLR, ITGA6 (tumor) | CD36, SRC, LYN, FYN, PTK2, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, CHRD, TLR4, CD9, HSD11B1, TLR6, MATK, ITGA2B, LDLR, ITGA6 (normal) |
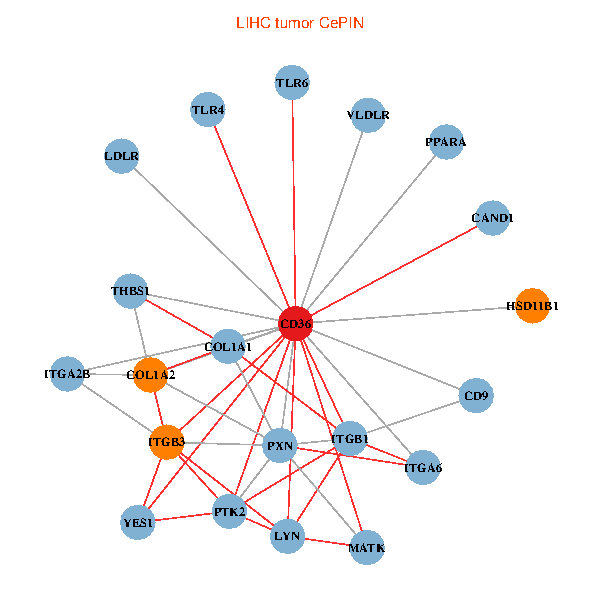 | 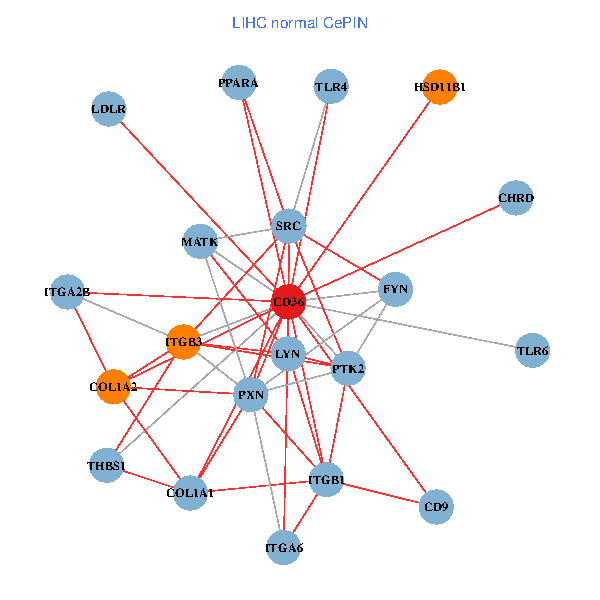 |
|
| LUAD (tumor) | LUAD (normal) |
| CD36, CAND1, SRC, LYN, YES1, FYN, PTK2, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, TLR4, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, ITGA6 (tumor) | CD36, CAND1, SRC, LYN, YES1, FYN, PTK2, THBS1, ITGB1, COL1A1, ITGB3, PPARA, TLR4, CD9, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, LDLR, ITGA6 (normal) |
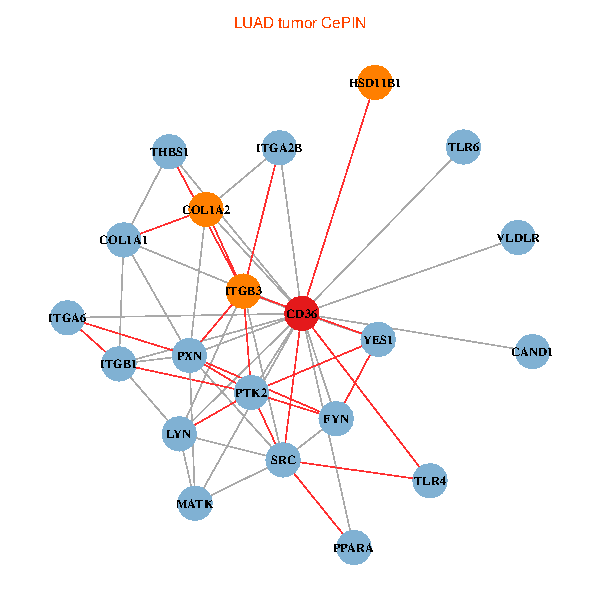 | 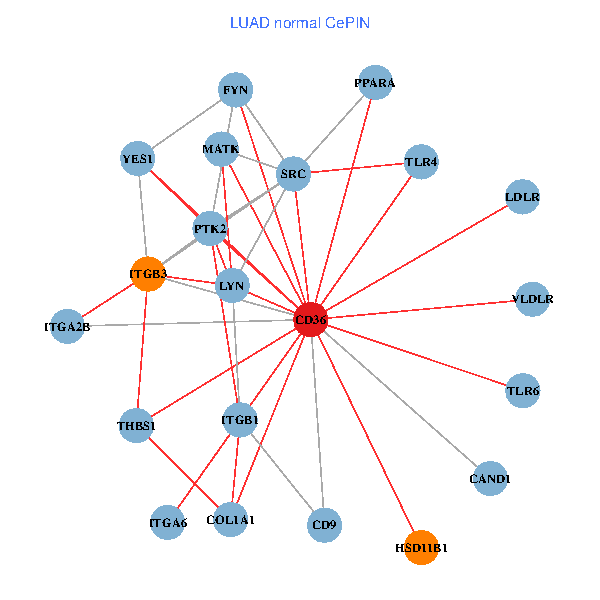 |
|
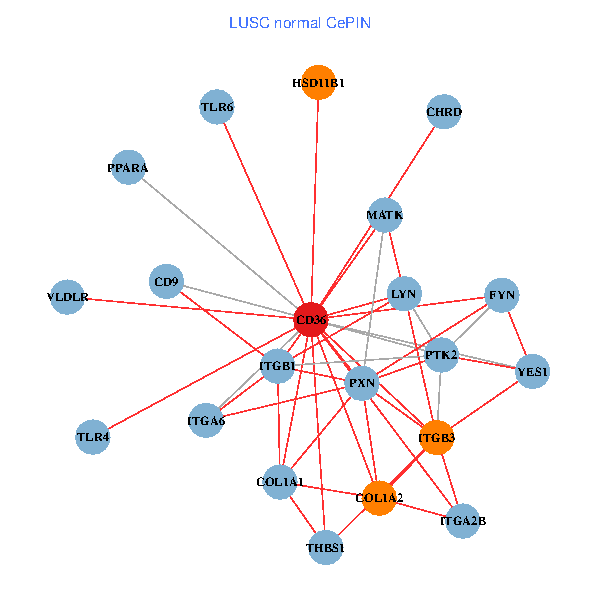
| LUSC (tumor) | LUSC (normal) |
| CD36, CAND1, SRC, LYN, YES1, FYN, PTK2, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, TLR4, CD9, TLR6, VLDLR, MATK, ITGA2B, LDLR, ITGA6 (tumor) | CD36, LYN, YES1, FYN, PTK2, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, CHRD, TLR4, CD9, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, ITGA6 (normal) |
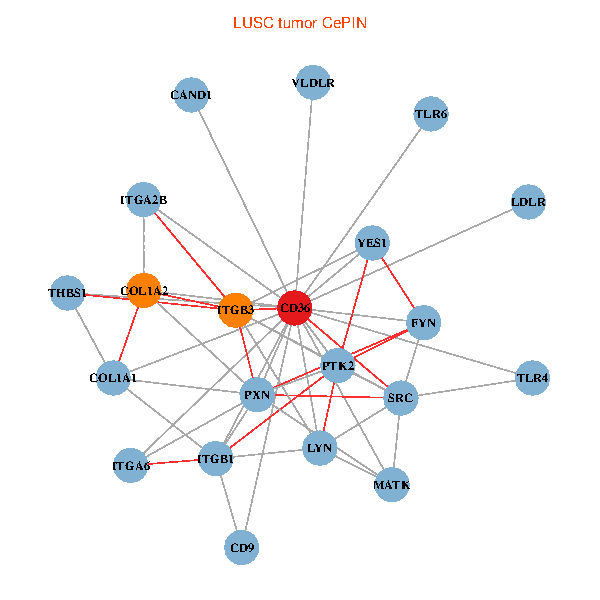 |  |
|
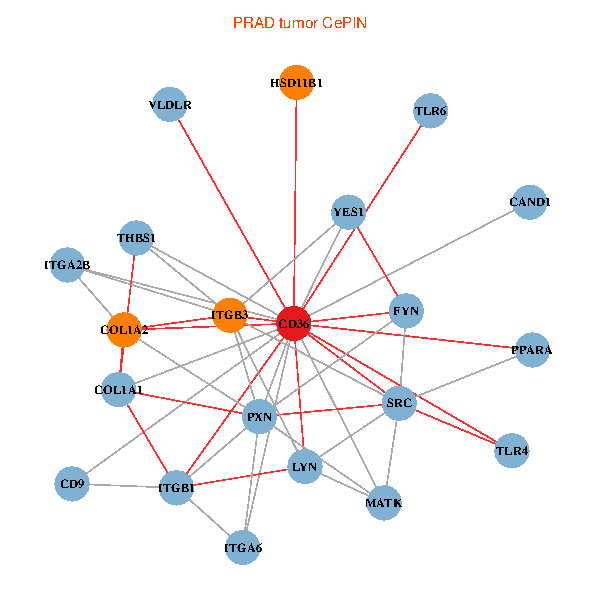
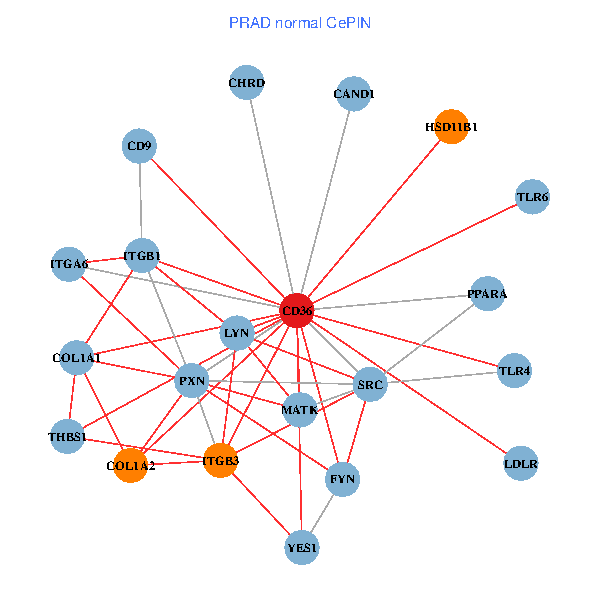
| PRAD (tumor) | PRAD (normal) |
| CD36, CAND1, SRC, LYN, YES1, FYN, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, TLR4, CD9, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, ITGA6 (tumor) | CD36, CAND1, SRC, LYN, YES1, FYN, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, CHRD, TLR4, CD9, HSD11B1, TLR6, MATK, LDLR, ITGA6 (normal) |
 |  |
|
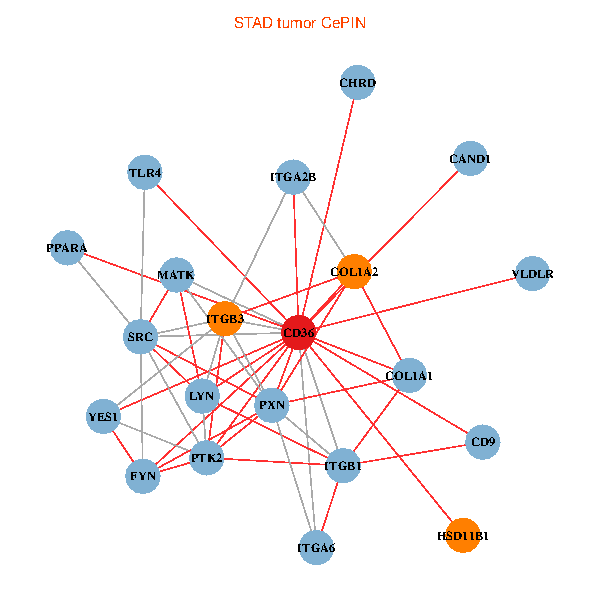
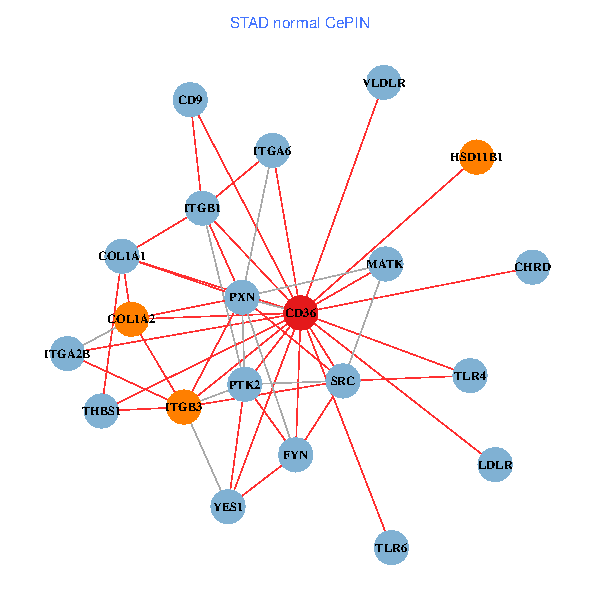
| STAD (tumor) | STAD (normal) |
| CD36, CAND1, SRC, LYN, YES1, FYN, PTK2, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, CHRD, TLR4, CD9, HSD11B1, VLDLR, MATK, ITGA2B, ITGA6 (tumor) | CD36, SRC, YES1, FYN, PTK2, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, CHRD, TLR4, CD9, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, LDLR, ITGA6 (normal) |
 |  |
|
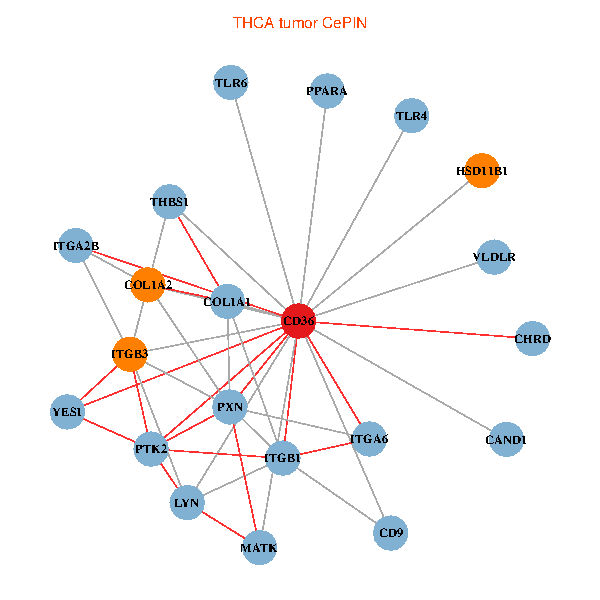
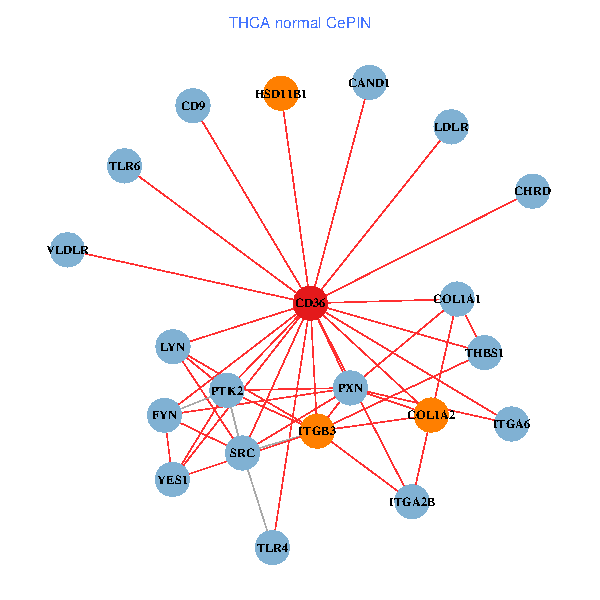
| THCA (tumor) | THCA (normal) |
| CD36, CAND1, LYN, YES1, PTK2, THBS1, ITGB1, COL1A1, ITGB3, COL1A2, PXN, PPARA, CHRD, TLR4, CD9, HSD11B1, TLR6, VLDLR, MATK, ITGA2B, ITGA6 (tumor) | CD36, CAND1, SRC, LYN, YES1, FYN, PTK2, THBS1, COL1A1, ITGB3, COL1A2, PXN, CHRD, TLR4, CD9, HSD11B1, TLR6, VLDLR, ITGA2B, LDLR, ITGA6 (normal) |
 |  |
|
| Disease ID | Disease name | # pubmeds | Source |
| umls:C0004153 | Atherosclerosis | 39 | BeFree,GAD,LHGDN |
| umls:C0003850 | Arteriosclerosis | 36 | BeFree,LHGDN |
| umls:C0028754 | Obesity | 30 | BeFree,GAD,RGD |
| umls:C0024530 | Malaria | 25 | BeFree,CTD_human,GAD,LHGDN |
| umls:C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 23 | BeFree,GAD,RGD |
| umls:C0524620 | Metabolic Syndrome X | 18 | BeFree,GAD,RGD |
| umls:C0011847 | Diabetes | 17 | BeFree |
| umls:C0011849 | Diabetes Mellitus | 17 | BeFree,LHGDN |
| umls:C1842090 | Platelet Glycoprotein IV Deficiency | 16 | BeFree,CLINVAR,CTD_human,MGD,UNIPROT |
| umls:C0020538 | Hypertensive disease | 11 | BeFree,CTD_human,GAD,LHGDN,RGD |
| umls:C0010054 | Coronary Arteriosclerosis | 9 | BeFree,GAD |
| umls:C0007222 | Cardiovascular Diseases | 8 | BeFree,LHGDN |
| umls:C0024535 | Malaria, Falciparum | 8 | BeFree,GAD |
| umls:C0010068 | Coronary heart disease | 7 | BeFree,CTD_human,LHGDN |
| umls:C0018799 | Heart Diseases | 7 | BeFree,LHGDN |
| umls:C1956346 | Coronary Artery Disease | 7 | BeFree,GAD |
| umls:C0019693 | HIV Infections | 6 | BeFree |
| umls:C0027051 | Myocardial Infarction | 6 | BeFree,GAD,LHGDN,RGD |
| umls:C0038454 | Cerebrovascular accident | 5 | BeFree |
| umls:C0242339 | Dyslipidemias | 5 | BeFree |
| umls:C0005779 | Blood Coagulation Disorders | 4 | BeFree |
| umls:C0015695 | Fatty Liver | 4 | BeFree |
| umls:C0020473 | Hyperlipidemia | 4 | BeFree,LHGDN |
| umls:C0024534 | Malaria, Cerebral | 4 | BeFree,GAD |
| umls:C0271650 | Impaired glucose tolerance | 4 | BeFree,CTD_human,LHGDN |
| umls:C0002395 | Alzheimer's Disease | 3 | BeFree |
| umls:C0006142 | Malignant neoplasm of breast | 3 | BeFree |
| umls:C0020443 | Hypercholesterolemia | 3 | BeFree |
| umls:C0020456 | Hyperglycemia | 3 | BeFree |
| umls:C0021655 | Insulin Resistance | 3 | CTD_human,GAD,RGD |
| umls:C0025202 | melanoma | 3 | BeFree,LHGDN |
| umls:C0027627 | Neoplasm Metastasis | 3 | BeFree |
| umls:C0032460 | Polycystic Ovary Syndrome | 3 | BeFree,LHGDN |
| umls:C0400966 | Non-alcoholic Fatty Liver Disease | 3 | BeFree |
| umls:C0596263 | Carcinogenesis | 3 | BeFree |
| umls:C0678222 | Breast Carcinoma | 3 | BeFree |
| umls:C2711227 | Steatohepatitis | 3 | BeFree |
| umls:C0002895 | Anemia, Sickle Cell | 2 | BeFree |
| umls:C0005586 | Bipolar Disorder | 2 | BeFree |
| umls:C0007102 | Malignant tumor of colon | 2 | BeFree |
| umls:C0007193 | Cardiomyopathy, Dilated | 2 | BeFree,CTD_human |
| umls:C0007194 | Hypertrophic Cardiomyopathy | 2 | BeFree,RGD |
| umls:C0009402 | Colorectal Carcinoma | 2 | BeFree |
| umls:C0011884 | Diabetic Retinopathy | 2 | BeFree |
| umls:C0014175 | Endometriosis | 2 | BeFree |
| umls:C0021390 | Inflammatory Bowel Diseases | 2 | BeFree |
| umls:C0023895 | Liver diseases | 2 | BeFree,GAD |
| umls:C0023903 | Liver neoplasms | 2 | BeFree |
| umls:C0025517 | Metabolic Diseases | 2 | BeFree |
| umls:C0029408 | Degenerative polyarthritis | 2 | BeFree,GAD |
| umls:C0085580 | Essential Hypertension | 2 | BeFree |
| umls:C0242383 | Age related macular degeneration | 2 | BeFree,GAD,LHGDN,RGD |
| umls:C0271084 | Exudative age-related macular degeneration | 2 | BeFree |
| umls:C0409959 | Osteoarthritis, Knee | 2 | BeFree,RGD |
| umls:C0699790 | Colon Carcinoma | 2 | BeFree |
| umls:C0878544 | Cardiomyopathies | 2 | BeFree |
| umls:C1527249 | Colorectal Cancer | 2 | BeFree,GAD |
| umls:C1800706 | Idiopathic Pulmonary Fibrosis | 2 | BeFree,LHGDN |
| umls:C1846502 | Mammographic Density | 2 | BeFree |
| umls:C1959635 | Parvovirus B19 (disease) | 2 | BeFree |
| umls:C2239176 | Liver carcinoma | 2 | BeFree |
| umls:C0002871 | Anemia | 1 | BeFree,GAD |
| umls:C0002962 | Angina Pectoris | 1 | GAD |
| umls:C0003811 | Cardiac Arrhythmia | 1 | RGD |
| umls:C0003873 | Rheumatoid Arthritis | 1 | BeFree |
| umls:C0005818 | Blood Platelet Disorders | 1 | BeFree |
| umls:C0007137 | Squamous cell carcinoma | 1 | BeFree |
| umls:C0011570 | Mental Depression | 1 | BeFree |
| umls:C0011581 | Depressive disorder | 1 | BeFree |
| umls:C0011615 | Dermatitis, Atopic | 1 | BeFree |
| umls:C0011853 | Diabetes Mellitus, Experimental | 1 | RGD |
| umls:C0011854 | Diabetes Mellitus, Insulin-Dependent | 1 | BeFree |
| umls:C0011881 | Diabetic Nephropathy | 1 | BeFree,RGD |
| umls:C0013595 | Eczema | 1 | BeFree |
| umls:C0016169 | pathologic fistula | 1 | BeFree |
| umls:C0017661 | IGA Glomerulonephritis | 1 | CTD_human |
| umls:C0017667 | Nodular glomerulosclerosis | 1 | BeFree |
| umls:C0018801 | Heart failure | 1 | BeFree |
| umls:C0018802 | Congestive heart failure | 1 | BeFree |
| umls:C0020175 | Hunger | 1 | BeFree |
| umls:C0020459 | Hyperinsulinism | 1 | BeFree |
| umls:C0020474 | Hyperlipidemia, Familial Combined | 1 | BeFree,LHGDN |
| umls:C0020557 | Hypertriglyceridemia | 1 | LHGDN |
| umls:C0021051 | Immunologic Deficiency Syndromes | 1 | CTD_human |
| umls:C0022661 | Kidney Failure, Chronic | 1 | GAD |
| umls:C0023348 | Leprosy, Lepromatous | 1 | BeFree |
| umls:C0023418 | leukemia | 1 | BeFree |
| umls:C0023462 | Acute Megakaryocytic Leukemias | 1 | LHGDN |
| umls:C0023467 | Leukemia, Myelocytic, Acute | 1 | BeFree |
| umls:C0023890 | Liver Cirrhosis | 1 | BeFree |
| umls:C0026847 | Spinal Muscular Atrophy | 1 | BeFree |
| umls:C0027720 | Nephrosis | 1 | RGD |
| umls:C0029463 | Osteosarcoma | 1 | BeFree,LHGDN |
| umls:C0032463 | Polycythemia Vera | 1 | BeFree |
| umls:C0032897 | Prader-Willi Syndrome | 1 | BeFree |
| umls:C0033027 | Preleukemia | 1 | BeFree |
| umls:C0034069 | Pulmonary Fibrosis | 1 | RGD |
| umls:C0035126 | Reperfusion Injury | 1 | CTD_human |
| umls:C0035304 | Retinal Degeneration | 1 | LHGDN |
| umls:C0037278 | Skin Diseases, Infectious | 1 | BeFree |
| umls:C0038160 | Staphylococcal Infections | 1 | CTD_human |
| umls:C0040034 | Thrombocytopenia | 1 | LHGDN |
| umls:C0041296 | Tuberculosis | 1 | BeFree |
| umls:C0043094 | Weight Gain | 1 | CTD_human |
| umls:C0079744 | Diffuse Large B-Cell Lymphoma | 1 | BeFree |
| umls:C0085159 | Seasonal Affective Disorder | 1 | BeFree |
| umls:C0151744 | Myocardial Ischemia | 1 | CTD_human |
| umls:C0153381 | Malignant neoplasm of mouth | 1 | BeFree |
| umls:C0162820 | Dermatitis, Allergic Contact | 1 | CTD_human |
| umls:C0205700 | Asymmetric Septal Hypertrophy | 1 | BeFree |
| umls:C0205824 | Liposarcoma, Dedifferentiated | 1 | BeFree |
| umls:C0206685 | Acinar Cell Carcinoma | 1 | BeFree |
| umls:C0220641 | Lip and Oral Cavity Carcinoma | 1 | BeFree |
| umls:C0238190 | Inclusion Body Myositis (disorder) | 1 | BeFree,LHGDN |
| umls:C0338656 | Impaired cognition | 1 | BeFree |
| umls:C0344388 | Platelet mean volume finding | 1 | GWASCAT |
| umls:C0349782 | Ischemic cardiomyopathy | 1 | BeFree |
| umls:C0442887 | Septal hypertrophy | 1 | BeFree |
| umls:C0455825 | Left ventricular mass | 1 | GAD |
| umls:C0518010 | body mass | 1 | GAD |
| umls:C0524910 | Hepatitis C, Chronic | 1 | BeFree |
| umls:C0577698 | Exercise-induced angina | 1 | BeFree |
| umls:C0741923 | cardiac event | 1 | BeFree |
| umls:C1271104 | Blood pressure finding | 1 | GAD |
| umls:C1272641 | Systemic arterial pressure | 1 | GAD |
| umls:C1332632 | Breast Liposarcoma | 1 | BeFree |
| umls:C1458155 | Mammary Neoplasms | 1 | LHGDN |
| umls:C1511789 | Desmoplastic | 1 | BeFree |
| umls:C1562908 | Leprosy, Paucibacillary | 1 | BeFree |
| umls:C1704436 | Peripheral Arterial Diseases | 1 | BeFree |
| umls:C1709246 | Non-Neoplastic Disorder | 1 | BeFree |
| umls:C1960636 | Dysglycemia | 1 | BeFree |
| umls:C2718067 | Alcoholic Steatohepatitis | 1 | BeFree |
| umls:C2930826 | Acute malaria | 1 | BeFree |
| umls:C3463824 | MYELODYSPLASTIC SYNDROME | 1 | BeFree |
| umls:C3665419 | Intracranial glioma | 1 | BeFree |
| umls:C3714514 | Infection | 1 | LHGDN |
| umls:C3714619 | Insulin resistance syndrome | 1 | BeFree |
| umls:C1855456 | PLASMODIUM FALCIPARUM BLOOD INFECTION LEVEL | 0 | CTD_human |
| umls:C1855457 | MALARIA, CEREBRAL, SUSCEPTIBILITY TO (finding) | 0 | CLINVAR |
| umls:C1867441 | Pterygium Of Conjunctiva And Cornea | 0 | MGD |
| umls:C1969379 | MALARIA, CEREBRAL, RESISTANCE TO | 0 | CLINVAR |
| umls:C1970441 | CORONARY HEART DISEASE, SUSCEPTIBILITY TO, 7 | 0 | CLINVAR |

 Gene summary
Gene summary  Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology having evidence of Inferred from Direct Assay (IDA) from Entrez  Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
 Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Gene isoform expressions across 28 cancer types (X-axis: cancer type and Y-axis: log2(norm_counts+1))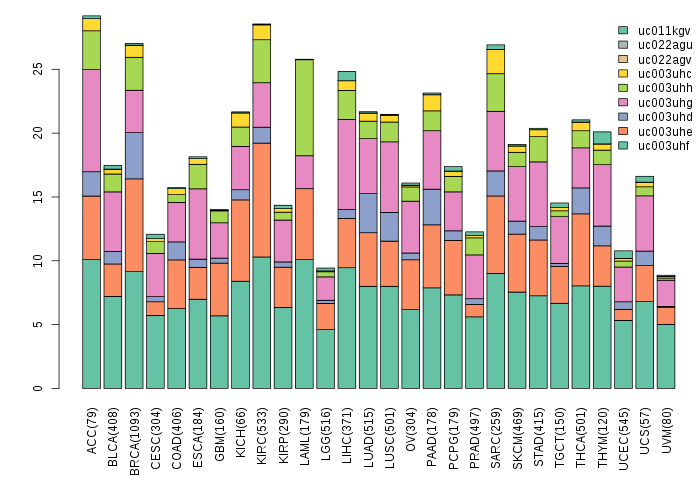
 Gene expressions across normal tissues of GTEx data
Gene expressions across normal tissues of GTEx data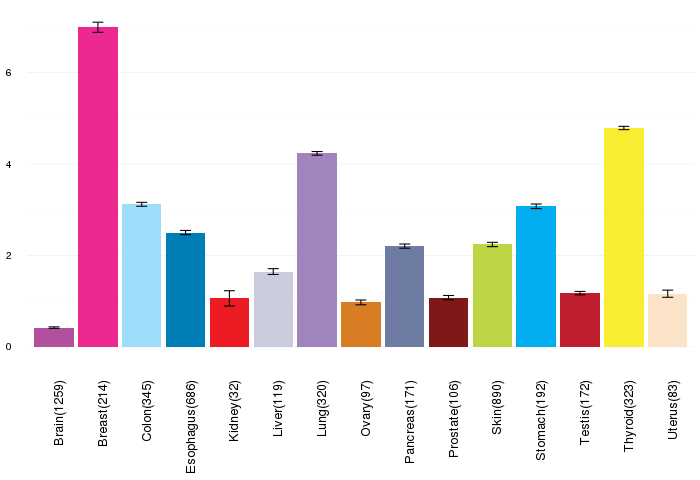
 Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))
Different expressions across 14 cancer types with more than 10 samples between matched tumors and normals (X-axis: cancer type and Y-axis: log2(norm_counts+1))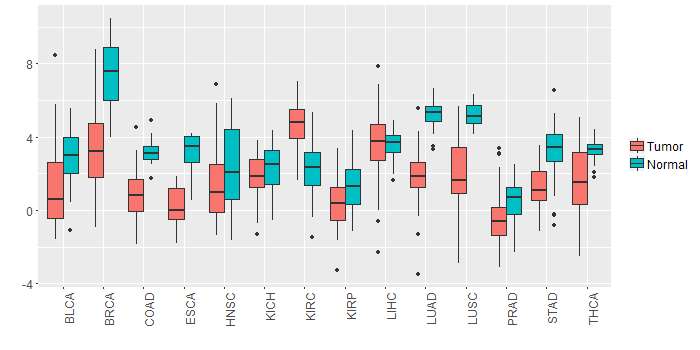
 Significantly anti-correlated miRNAs of TissGene across 28 cancer types
Significantly anti-correlated miRNAs of TissGene across 28 cancer types nsSNV counts per each loci.
nsSNV counts per each loci.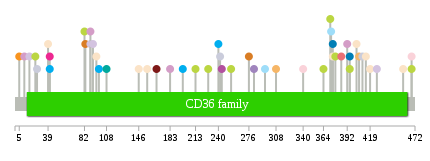

 Somatic nucleotide variants of TissGene across 28 cancer types
Somatic nucleotide variants of TissGene across 28 cancer types 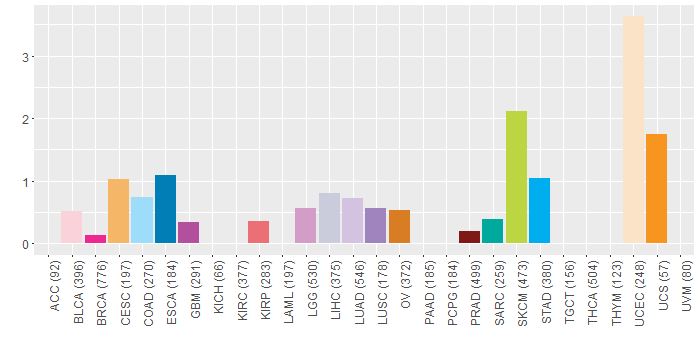
 Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)
Copy number variations of TissGene across 28 cancer types (X-axis: cancer type and Y-axis: % of CNV samples)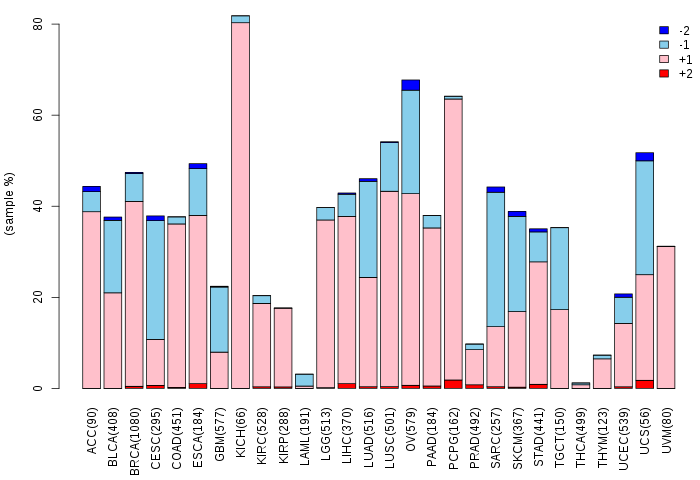
 Fusion genes including TissGene
Fusion genes including TissGene  Co-expressed gene networks based on protein-protein interaction data (CePIN)
Co-expressed gene networks based on protein-protein interaction data (CePIN) Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types
Kaplan-Meier plots with logrank tests of overall survival (OS) using 28 cancer types 
 Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types
Kaplan-Meier plots with logrank test of relapse free survival (RFS) using 28 cancer types 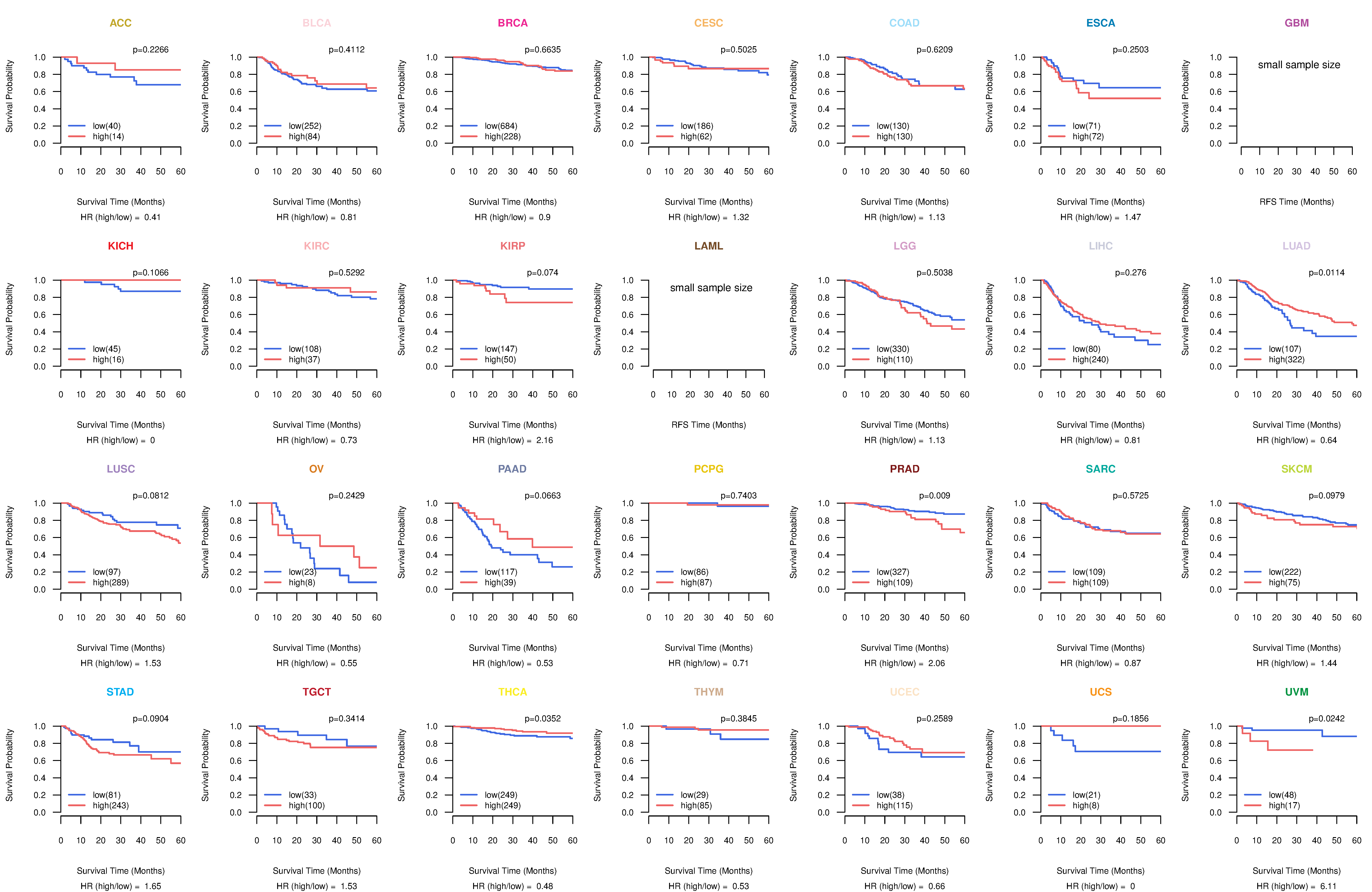
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of overall survival (OS) using 28 cancer types 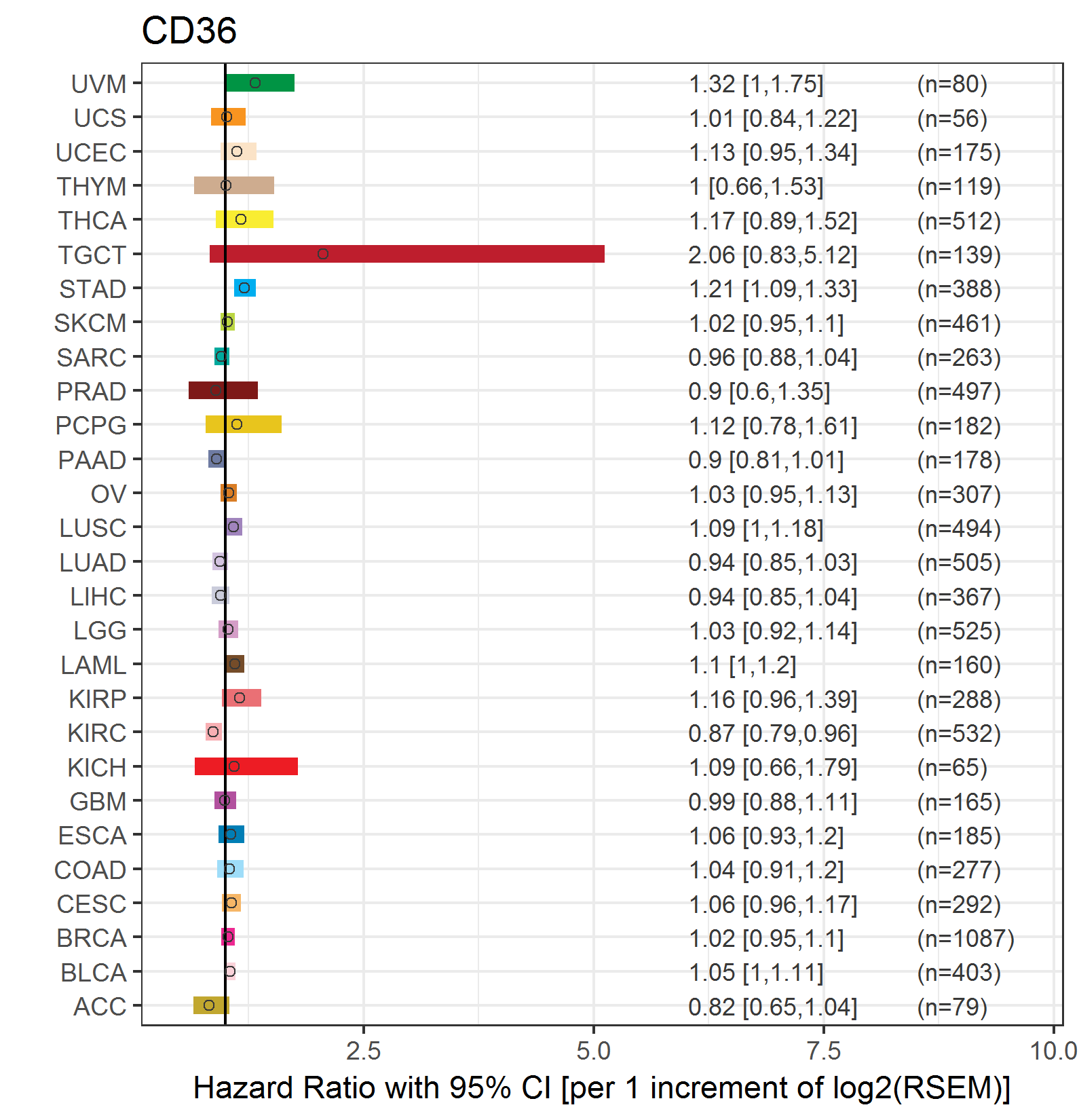
 Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types
Forest plot of Cox proportional hazard ratio (HR) and 95% CI of relapse free survival (RFS) using 28 cancer types 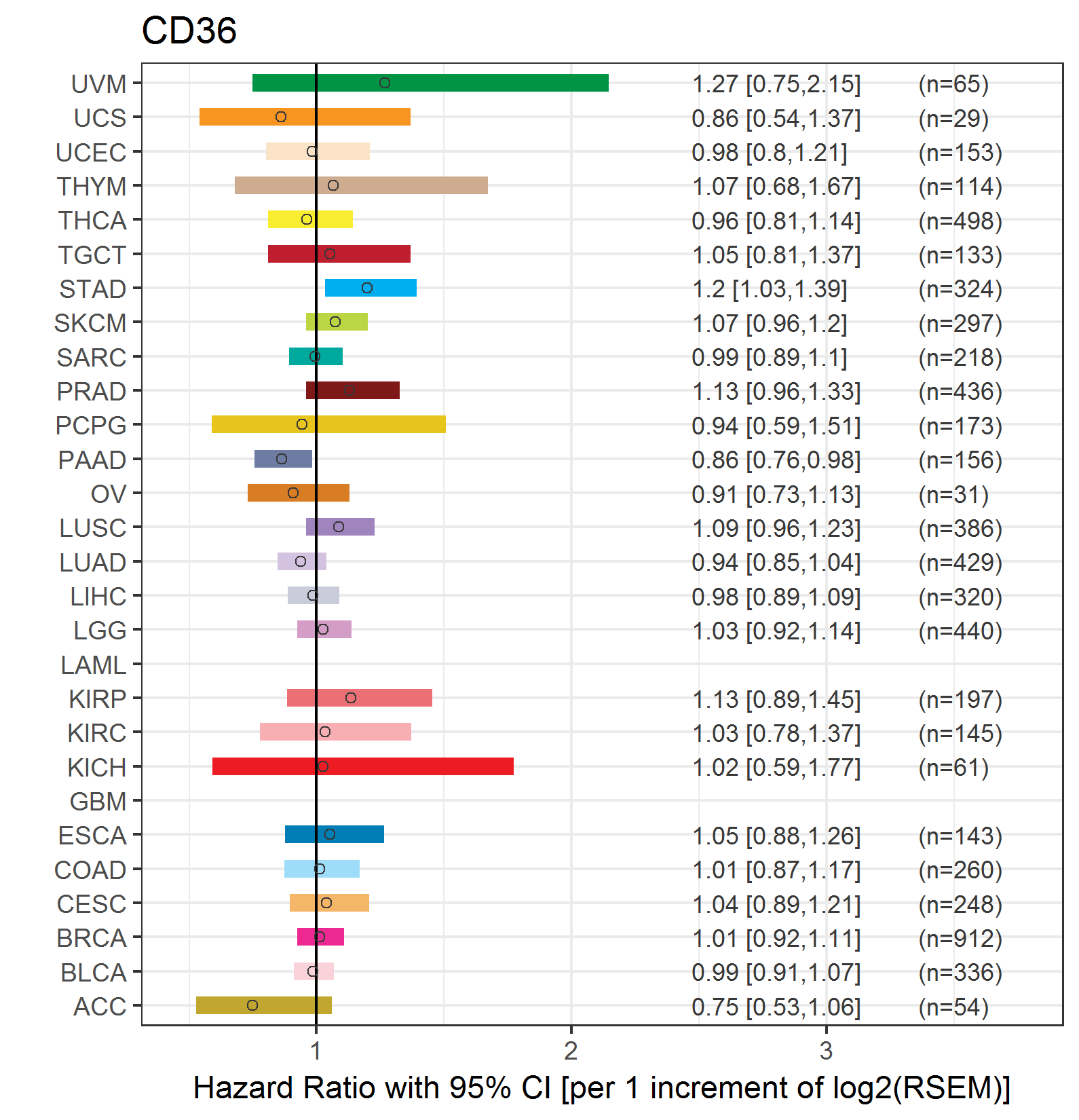
 Drug information targeting TissGene
Drug information targeting TissGene  Disease information associated with TissGene
Disease information associated with TissGene 
























