|
||||||||||||||||||||
| |
| Phenotypic Information (metabolism pathway, cancer, disease, phenome) |
| |
| |
| Gene-Gene Network Information: Co-Expression Network, Interacting Genes & KEGG |
| |
|
| Gene Summary for COMT |
| Basic gene info. | Gene symbol | COMT |
| Gene name | catechol-O-methyltransferase | |
| Synonyms | HEL-S-98n | |
| Cytomap | UCSC genome browser: 22q11.21 | |
| Genomic location | chr22 :19939044-19957498 | |
| Type of gene | protein-coding | |
| RefGenes | NM_000754.3, NM_001135161.1,NM_001135162.1,NM_007310.2, | |
| Ensembl id | ENSG00000093010 | |
| Description | catechol O-methyltransferaseepididymis secretory sperm binding protein Li 98n | |
| Modification date | 20141222 | |
| dbXrefs | MIM : 116790 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000093010 | ||
| HPRD : 00284 | ||
| Vega : OTTHUMG00000150529 | ||
| Protein | UniProt: P21964 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_COMT | |
| BioGPS: 1312 | ||
| Gene Expression Atlas: ENSG00000093010 | ||
| The Human Protein Atlas: ENSG00000093010 | ||
| Pathway | NCI Pathway Interaction Database: COMT | |
| KEGG: COMT | ||
| REACTOME: COMT | ||
| ConsensusPathDB | ||
| Pathway Commons: COMT | ||
| Metabolism | MetaCyc: COMT | |
| HUMANCyc: COMT | ||
| Regulation | Ensembl's Regulation: ENSG00000093010 | |
| miRBase: chr22 :19,939,044-19,957,498 | ||
| TargetScan: NM_000754 | ||
| cisRED: ENSG00000093010 | ||
| Context | iHOP: COMT | |
| cancer metabolism search in PubMed: COMT | ||
| UCL Cancer Institute: COMT | ||
| Assigned class in ccmGDB | A - This gene has a literature evidence and it belongs to cancer gene. | |
| References showing role of COMT in cancer cell metabolism | 1. Swift-Scanlan T, Smith CT, Bardowell SA, Boettiger CA (2014) Comprehensive interrogation of CpG island methylation in the gene encoding COMT, a key estrogen and catecholamine regulator. BMC Med Genomics 7: 5. doi: 10.1186/1755-8794-7-5. pmid: 3910242. go to article 2. Yager JD (2015) Mechanisms of estrogen carcinogenesis: The role of E2/E1-quinone metabolites suggests new approaches to preventive intervention - A review. Steroids 99: 56-60. doi: 10.1016/j.steroids.2014.08.006. pmid: 4339663. go to article 3. Hevir-Kene N, Rizner TL (2015) The endometrial cancer cell lines Ishikawa and HEC-1A, and the control cell line HIEEC, differ in expression of estrogen biosynthetic and metabolic genes, and in androstenedione and estrone-sulfate metabolism. Chem Biol Interact 234: 309-319. doi: 10.1016/j.cbi.2014.11.015. go to article | |
| Top |
| Phenotypic Information for COMT(metabolism pathway, cancer, disease, phenome) |
| Cancer | CGAP: COMT |
| Familial Cancer Database: COMT | |
| * This gene is included in those cancer gene databases. |
|
|
|
|
|
| . | ||||||||||||||
Oncogene 1 | Significant driver gene in | |||||||||||||||||||
| cf) number; DB name 1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 2 Tumor Suppressor gene; https://bioinfo.uth.edu/TSGene/, 3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html, 4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php, 1Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/ |
| KEGG_TYROSINE_METABOLISM | |
| OMIM | 103780; phenotype. 116790; gene+phenotype. |
| Orphanet | 240863; Cisplatin toxicity. 240999; Susceptibility to deafness due to cisplatin treatment. 3140; Schizophrenia. 567; 22q11.2 deletion syndrome. |
| Disease | KEGG Disease: COMT |
| MedGen: COMT (Human Medical Genetics with Condition) | |
| ClinVar: COMT | |
| Phenotype | MGI: COMT (International Mouse Phenotyping Consortium) |
| PhenomicDB: COMT | |
| Mutations for COMT |
| * Under tables are showing count per each tissue to give us broad intuition about tissue specific mutation patterns.You can go to the detailed page for each mutation database's web site. |
| - Statistics for Tissue and Mutation type | Top |
 |
| - For Inter-chromosomal Variations |
| There's no inter-chromosomal structural variation. |
| - For Intra-chromosomal Variations |
| * Intra-chromosomal variantions includes 'intrachromosomal amplicon to amplicon', 'intrachromosomal amplicon to non-amplified dna', 'intrachromosomal deletion', 'intrachromosomal fold-back inversion', 'intrachromosomal inversion', 'intrachromosomal tandem duplication', 'Intrachromosomal unknown type', 'intrachromosomal with inverted orientation', 'intrachromosomal with non-inverted orientation'. |
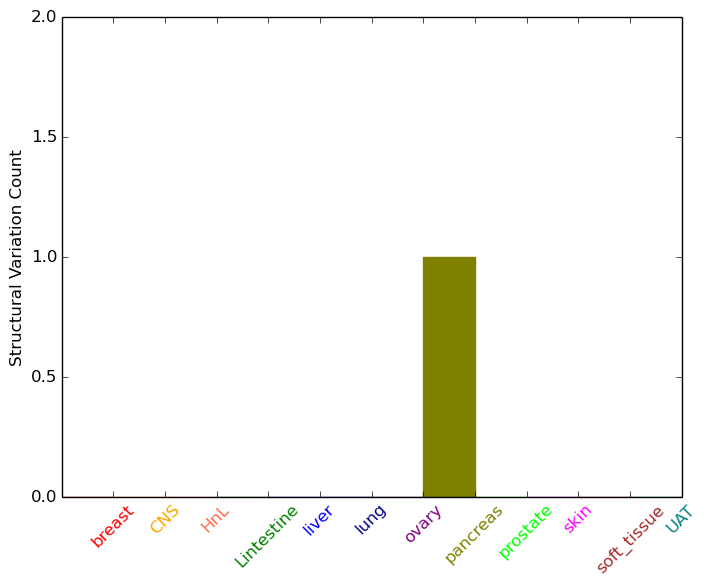 |
| Sample | Symbol_a | Chr_a | Start_a | End_a | Symbol_b | Chr_b | Start_b | End_b |
| pancreas | COMT | chr22 | 19949804 | 19949824 | TXNRD2 | chr22 | 19926916 | 19926936 |
| cf) Tissue number; Tissue name (1;Breast, 2;Central_nervous_system, 3;Haematopoietic_and_lymphoid_tissue, 4;Large_intestine, 5;Liver, 6;Lung, 7;Ovary, 8;Pancreas, 9;Prostate, 10;Skin, 11;Soft_tissue, 12;Upper_aerodigestive_tract) |
| * From mRNA Sanger sequences, Chitars2.0 arranged chimeric transcripts. This table shows COMT related fusion information. |
| ID | Head Gene | Tail Gene | Accession | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a |
| CB529248 | COMT | 16 | 605 | 22 | 19951714 | 19956539 | NICN1 | 601 | 689 | 3 | 49461613 | 49461701 | |
| BG259212 | IQSEC1 | 327 | 346 | 3 | 12942516 | 12942535 | COMT | 334 | 357 | 22 | 19946481 | 19946506 | |
| BI053757 | COMT | 24 | 87 | 22 | 19956314 | 19956377 | MAP7D1 | 81 | 457 | 1 | 36644863 | 36645663 | |
| AI360037 | COMT | 1 | 356 | 22 | 19957420 | 19957775 | PCBP2 | 350 | 465 | 12 | 53849140 | 53849736 | |
| BI053724 | COMT | 12 | 74 | 22 | 19956314 | 19956377 | MAP7D1 | 68 | 444 | 1 | 36644863 | 36645663 | |
| AW340773 | COMT | 5 | 340 | 22 | 19957168 | 19957503 | THBS3 | 337 | 426 | 1 | 155177604 | 155177693 | |
| AI298713 | COMT | 9 | 71 | 22 | 19951222 | 19951284 | COMT | 67 | 497 | 22 | 19951702 | 19956367 | |
| Top |
| There's no copy number variation information in COSMIC data for this gene. |
| Top |
|
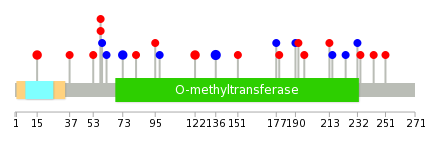 |
| Top |
| Stat. for Non-Synonymous SNVs (# total SNVs=7) | (# total SNVs=5) |
 | 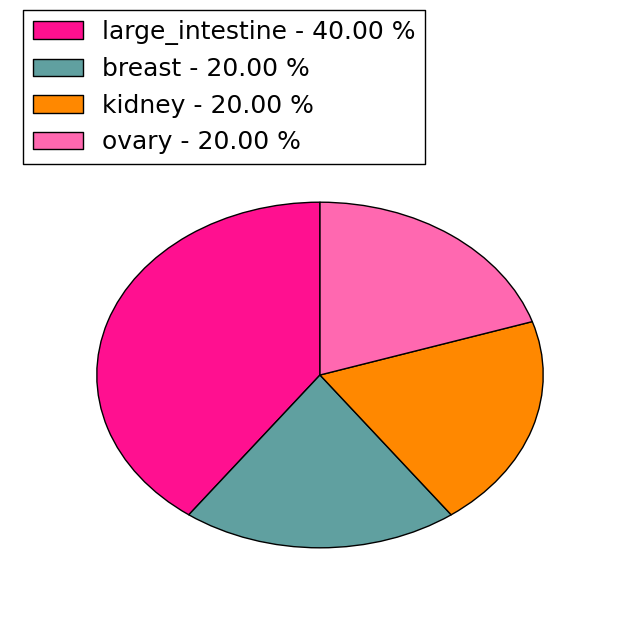 |
(# total SNVs=1) | (# total SNVs=0) |
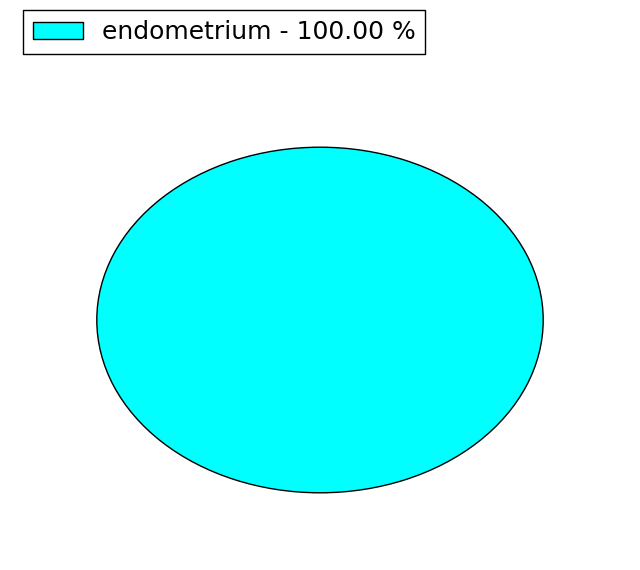 |
| Top |
| * When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site,primary_histology,mutation(aa),pubmedID. |
| GRCh37 position | Mutation(aa) | Unique sampleID count |
| chr22:19951207-19951207 | p.L136L | 3 |
| chr22:19951164-19951164 | p.S122L | 2 |
| chr22:19950093-19950093 | p.G15V | 2 |
| chr22:19950221-19950221 | p.R58C | 2 |
| chr22:19950268-19950268 | p.Q73Q | 2 |
| chr22:19951793-19951793 | p.R196W | 1 |
| chr22:19950334-19950334 | p.K95N | 1 |
| chr22:19956054-19956054 | p.? | 1 |
| chr22:19951093-19951093 | p.K98K | 1 |
| chr22:19956080-19956080 | p.G213R | 1 |
| Top |
|
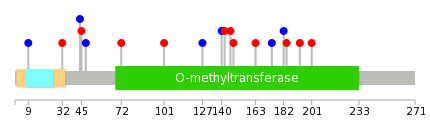 |
| Point Mutation/ Tissue ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| # sample | 4 | 1 | 1 | 1 | 3 | 6 | 2 | |||||||||||||
| # mutation | 4 | 1 | 1 | 1 | 3 | 6 | 2 | |||||||||||||
| nonsynonymous SNV | 2 | 2 | 5 | 2 | ||||||||||||||||
| synonymous SNV | 2 | 1 | 1 | 1 | 1 | 1 |
| cf) Tissue ID; Tissue type (1; BLCA[Bladder Urothelial Carcinoma], 2; BRCA[Breast invasive carcinoma], 3; CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], 4; COAD[Colon adenocarcinoma], 5; GBM[Glioblastoma multiforme], 6; Glioma Low Grade, 7; HNSC[Head and Neck squamous cell carcinoma], 8; KICH[Kidney Chromophobe], 9; KIRC[Kidney renal clear cell carcinoma], 10; KIRP[Kidney renal papillary cell carcinoma], 11; LAML[Acute Myeloid Leukemia], 12; LUAD[Lung adenocarcinoma], 13; LUSC[Lung squamous cell carcinoma], 14; OV[Ovarian serous cystadenocarcinoma ], 15; PAAD[Pancreatic adenocarcinoma], 16; PRAD[Prostate adenocarcinoma], 17; SKCM[Skin Cutaneous Melanoma], 18:STAD[Stomach adenocarcinoma], 19:THCA[Thyroid carcinoma], 20:UCEC[Uterine Corpus Endometrial Carcinoma]) |
| Top |
| * We represented just top 10 SNVs. When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site, primary_histology, mutation(aa), pubmedID. |
| Genomic Position | Mutation(aa) | Unique sampleID count |
| chr22:19951777 | p.L148M,COMT | 1 |
| chr22:19951781 | p.G163R,COMT | 1 |
| chr22:19951793 | p.P174P,COMT | 1 |
| chr22:19950226 | p.H182H,COMT | 1 |
| chr22:19951799 | p.R184C,COMT | 1 |
| chr22:19950294 | p.I9I,COMT | 1 |
| chr22:19956080 | p.H193Y,COMT | 1 |
| chr22:19950331 | p.Y32C,COMT | 1 |
| chr22:19956115 | p.R201M,COMT | 1 |
| chr22:19950334 | p.D44D,COMT | 1 |
| * Copy number data were extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered on Jan-05-2015. Function ProcessCNAData in TCGA-Assembler package was used to obtain gene-level copy number value which is calculated as the average copy number of the genomic region of a gene. |
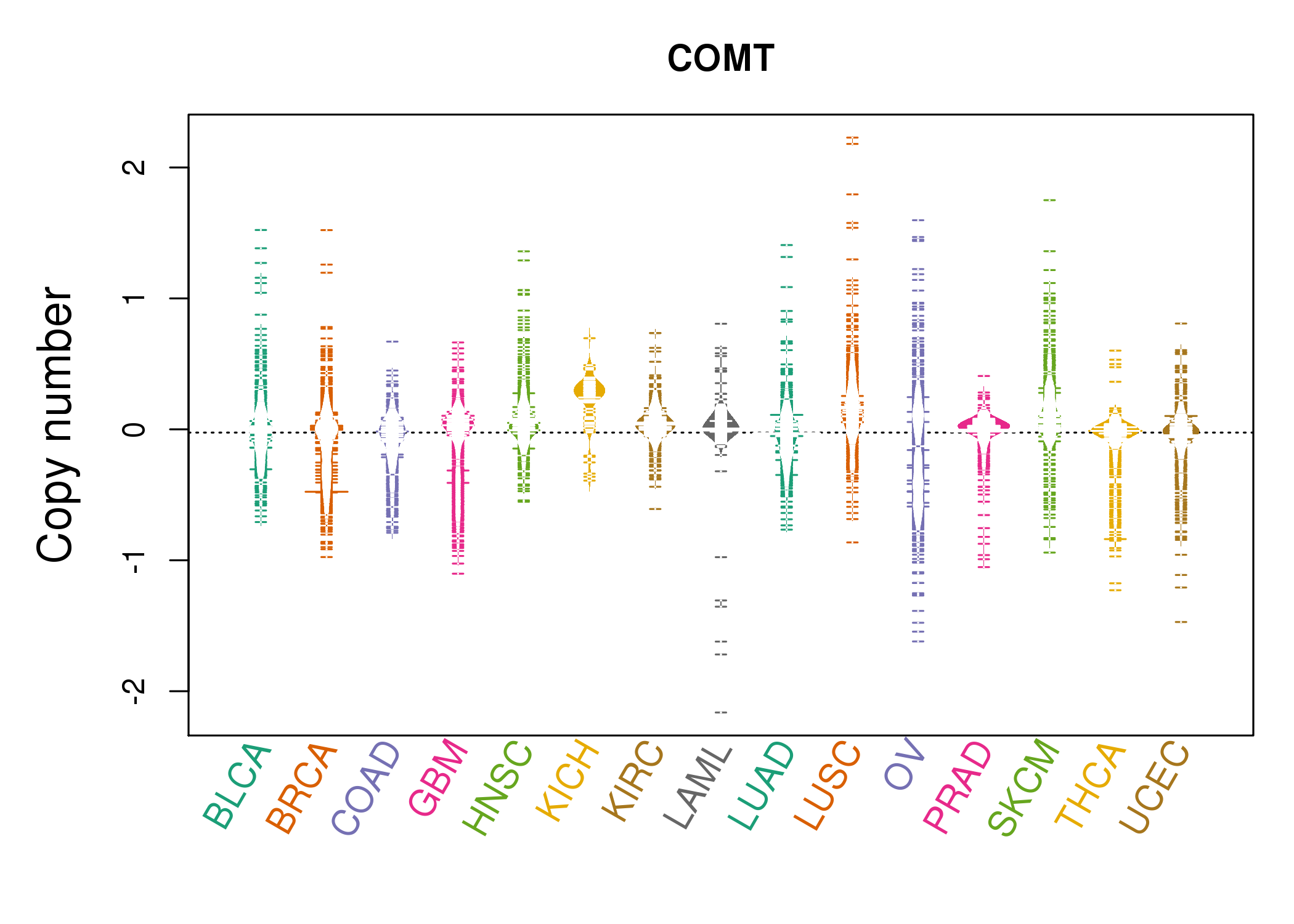 |
| cf) Tissue ID[Tissue type]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma], KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ], PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma] |
| Top |
| Gene Expression for COMT |
| * CCLE gene expression data were extracted from CCLE_Expression_Entrez_2012-10-18.res: Gene-centric RMA-normalized mRNA expression data. |
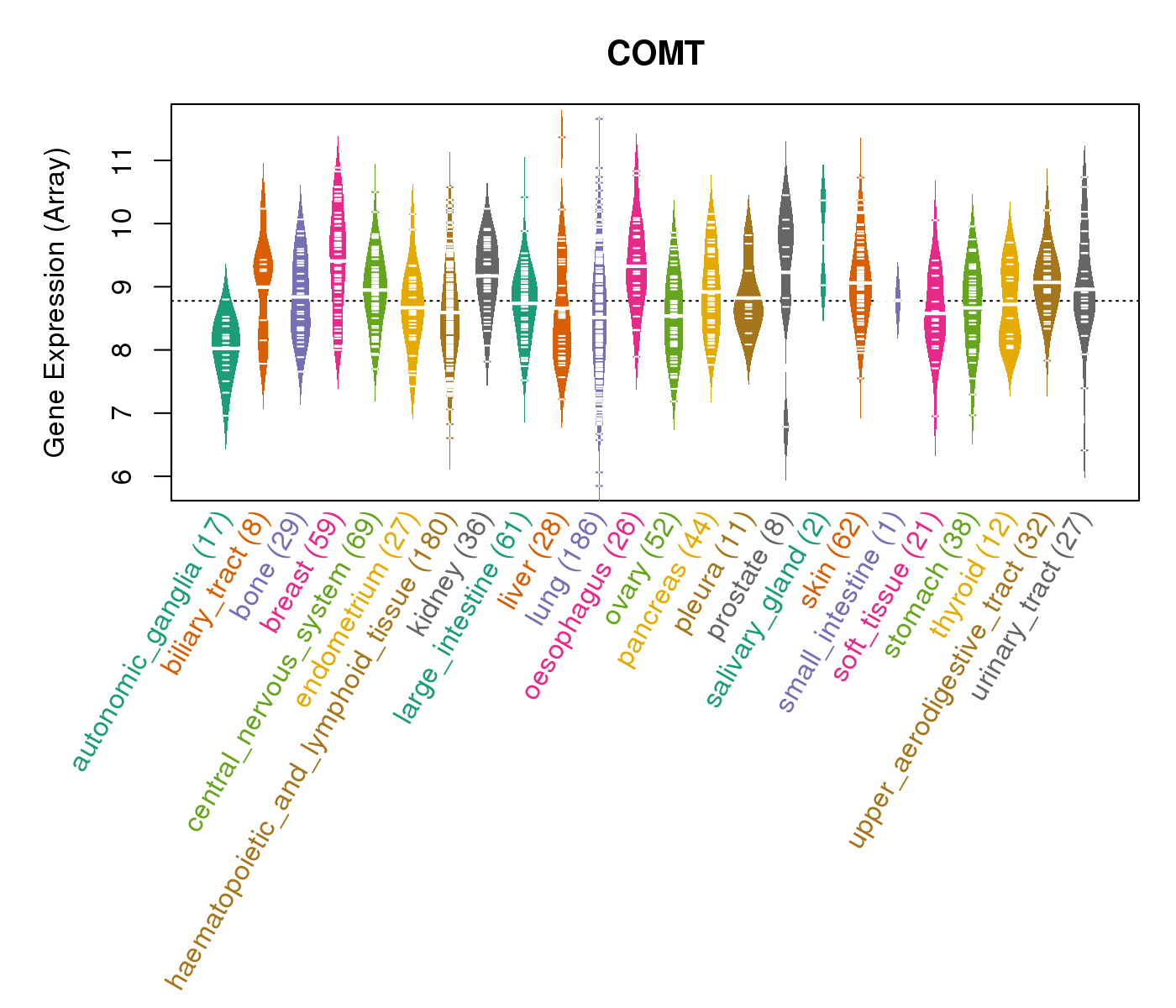 |
| * Normalized gene expression data of RNASeqV2 was extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered at Jan-05-2015. Only eight cancer types have enough normal control samples for differential expression analysis. (t test, adjusted p<0.05 (using Benjamini-Hochberg FDR)) |
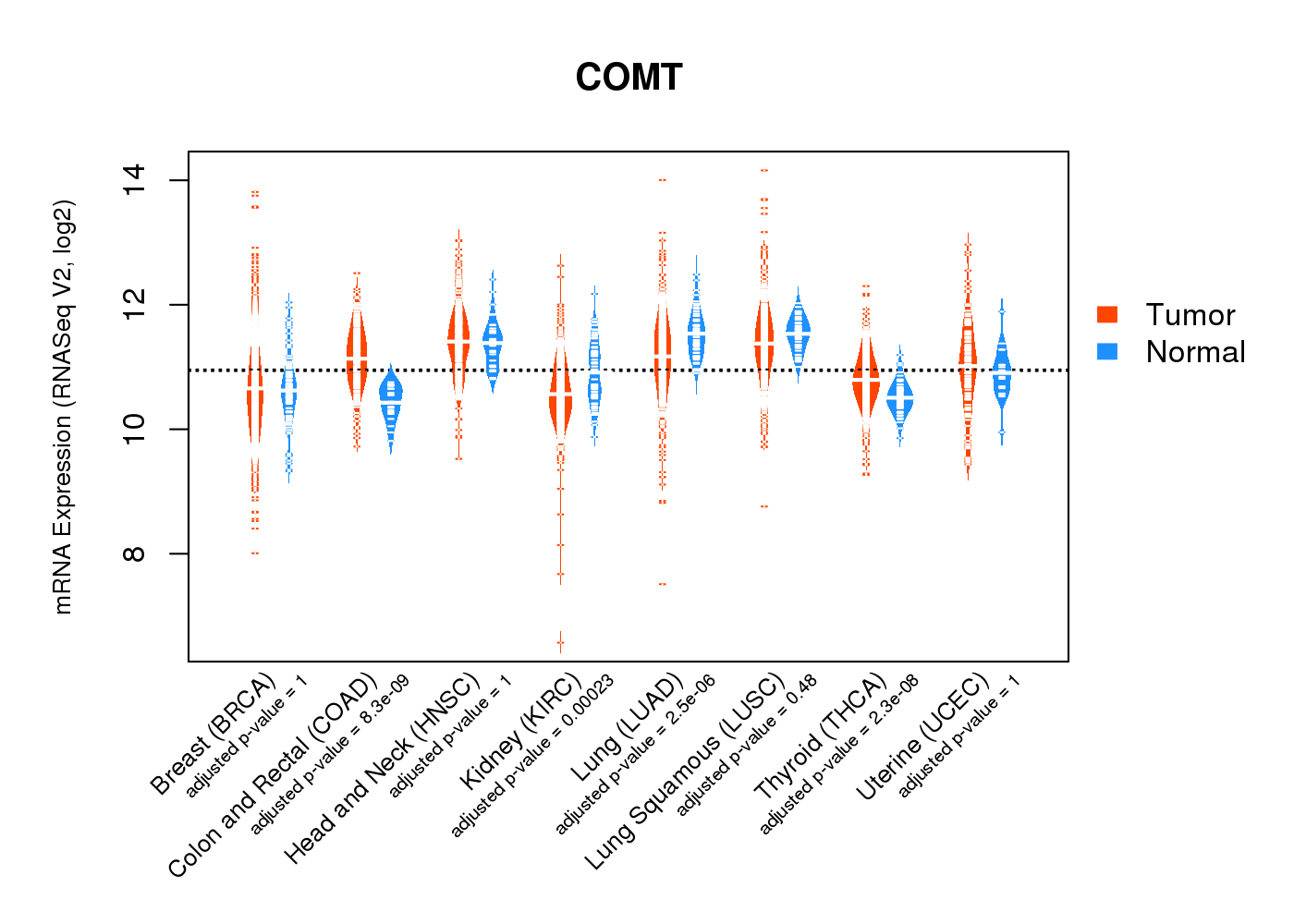 |
| Top |
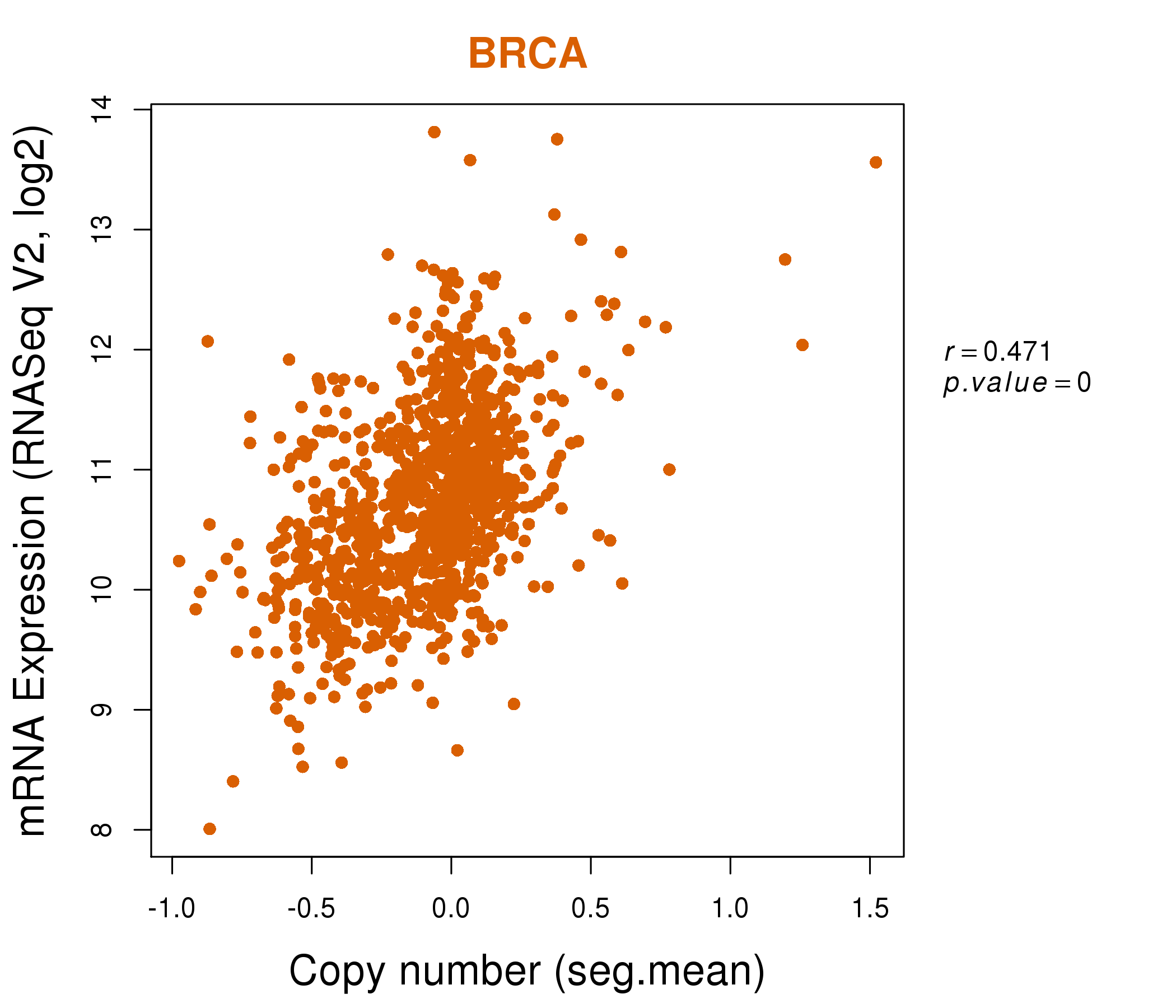
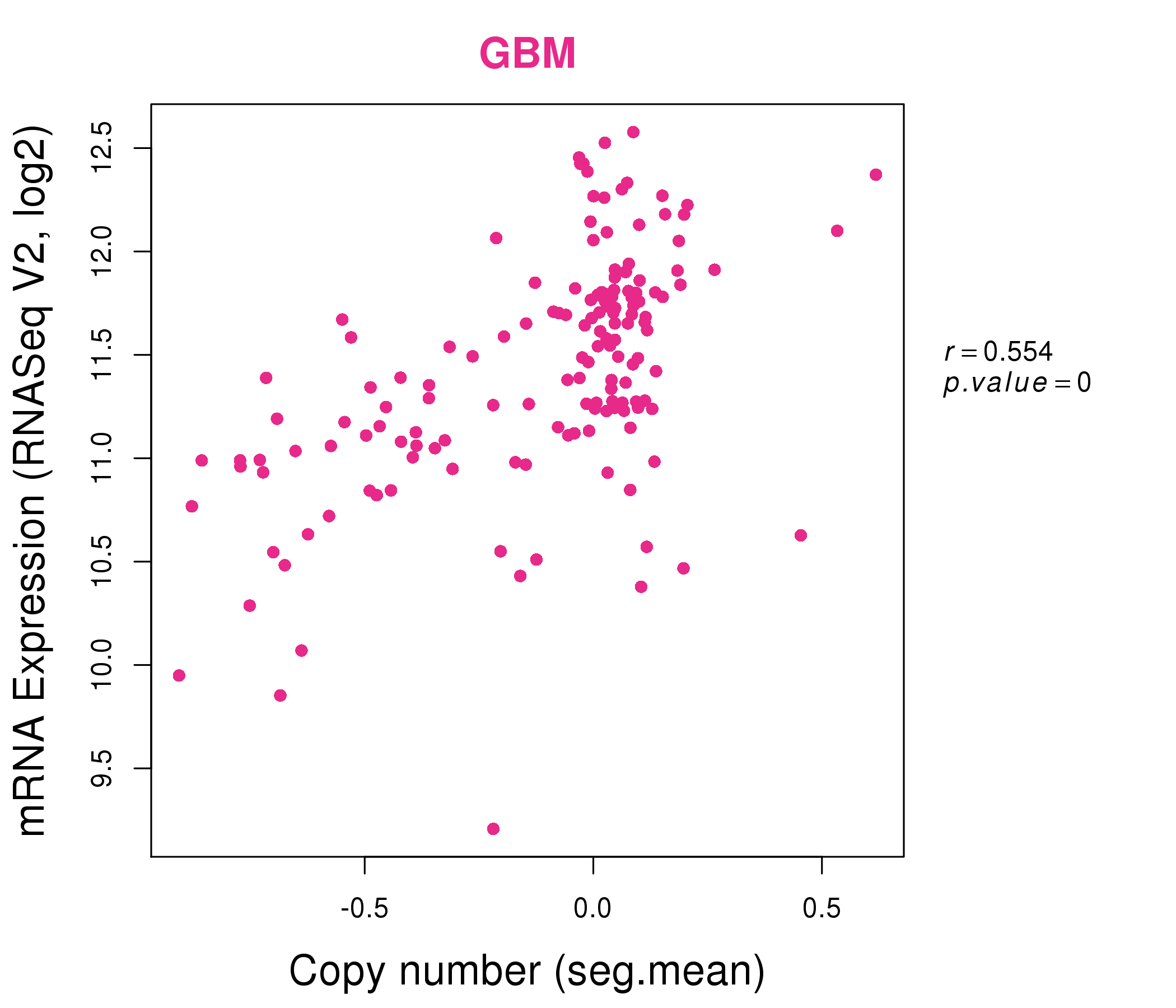
| * This plots show the correlation between CNV and gene expression. |
: Open all plots for all cancer types
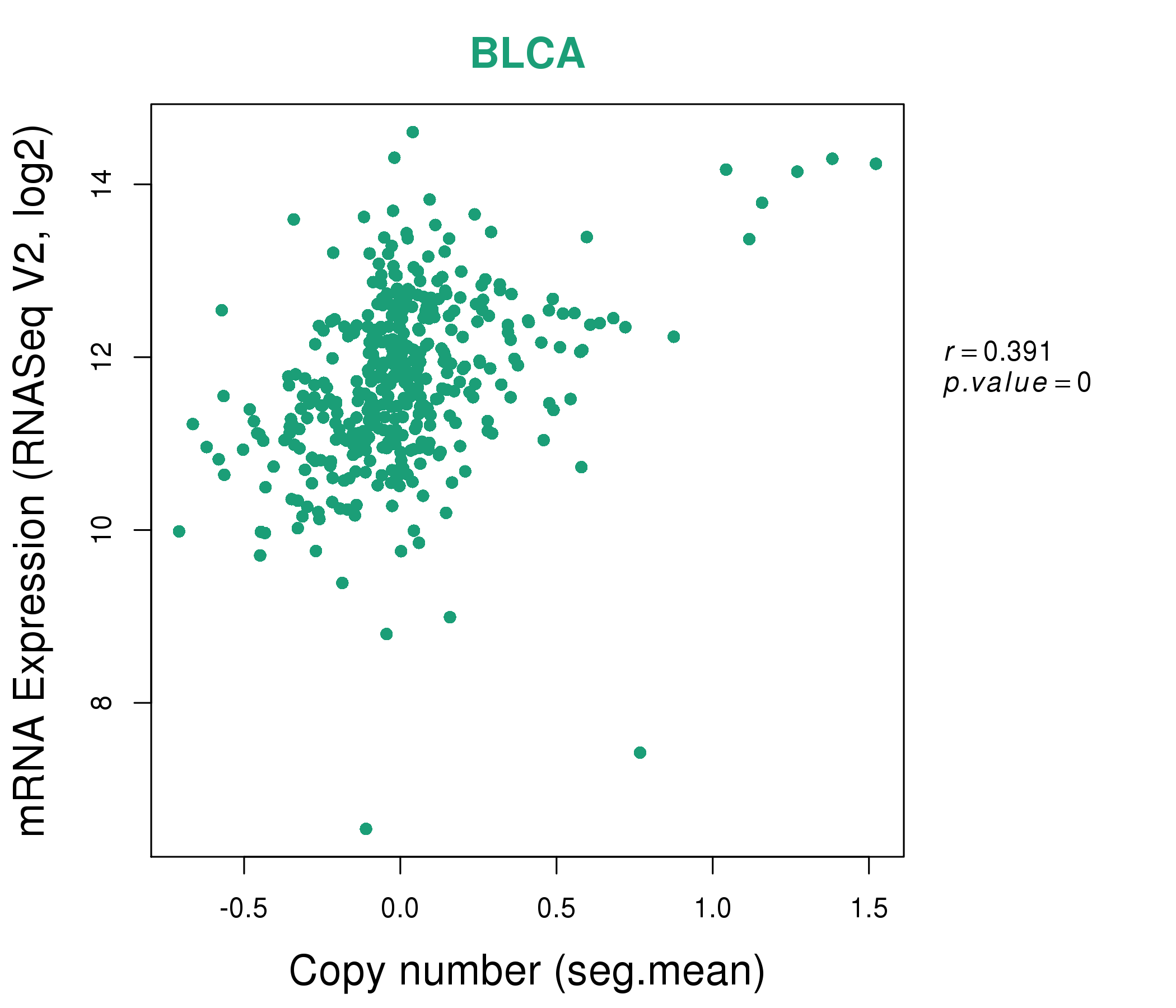 |
|
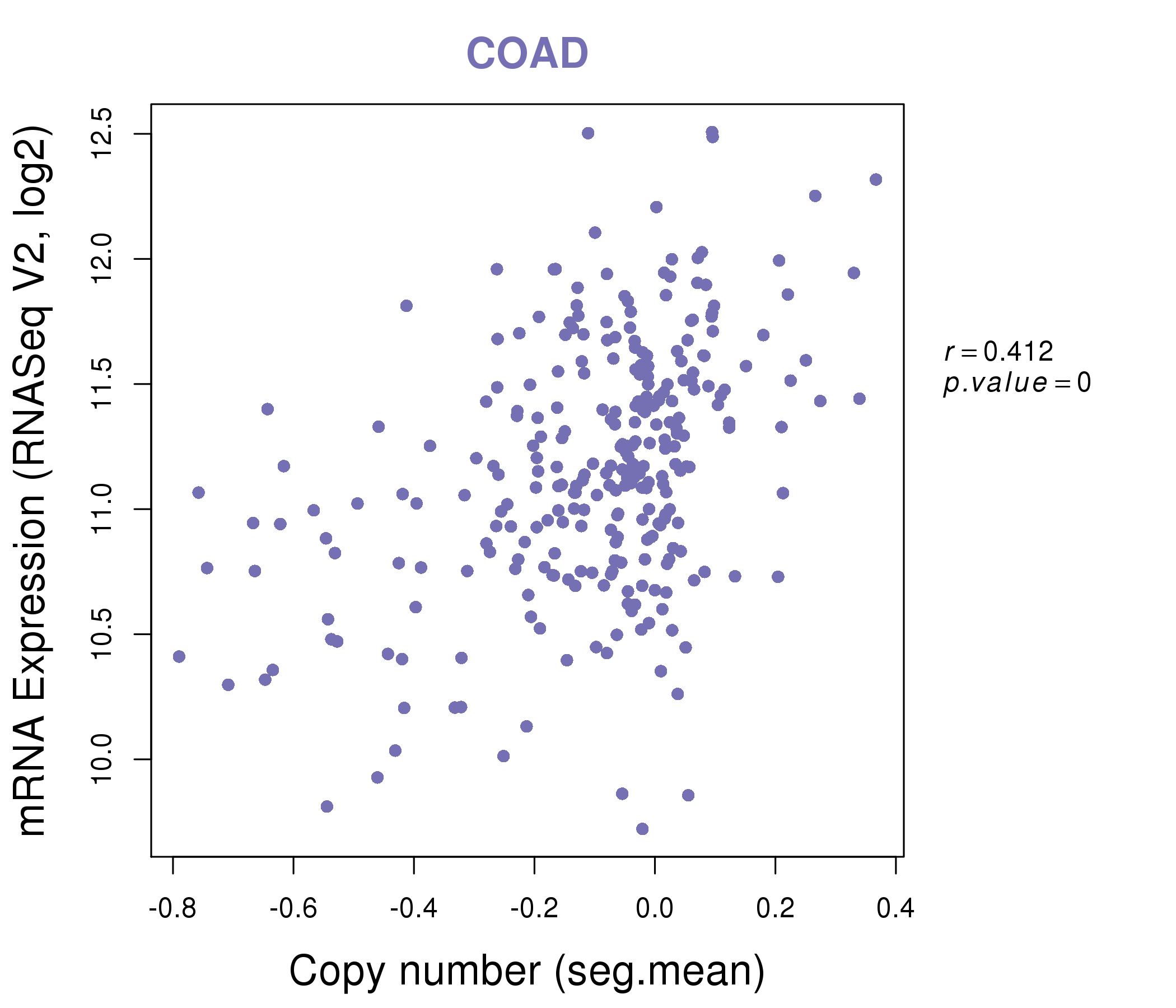 |
|
| Top |
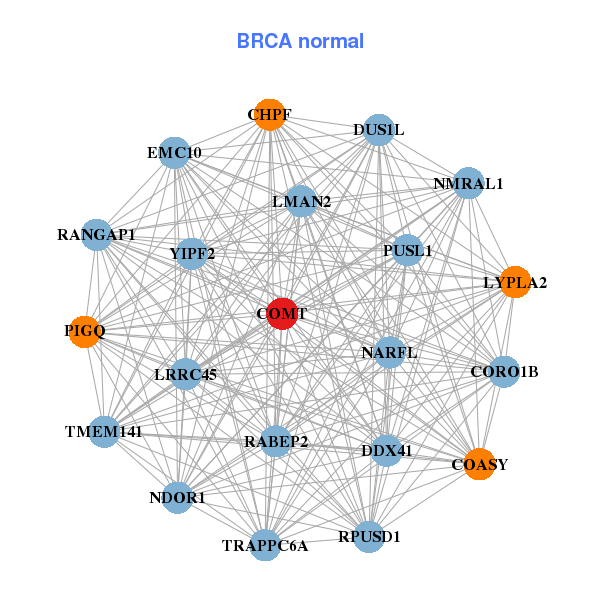
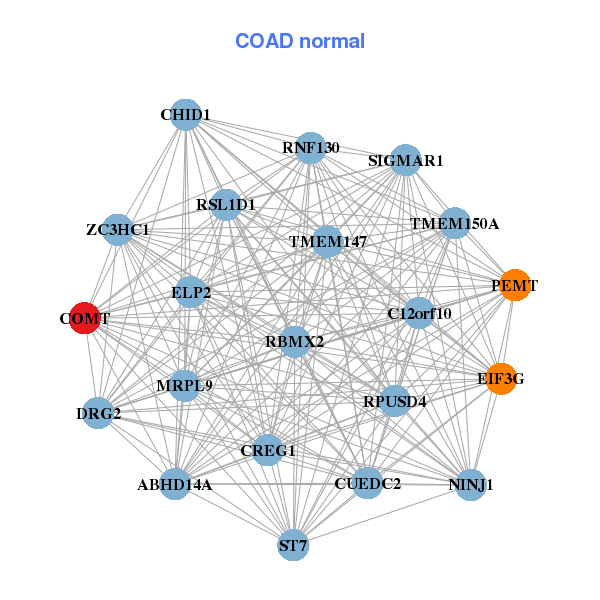
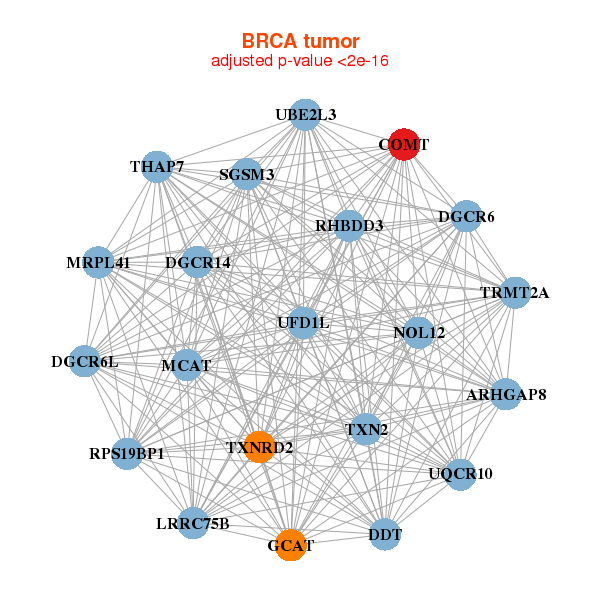
| Gene-Gene Network Information |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
 |
| ||||
| ARHGAP8,LRRC75B,COMT,DDT,DGCR14,DGCR6,DGCR6L, GCAT,MCAT,MRPL41,NOL12,RHBDD3,RPS19BP1,SGSM3, THAP7,TRMT2A,TXN2,TXNRD2,UBE2L3,UFD1L,UQCR10 | EMC10,CHPF,COASY,COMT,CORO1B,DDX41,DUS1L, LMAN2,LRRC45,LYPLA2,NARFL,NDOR1,NMRAL1,PIGQ, PUSL1,RABEP2,RANGAP1,RPUSD1,TMEM141,TRAPPC6A,YIPF2 | ||||
 |
| ||||
| ADSL,ARL2,BID,COMT,DRG1,MIF,MPST, MRPL40,NHP2L1,NIPSNAP1,PLA2G12B,POLR2F,RPS19BP1,SLC25A1, SLC39A5,SNRPD3,THAP7,TRMT2A,UQCR10,YDJC,ZMAT5 | ABHD14A,C12orf10,CHID1,COMT,CREG1,CUEDC2,DRG2, EIF3G,ELP2,MRPL9,NINJ1,PEMT,RBMX2,RNF130, RPUSD4,RSL1D1,SIGMAR1,ST7,TMEM147,TMEM150A,ZC3HC1 |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
| Top |
: Open all interacting genes' information including KEGG pathway for all interacting genes from DAVID
| Top |
| Pharmacological Information for COMT |
| DB Category | DB Name | DB's ID and Url link |
| Chemistry | BindingDB | P21964; -. |
| Chemistry | ChEMBL | CHEMBL2023; -. |
| Chemistry | GuidetoPHARMACOLOGY | 2472; -. |
| Organism-specific databases | PharmGKB | PA117; -. |
| Organism-specific databases | CTD | 1312; -. |
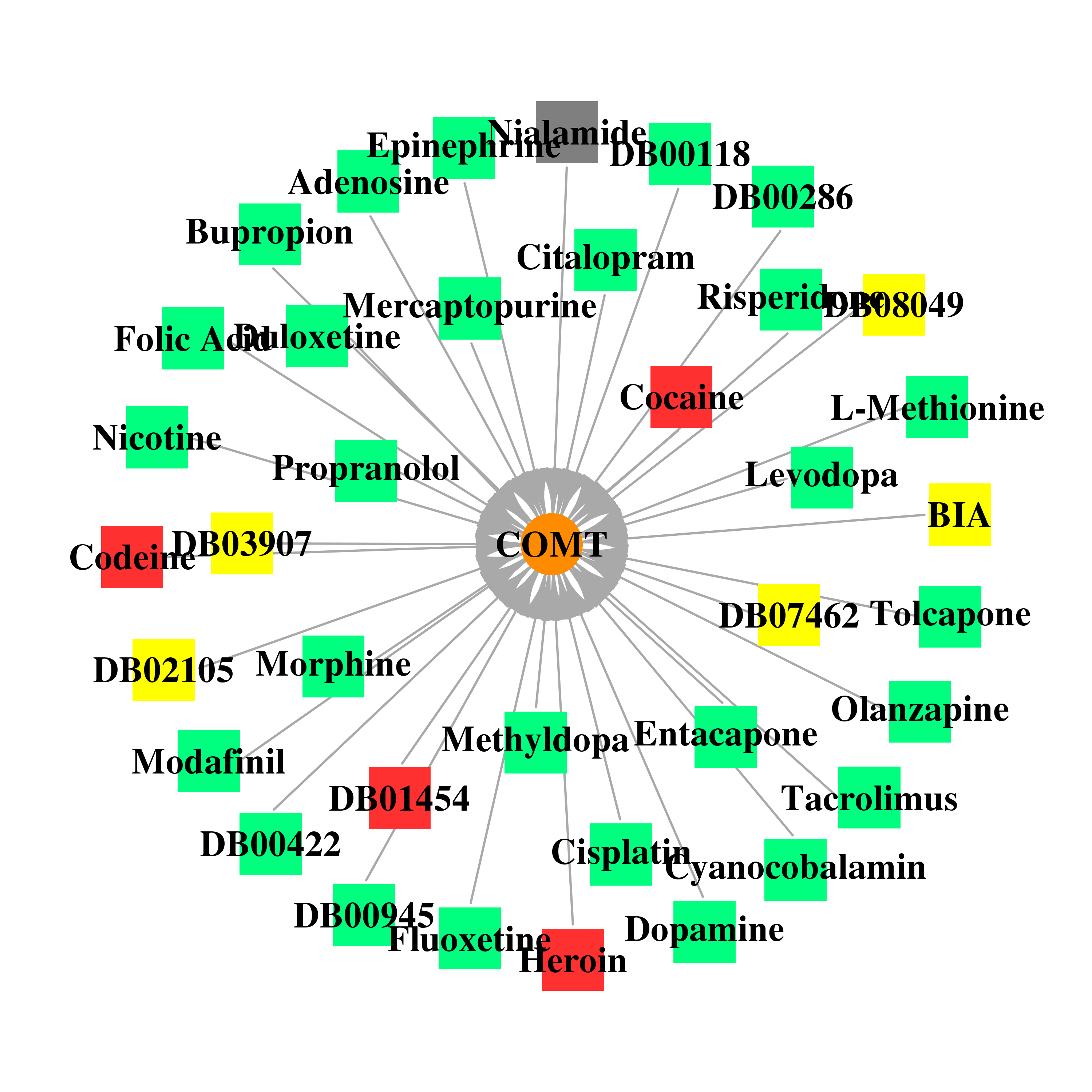
| * Gene Centered Interaction Network. |
 |
| * Drug Centered Interaction Network. |
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB00118 | catechol-O-methyltransferase | approved; nutraceutical | S-Adenosylmethionine | 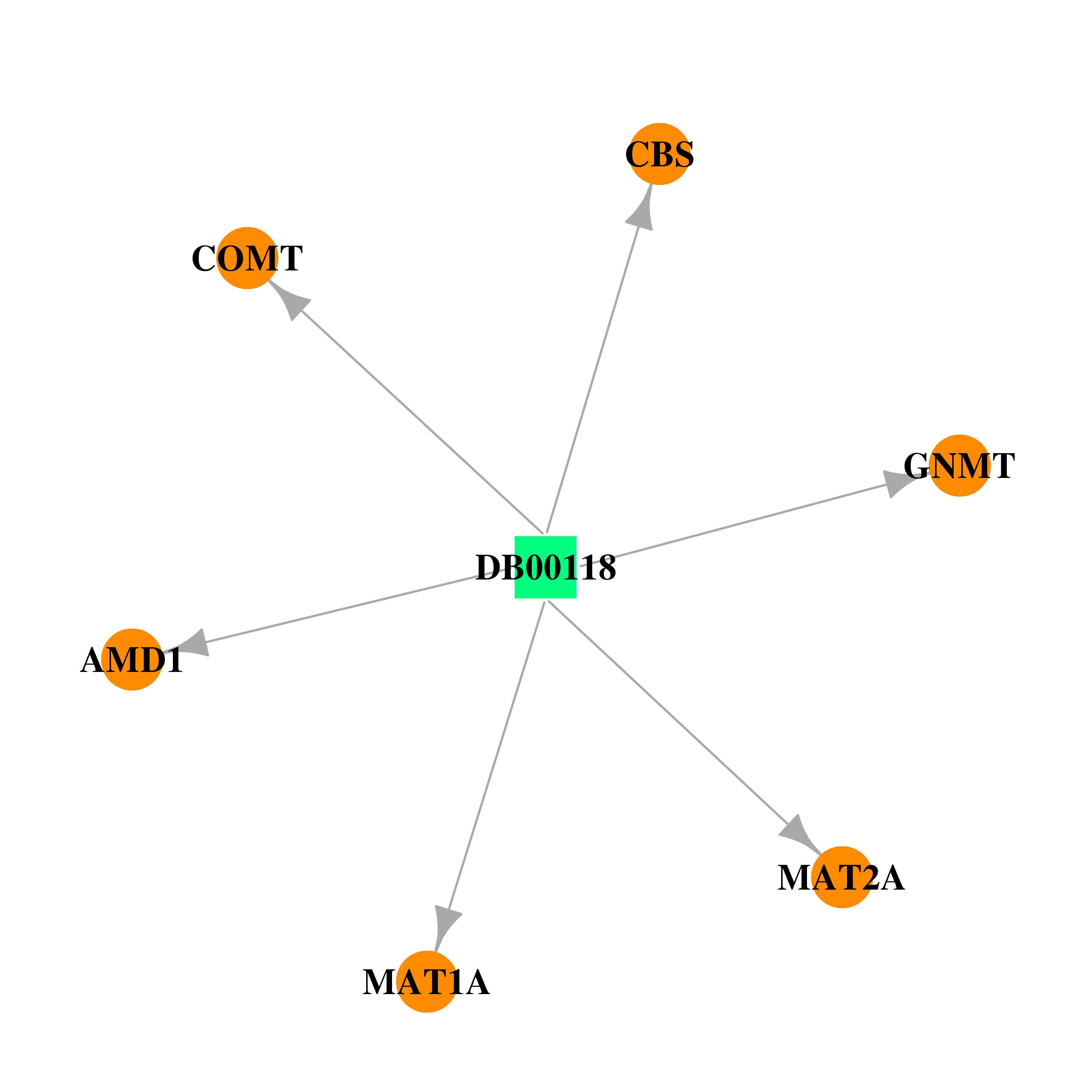 |  |
| DB00323 | catechol-O-methyltransferase | approved; withdrawn | Tolcapone | 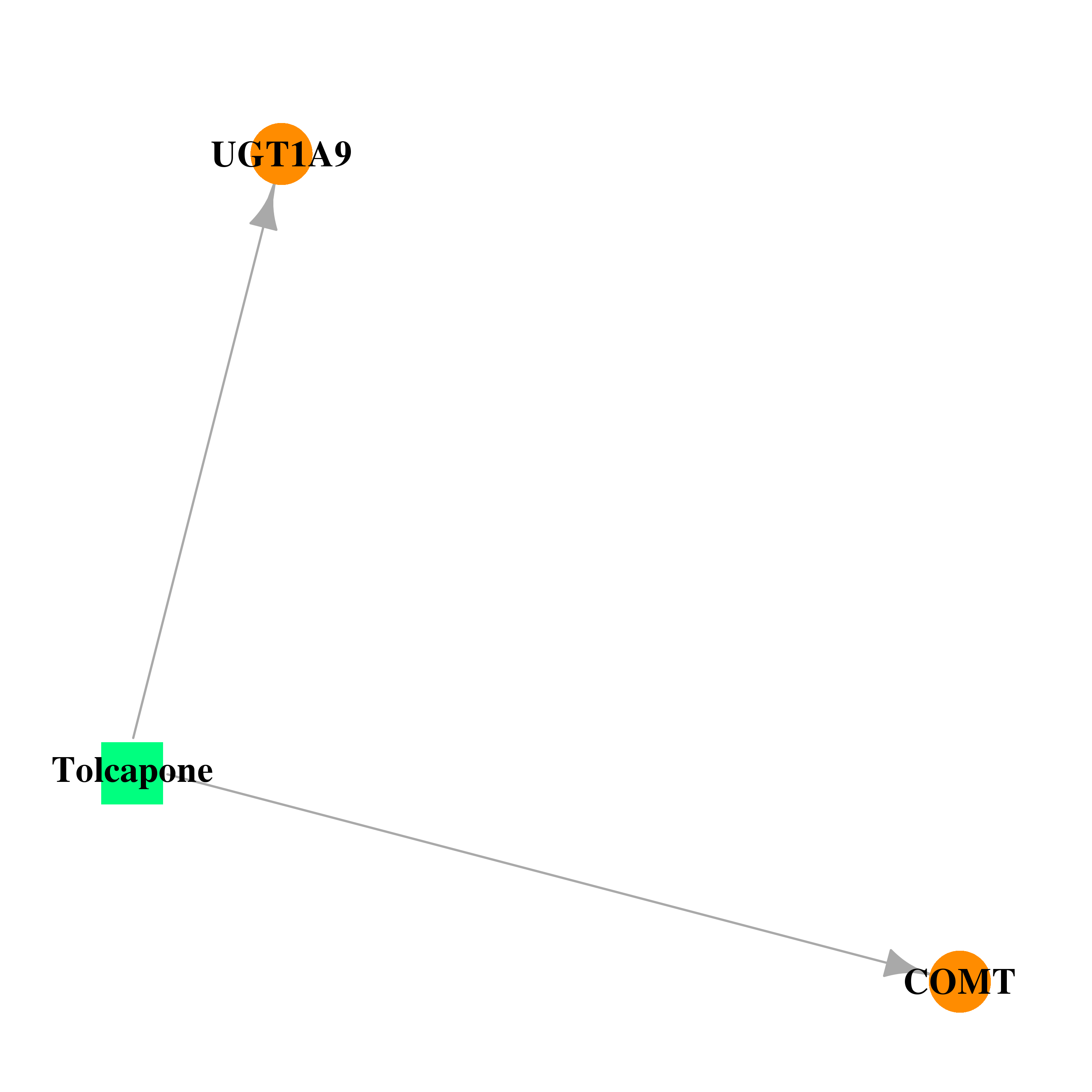 |  |
| DB00494 | catechol-O-methyltransferase | approved; investigational | Entacapone |  |  |
| DB02105 | catechol-O-methyltransferase | experimental | 3,5-Dinitrocatechol |  |  |
| DB03336 | catechol-O-methyltransferase | experimental | BIA |  |  |
| DB03907 | catechol-O-methyltransferase | experimental | N-{3-[5-(6-Amino-Purin-9-Yl)-3,4-Dihydroxy-Tetrahydro-Furan-2-Yl]-Allyl}-2,3-Dihydroxy-5-Nitro-Benzamide |  |  |
| DB04820 | catechol-O-methyltransferase | withdrawn | Nialamide | 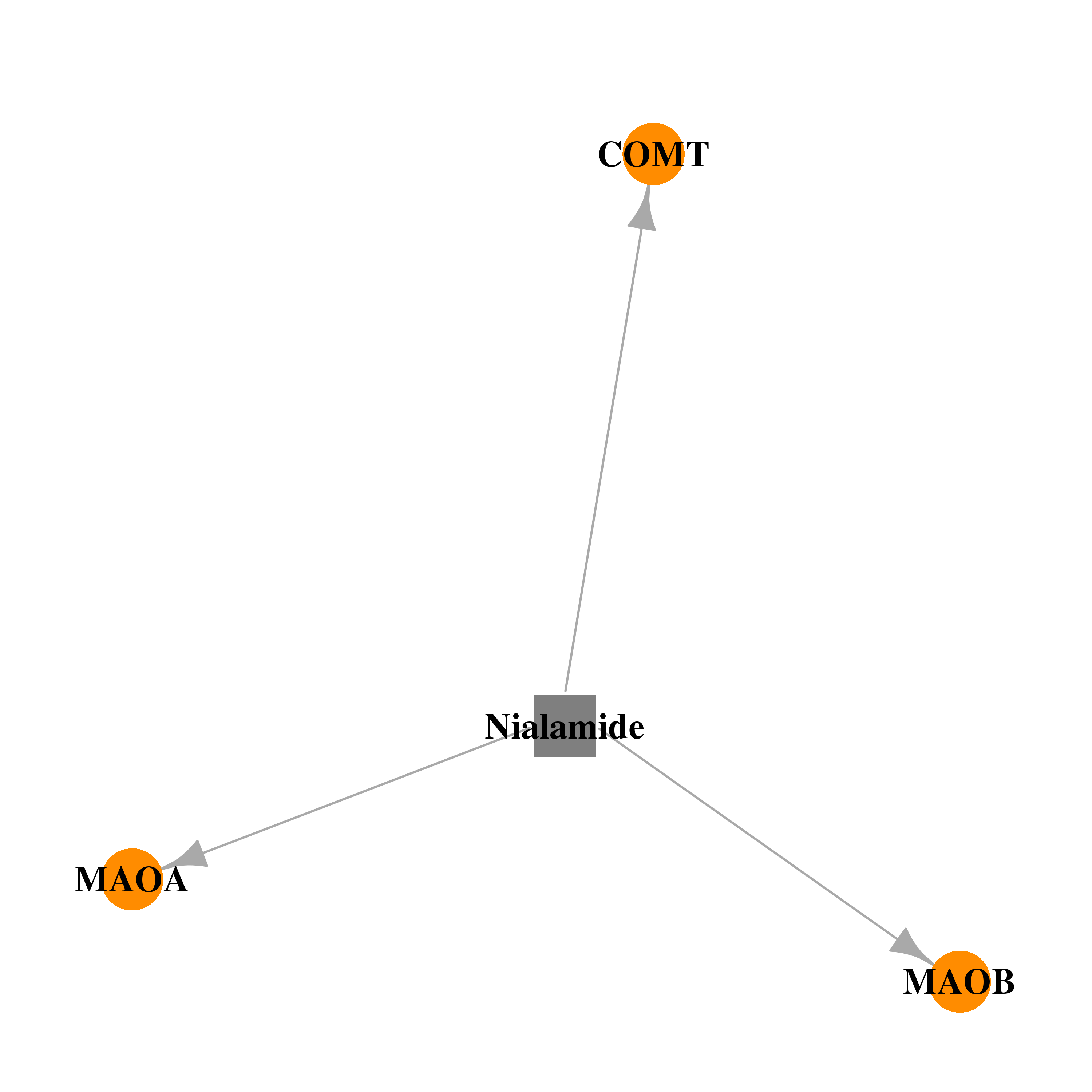 |  |
| DB07462 | catechol-O-methyltransferase | experimental | (3,4-DIHYDROXY-2-NITROPHENYL)(PHENYL)METHANONE | 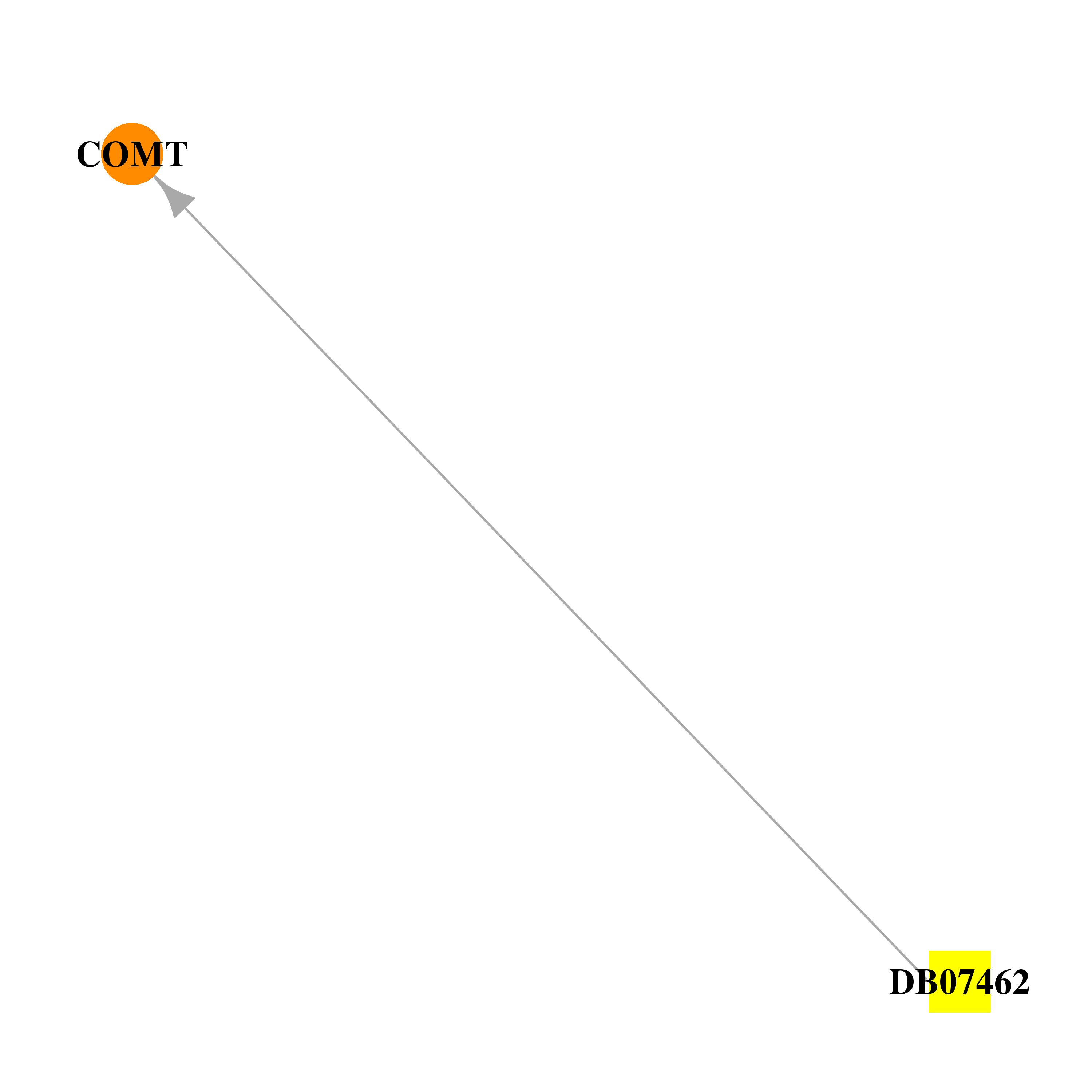 | 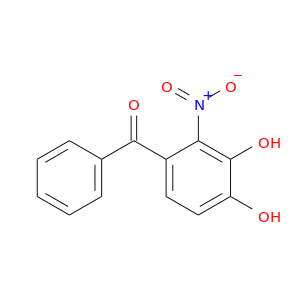 |
| DB08049 | catechol-O-methyltransferase | experimental | 7,8-dihydroxy-4-phenyl-2H-chromen-2-one |  | 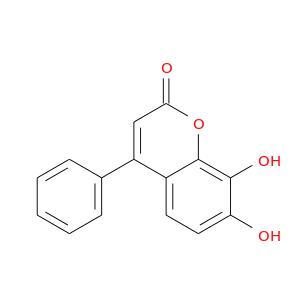 |
| DB00286 | catechol-O-methyltransferase | approved | Conjugated Estrogens | 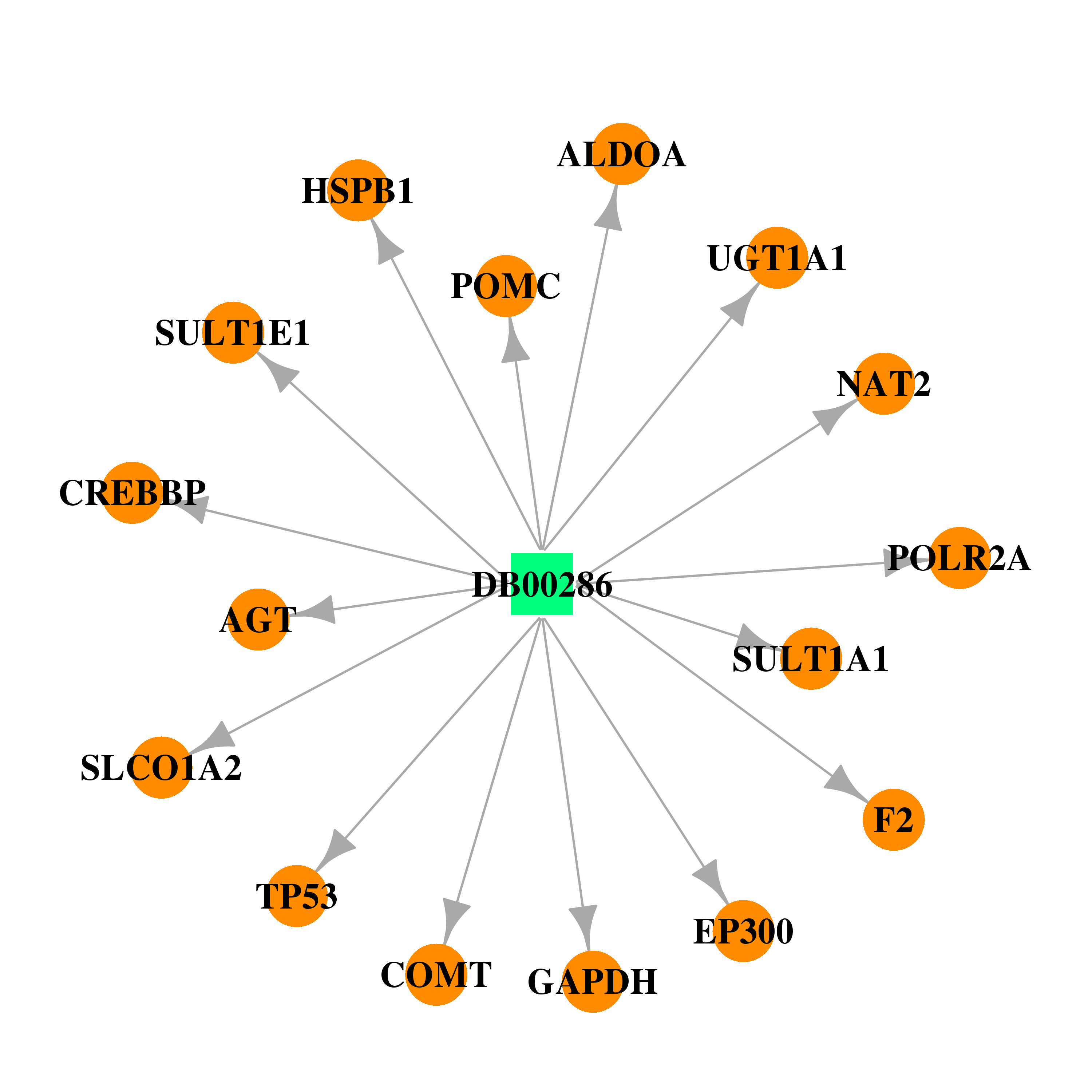 |  |
| DB00571 | catechol-O-methyltransferase | approved; investigational | Propranolol | 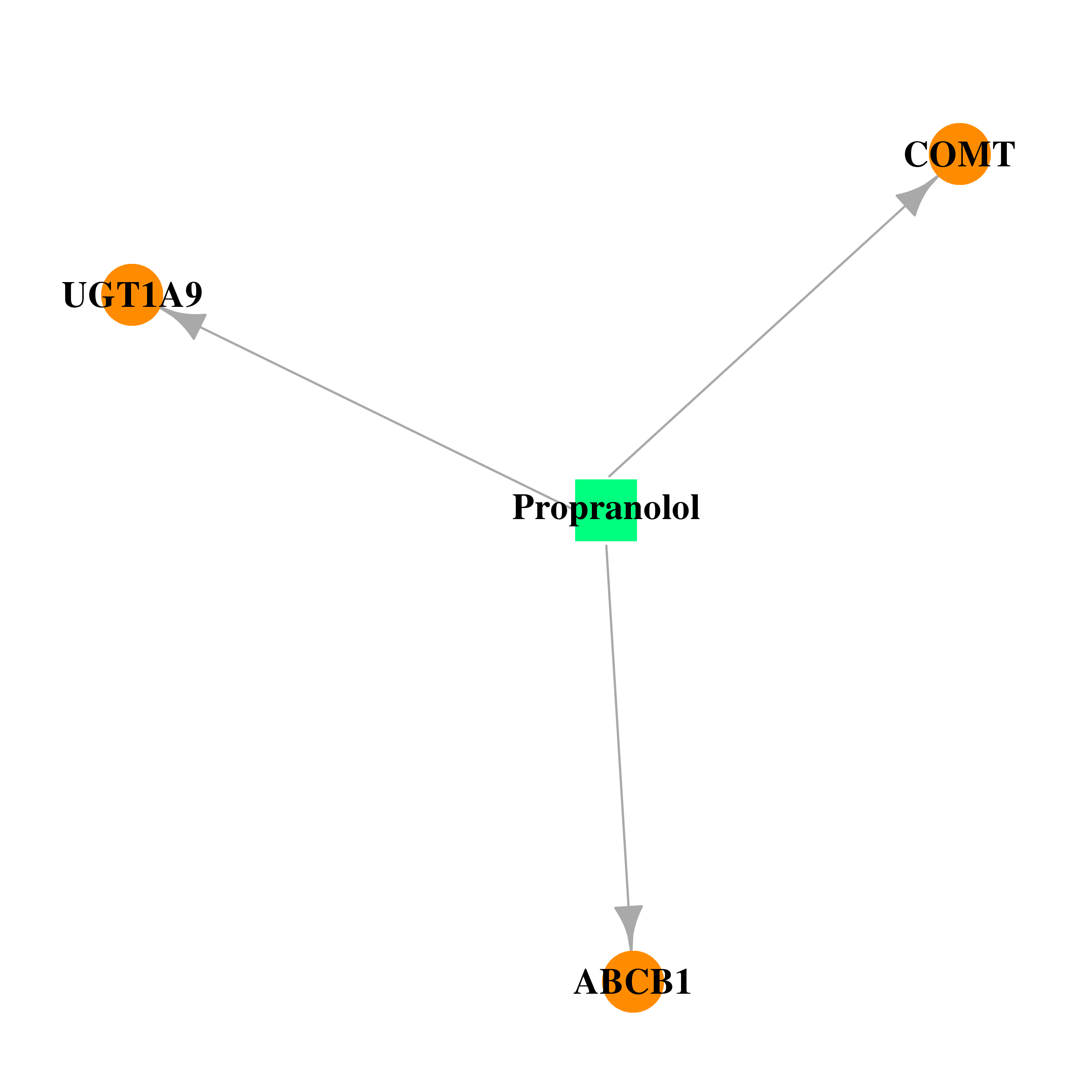 |  |
| DB00215 | catechol-O-methyltransferase | approved | Citalopram |  |  |
| DB00472 | catechol-O-methyltransferase | approved | Fluoxetine | 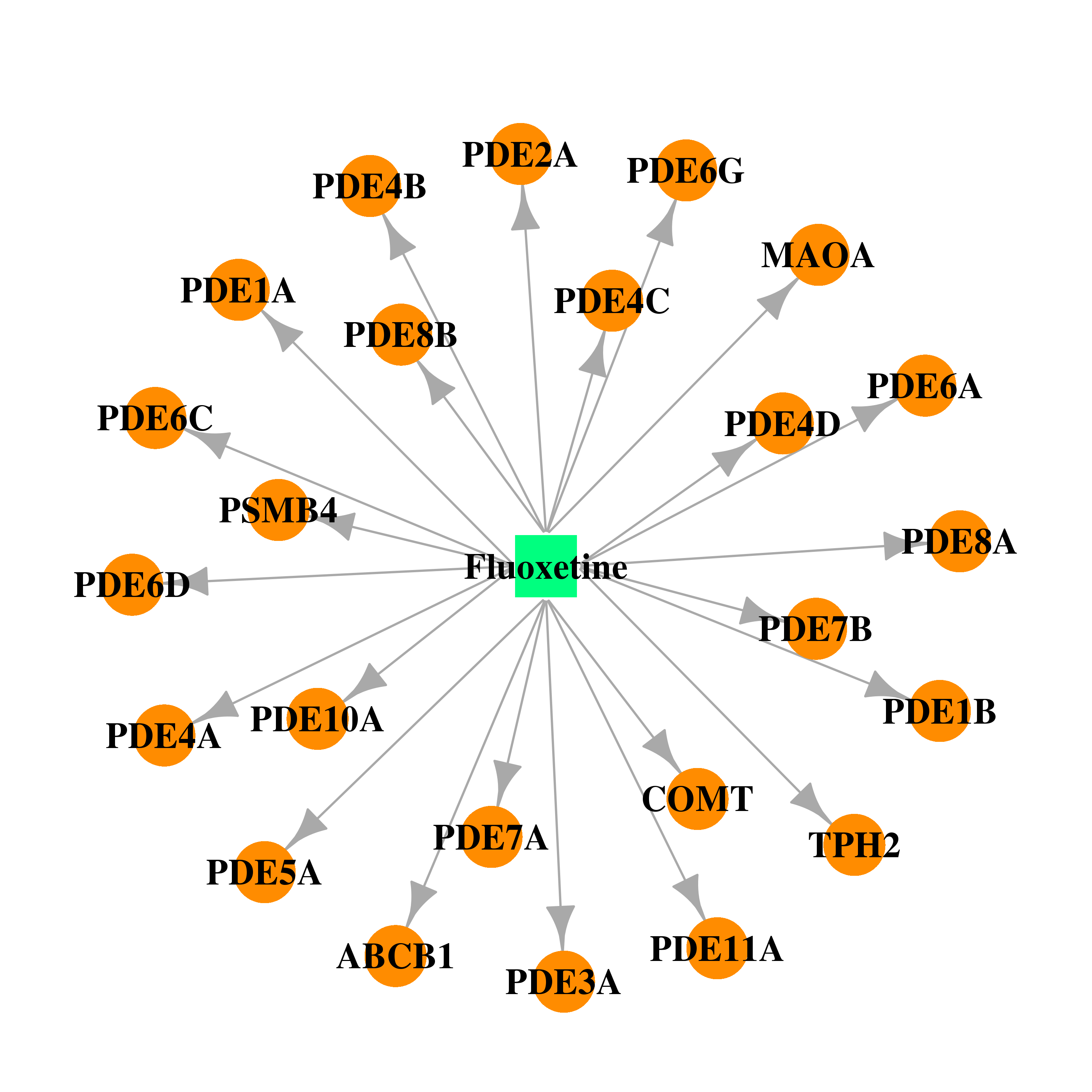 |  |
| DB00988 | catechol-O-methyltransferase | approved | Dopamine | 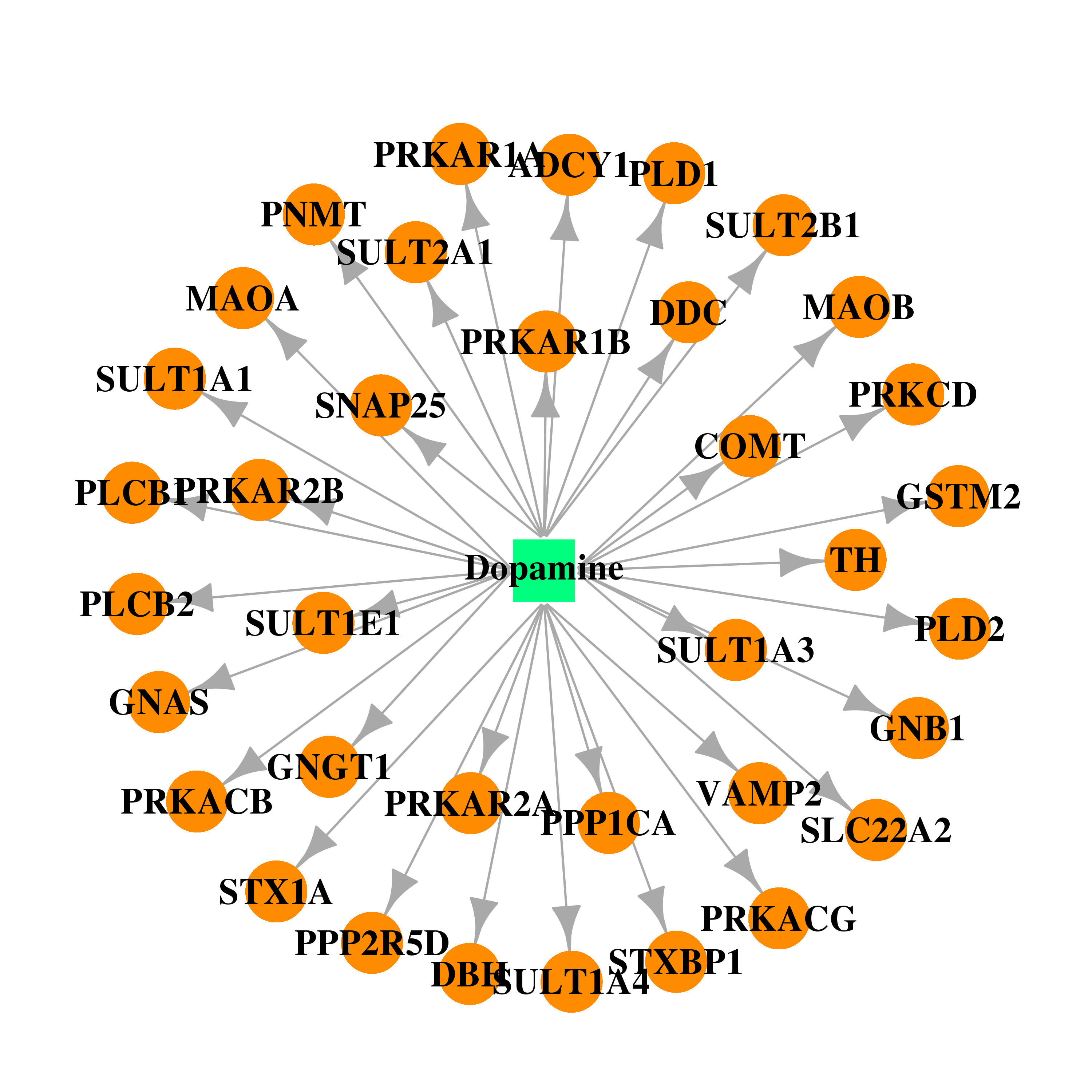 |  |
| DB00318 | catechol-O-methyltransferase | illicit; approved | Codeine | 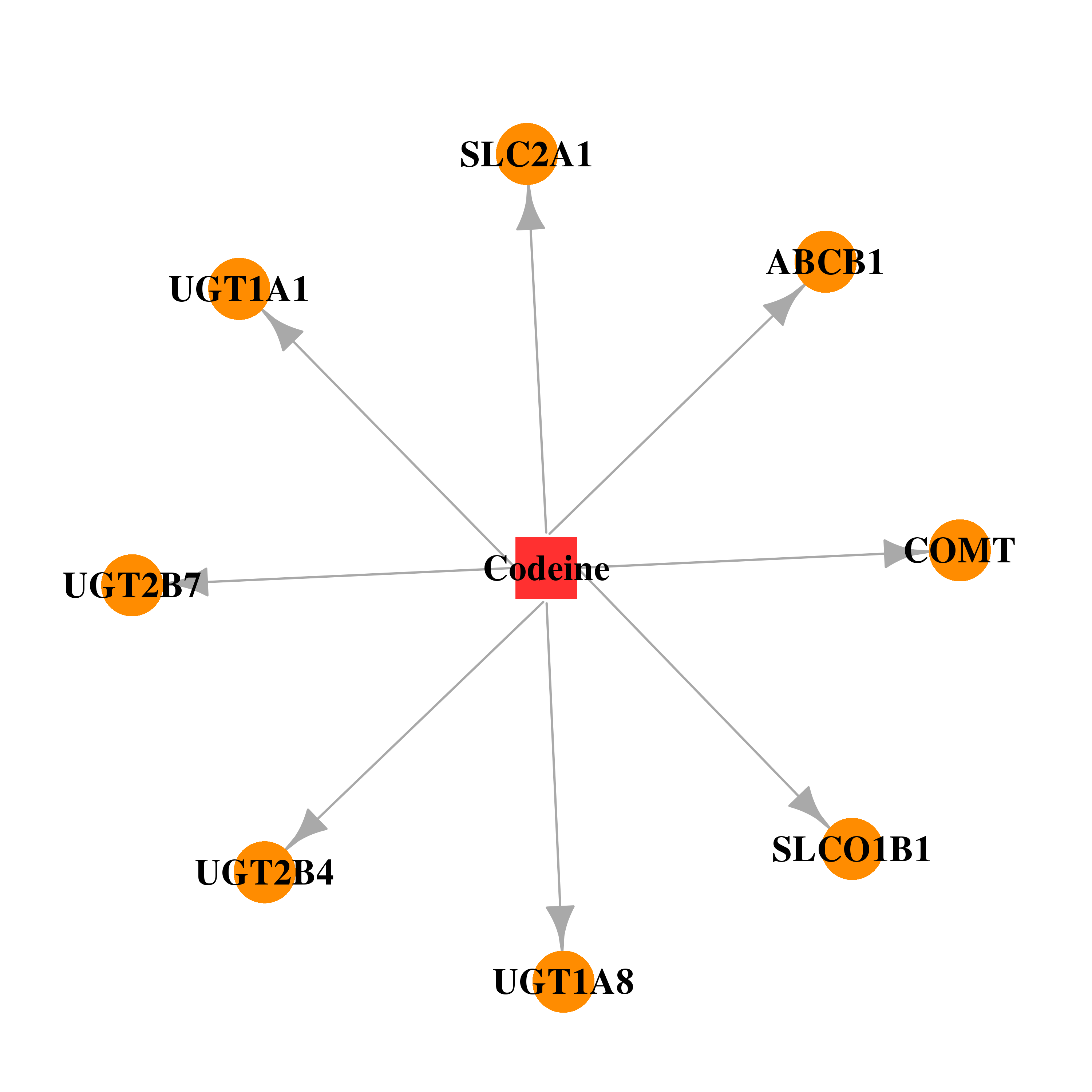 |  |
| DB00907 | catechol-O-methyltransferase | illicit; approved | Cocaine | 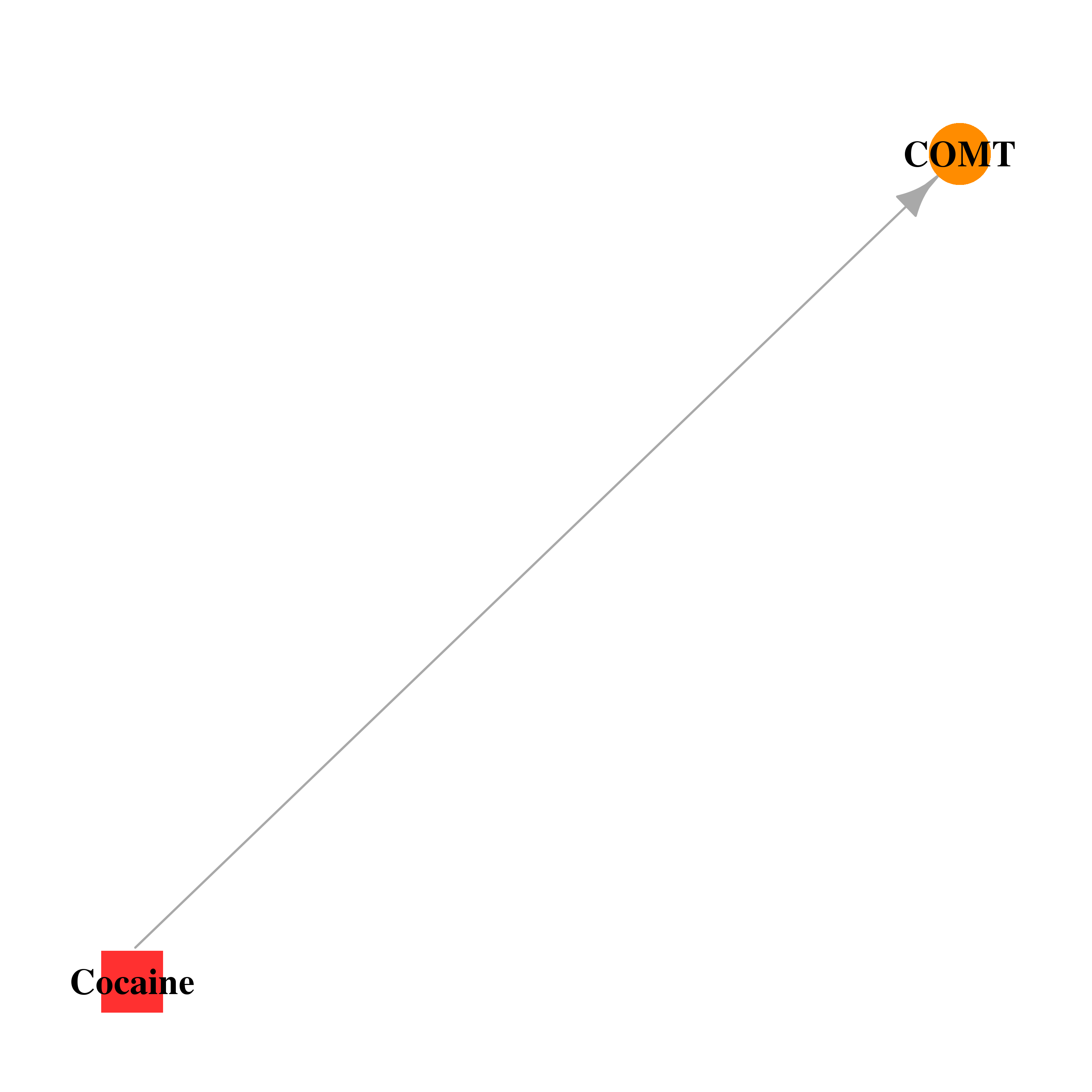 |  |
| DB00476 | catechol-O-methyltransferase | approved | Duloxetine |  |  |
| DB00864 | catechol-O-methyltransferase | approved; investigational | Tacrolimus |  |  |
| DB00640 | catechol-O-methyltransferase | approved; investigational | Adenosine | 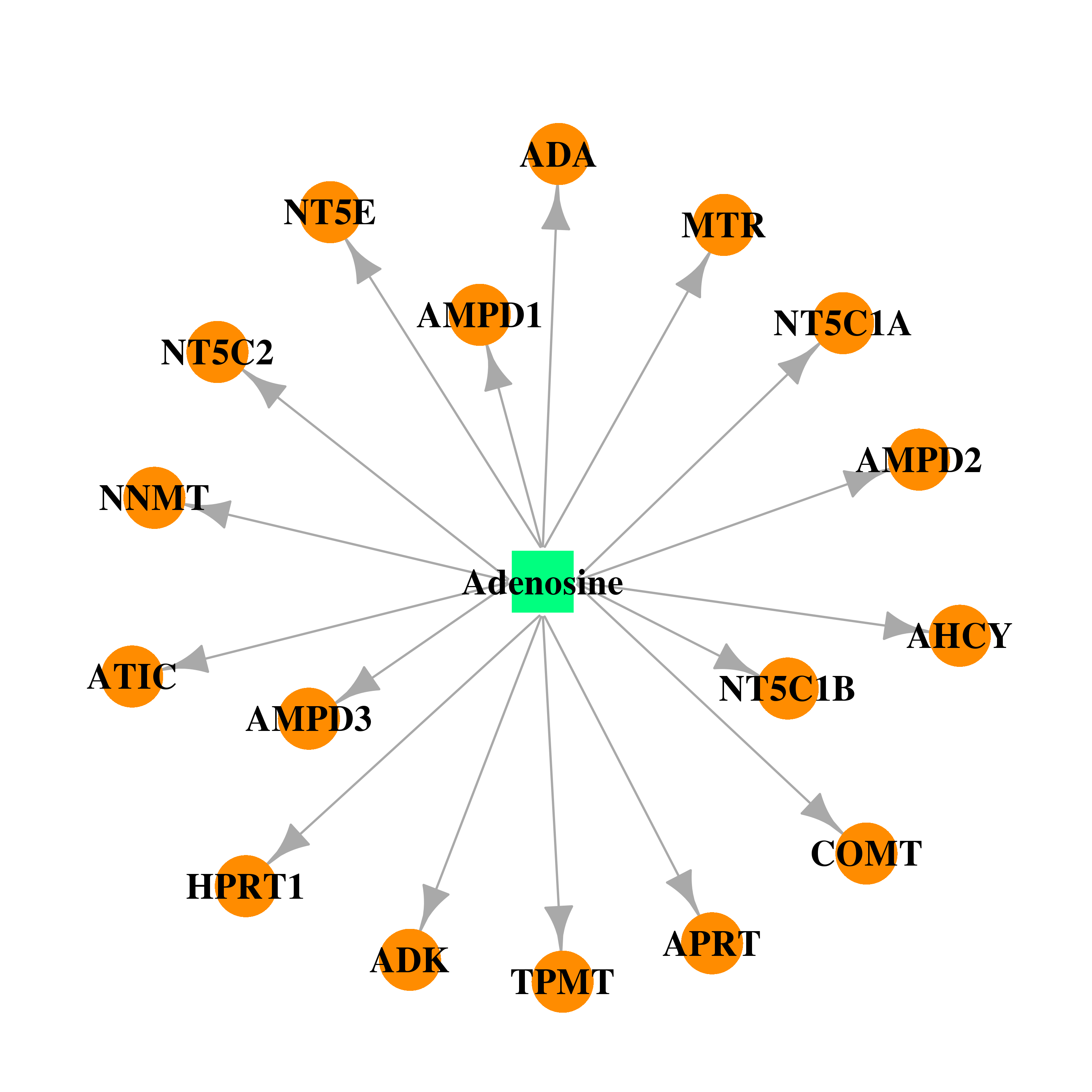 |  |
| DB00134 | catechol-O-methyltransferase | approved; nutraceutical | L-Methionine |  |  |
| DB01156 | catechol-O-methyltransferase | approved | Bupropion | 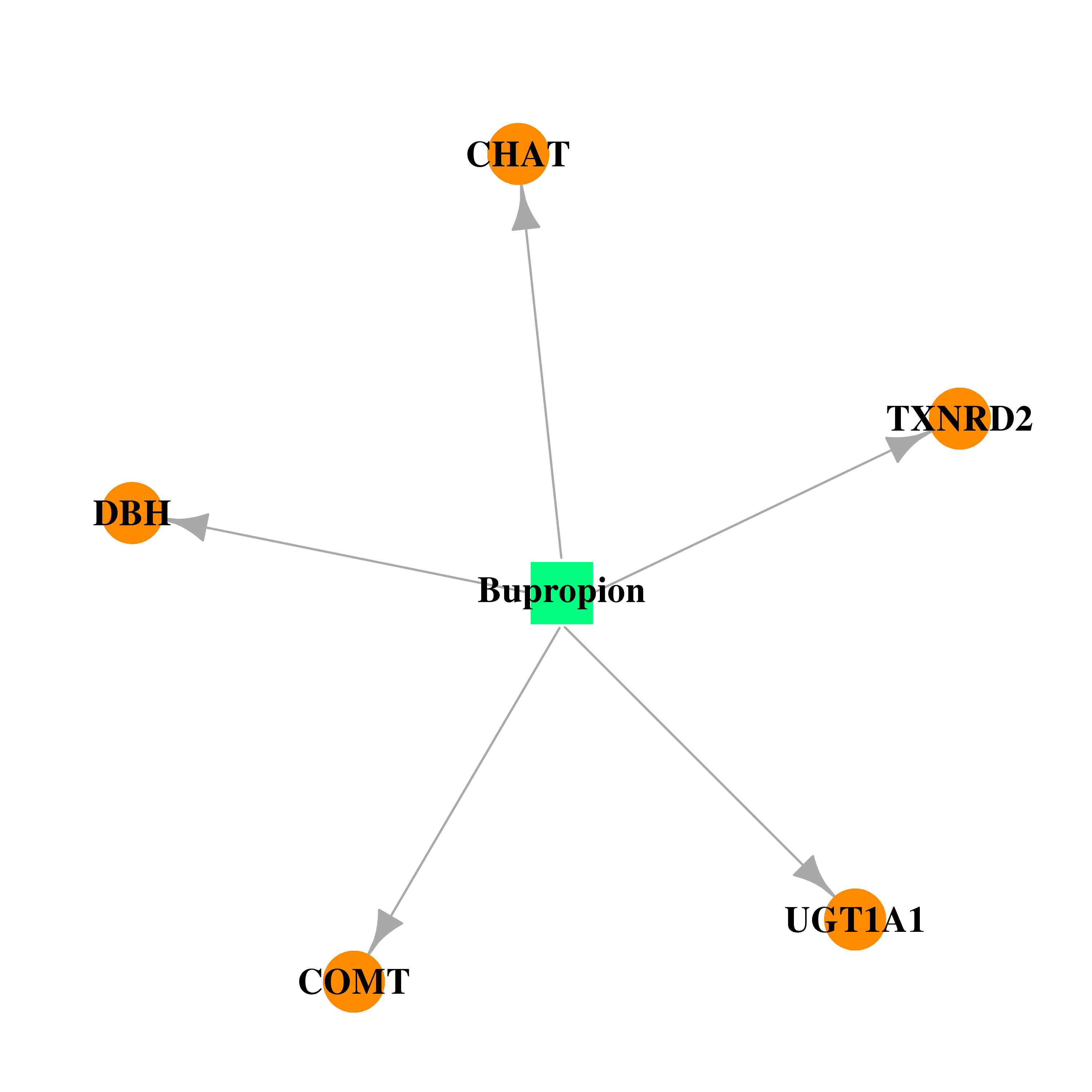 |  |
| DB00334 | catechol-O-methyltransferase | approved; investigational | Olanzapine | 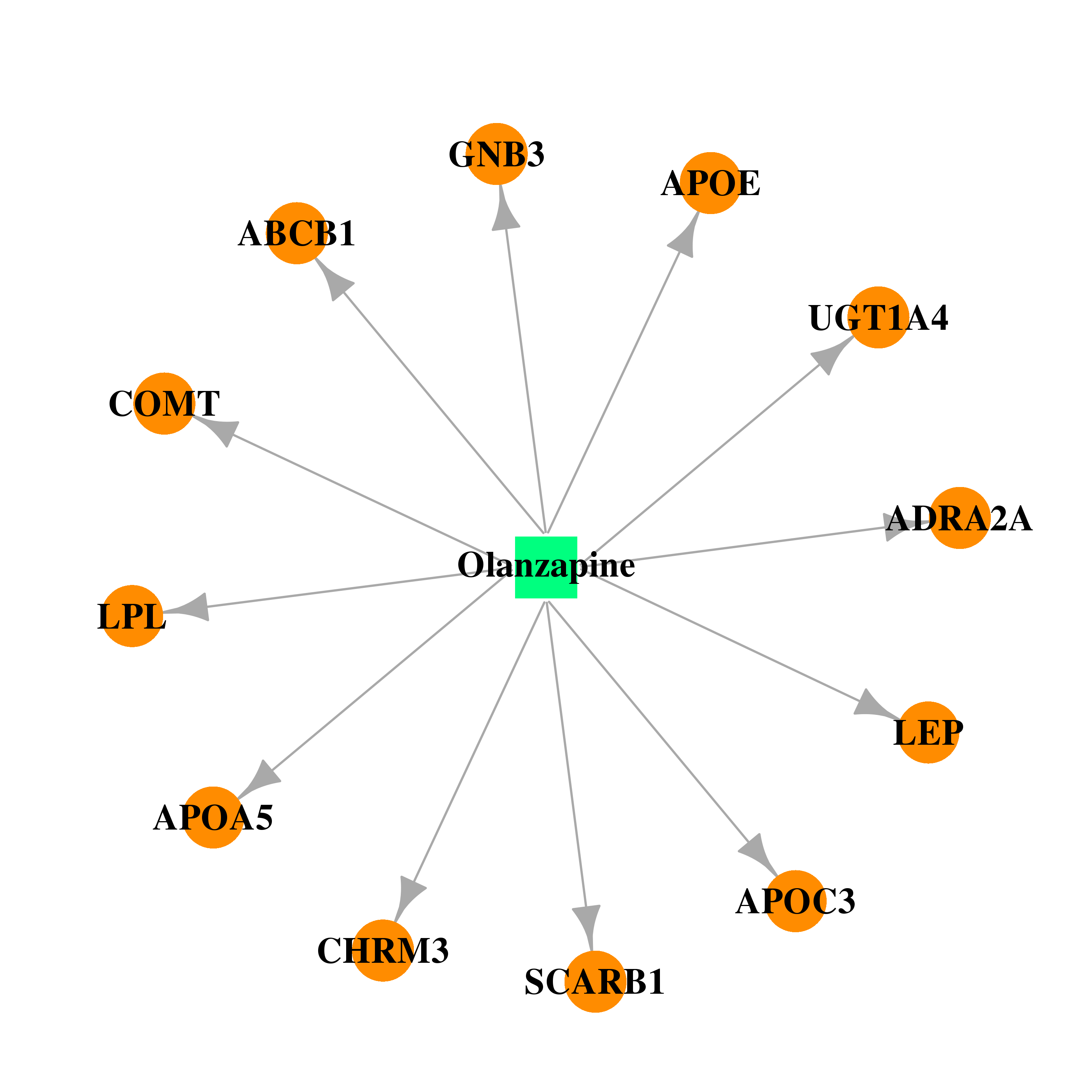 | 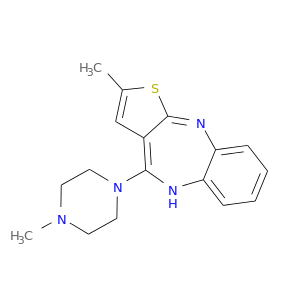 |
| DB01452 | catechol-O-methyltransferase | illicit; experimental | Heroin |  | 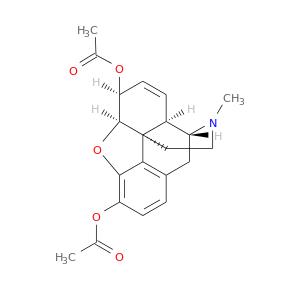 |
| DB00734 | catechol-O-methyltransferase | approved; investigational | Risperidone | 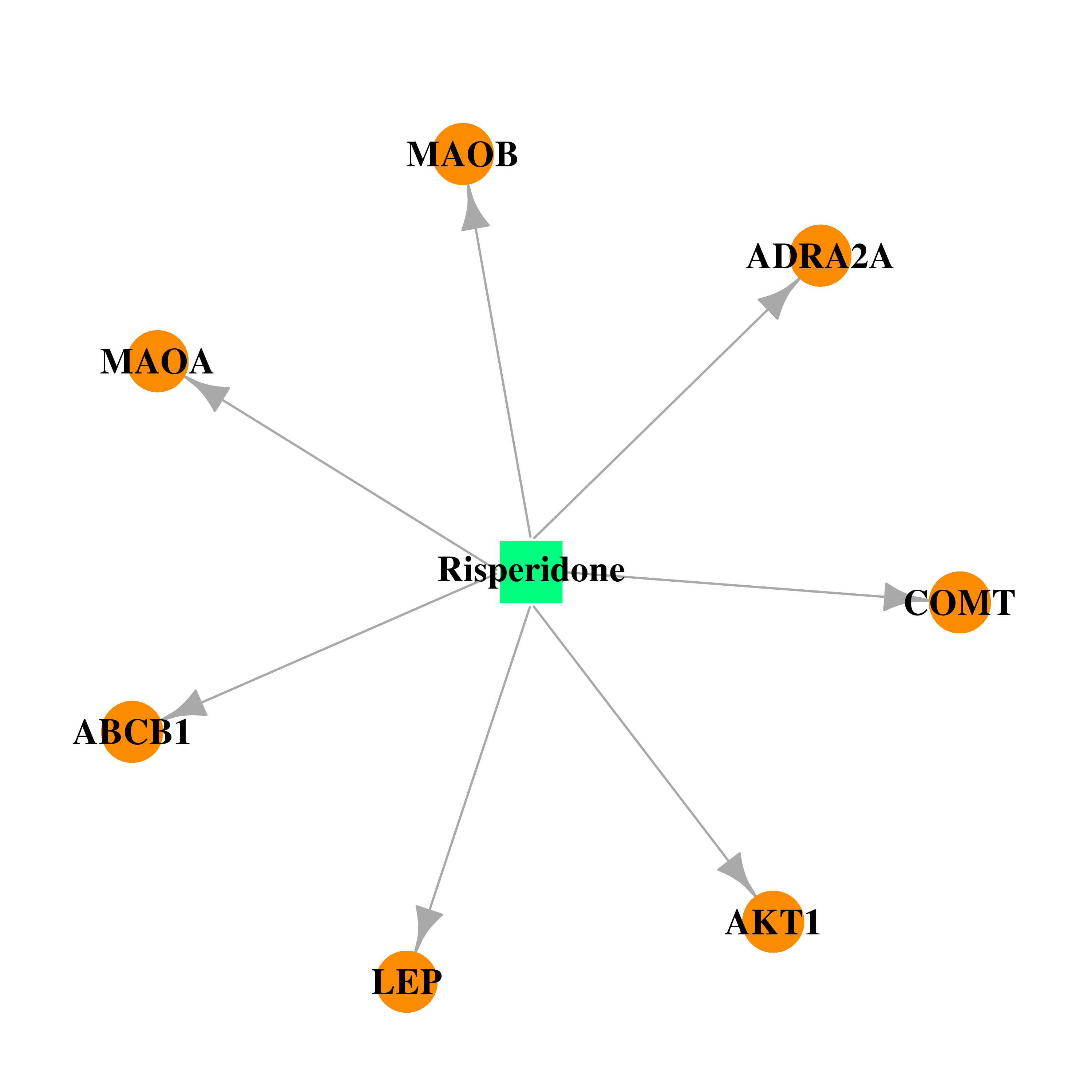 | 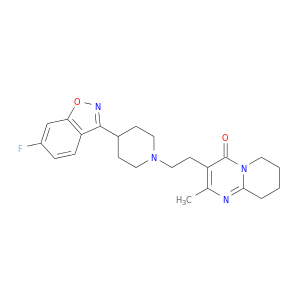 |
| DB00295 | catechol-O-methyltransferase | approved; investigational | Morphine | 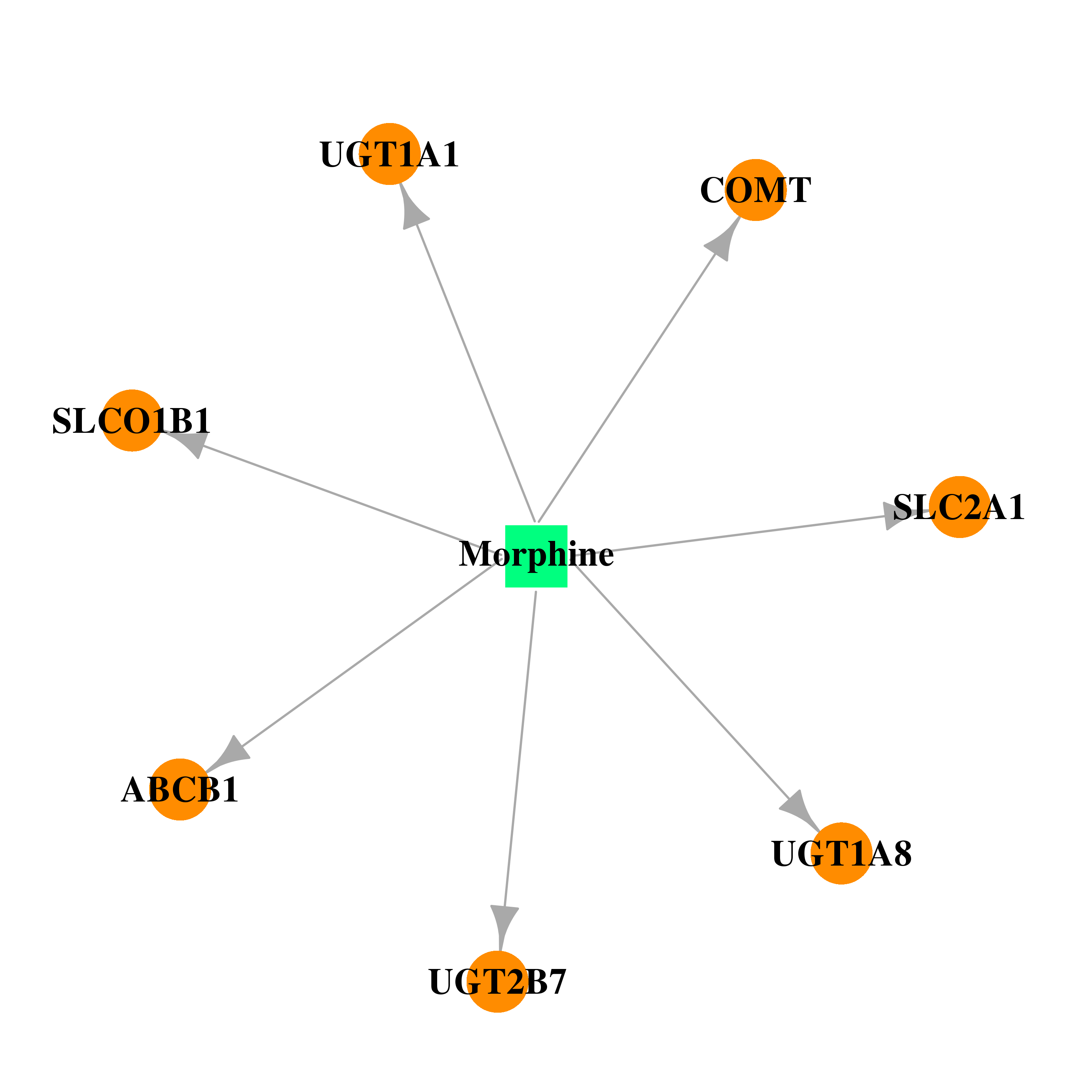 |  |
| DB00745 | catechol-O-methyltransferase | approved; investigational | Modafinil |  | 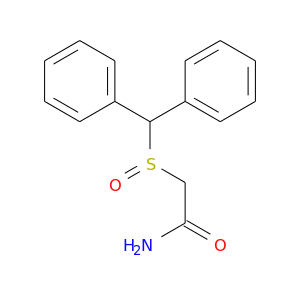 |
| DB00115 | catechol-O-methyltransferase | approved; nutraceutical | Cyanocobalamin |  | 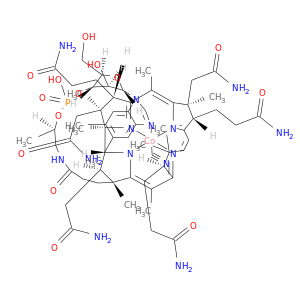 |
| DB00158 | catechol-O-methyltransferase | approved; nutraceutical | Folic Acid | 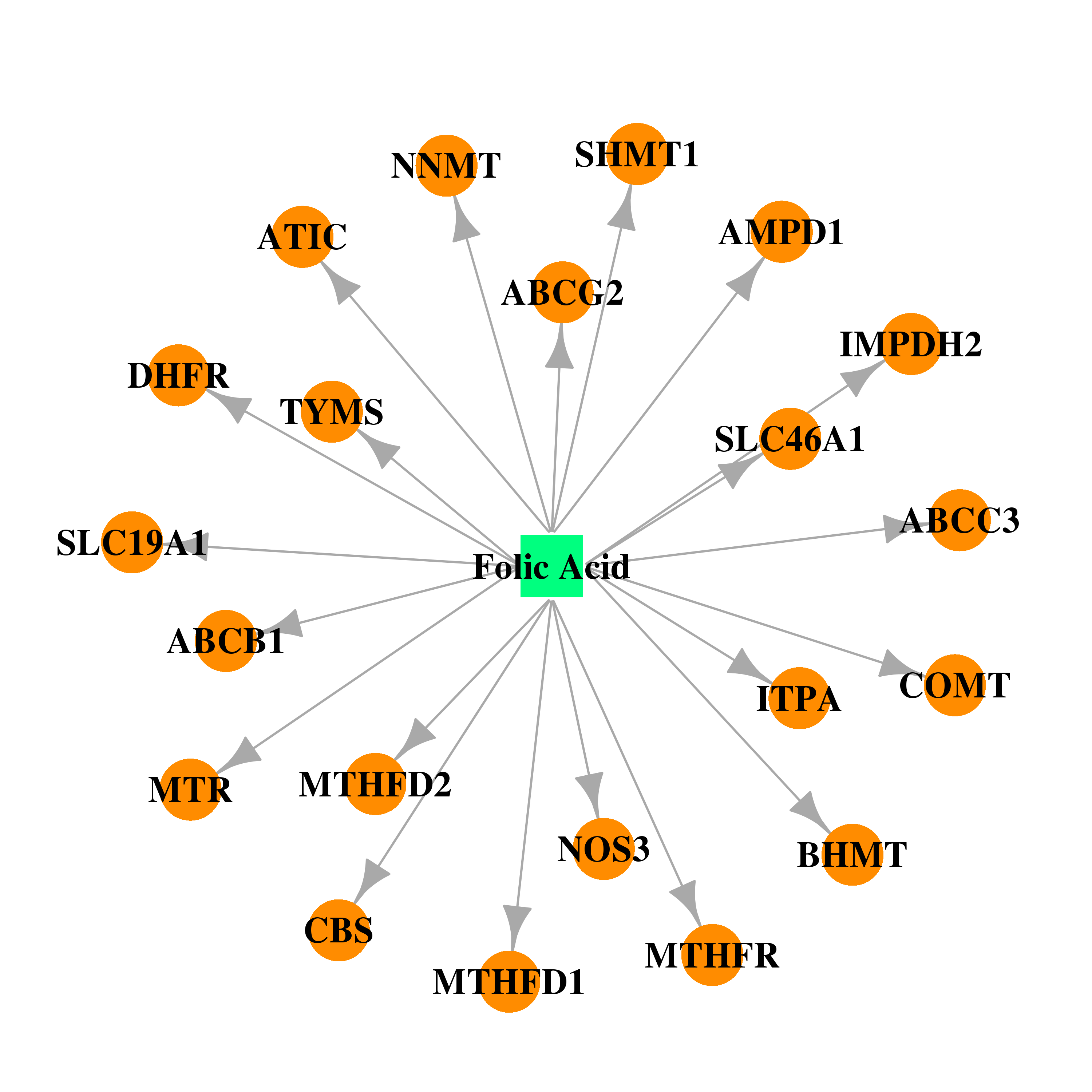 |  |
| DB01454 | catechol-O-methyltransferase | illicit; experimental | 3,4-Methylenedioxymethamphetamine | 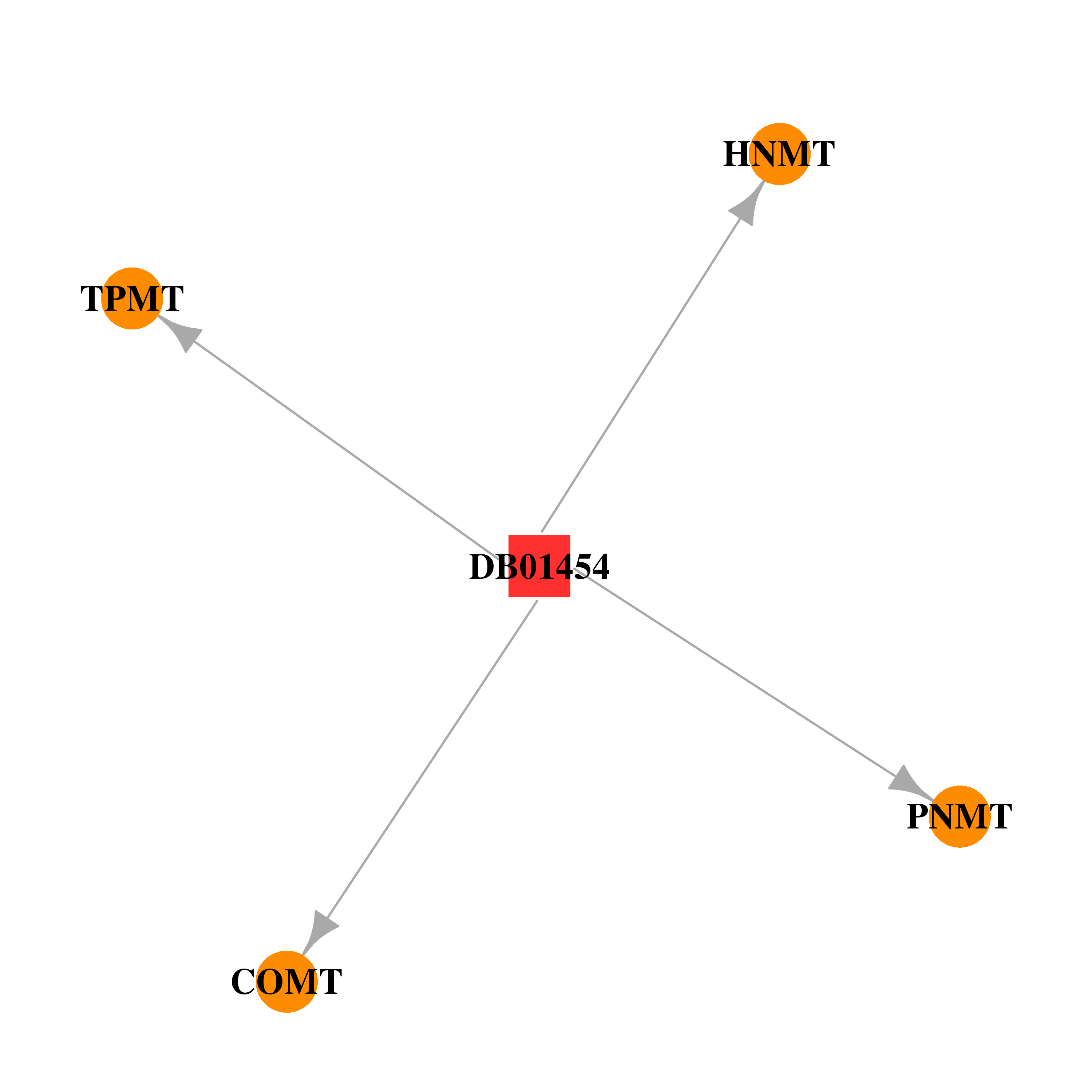 |  |
| DB01235 | catechol-O-methyltransferase | approved | Levodopa | 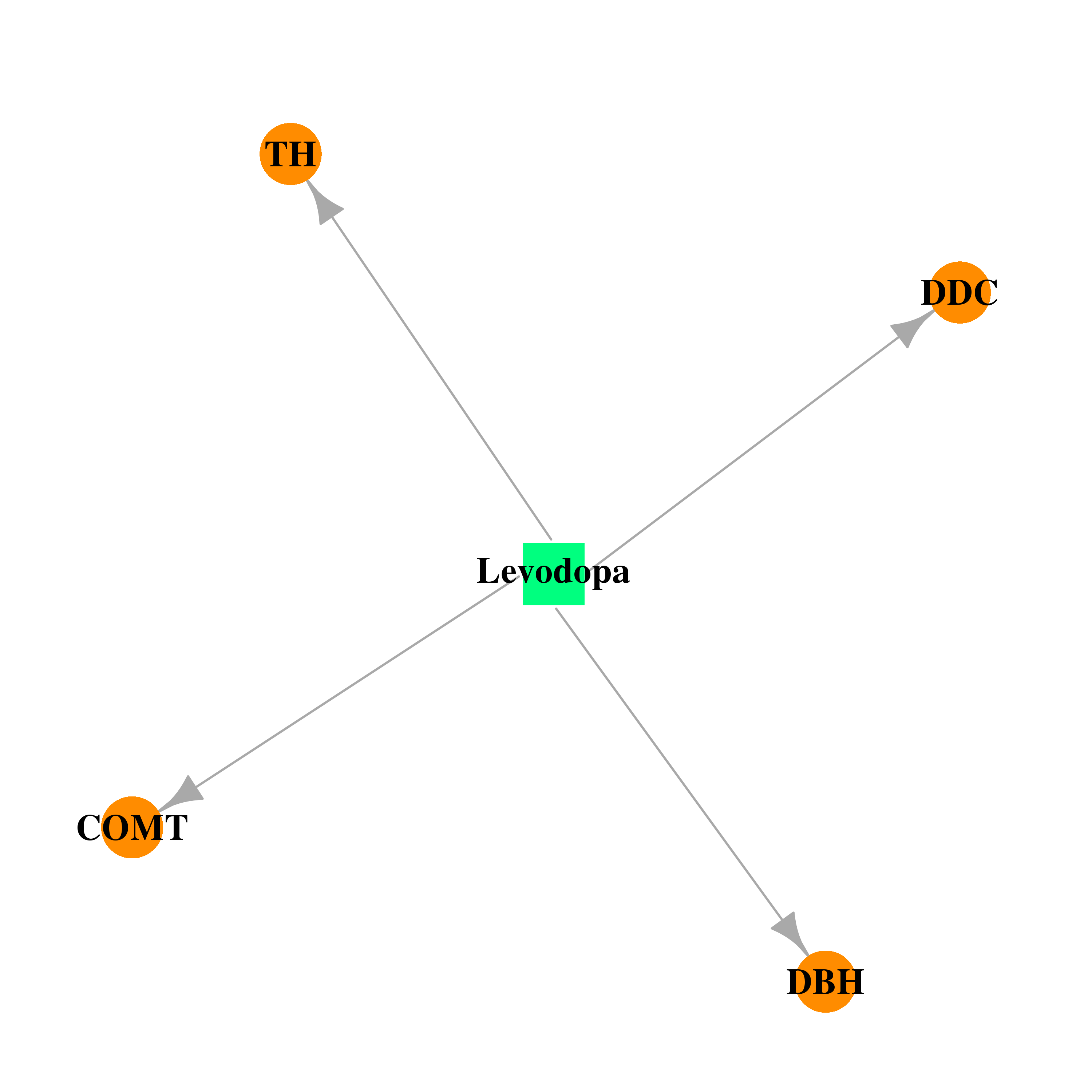 |  |
| DB00668 | catechol-O-methyltransferase | approved | Epinephrine | 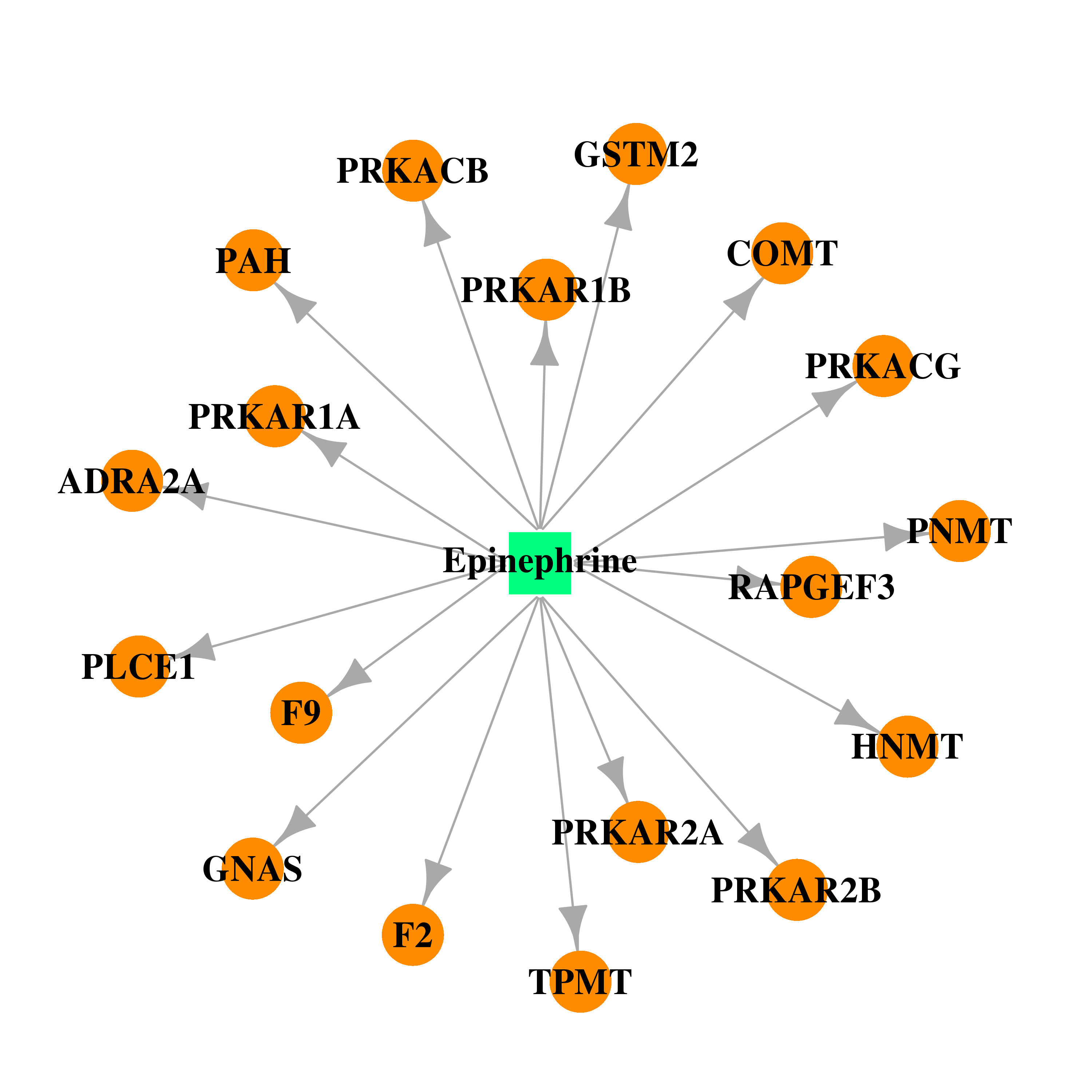 |  |
| DB01033 | catechol-O-methyltransferase | approved | Mercaptopurine | 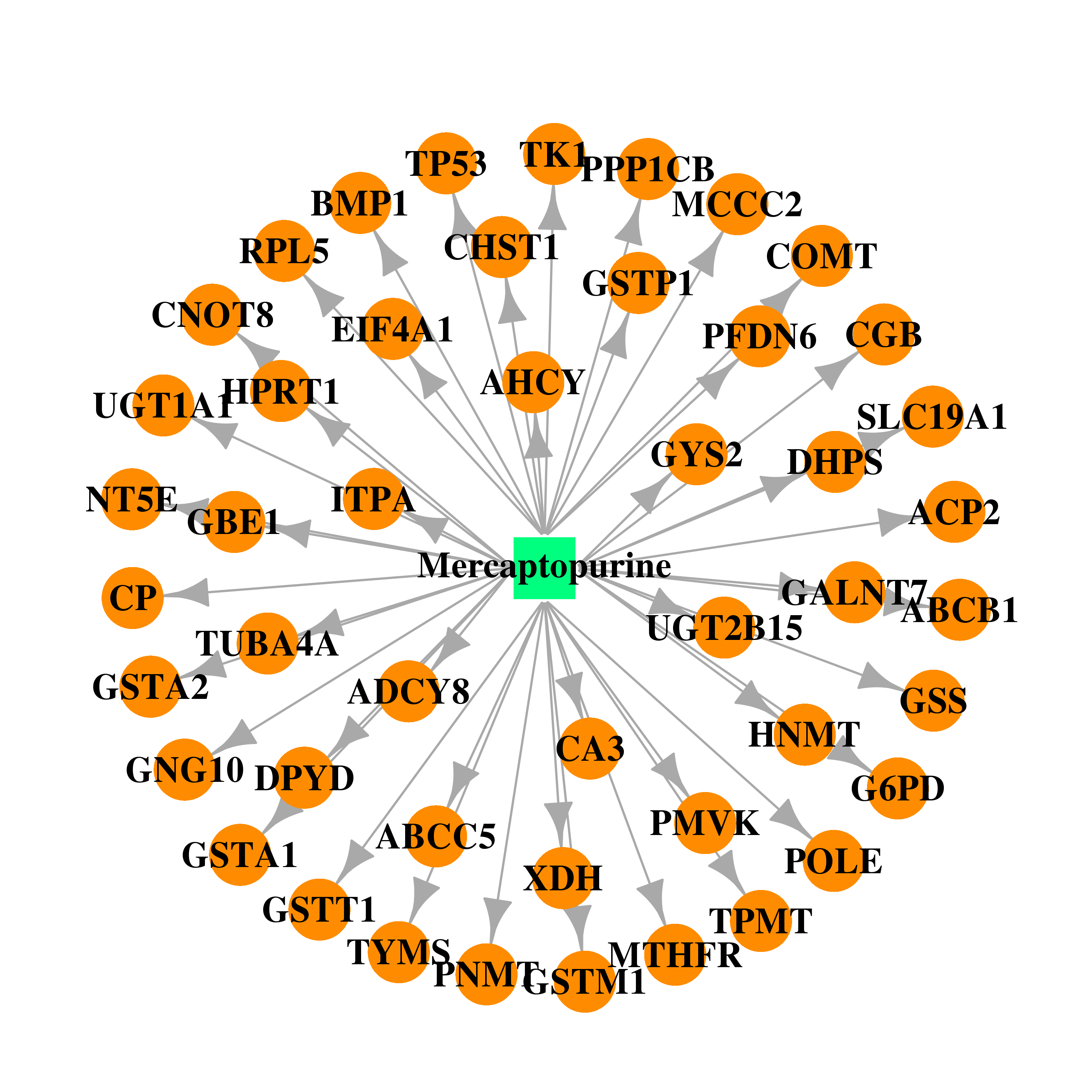 | 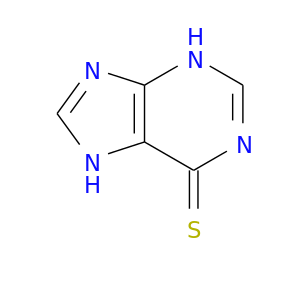 |
| DB00945 | catechol-O-methyltransferase | approved | Acetylsalicylic acid | 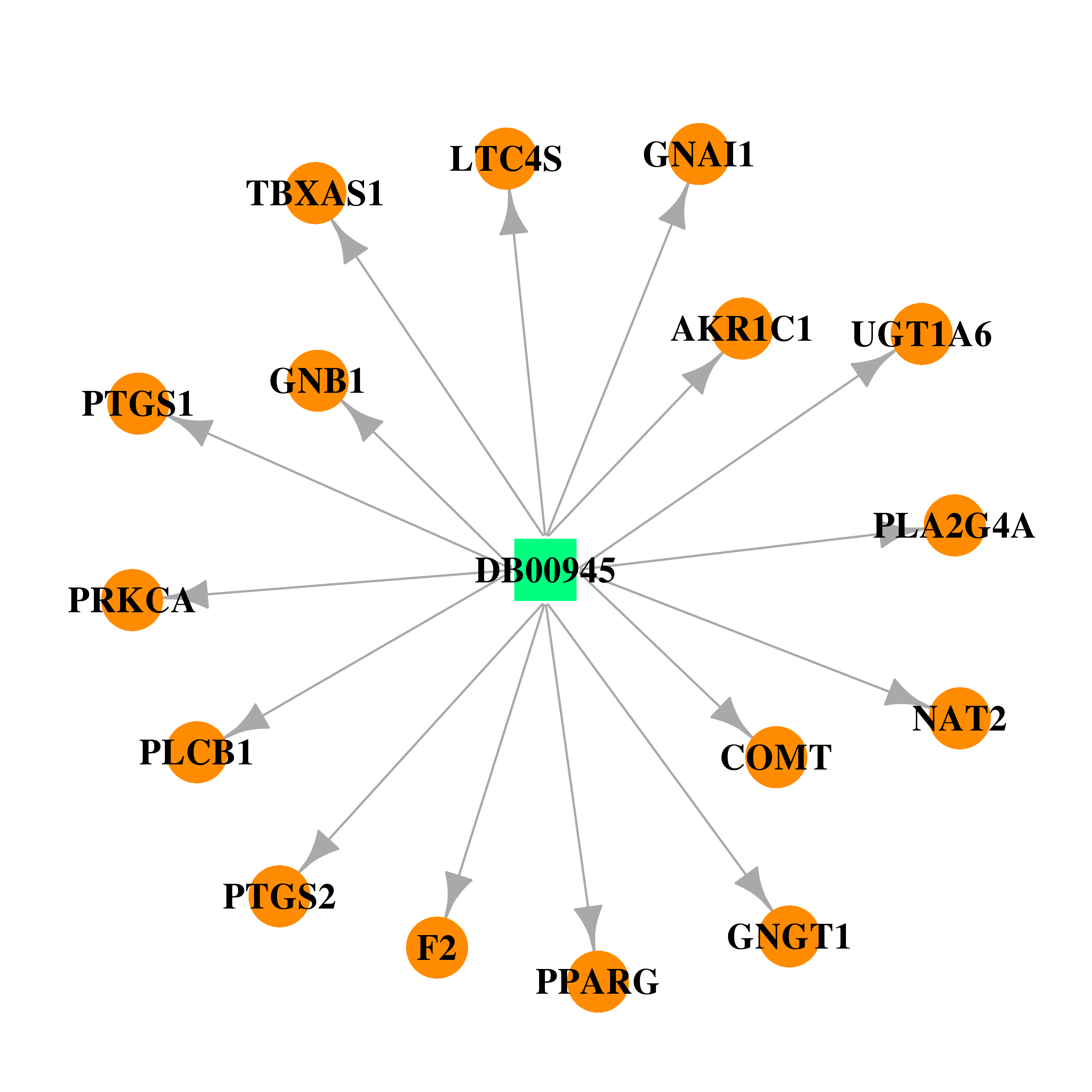 | 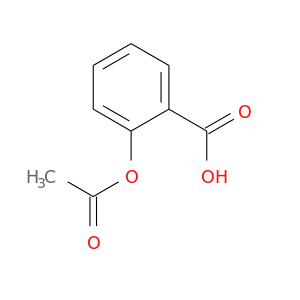 |
| DB00968 | catechol-O-methyltransferase | approved | Methyldopa | 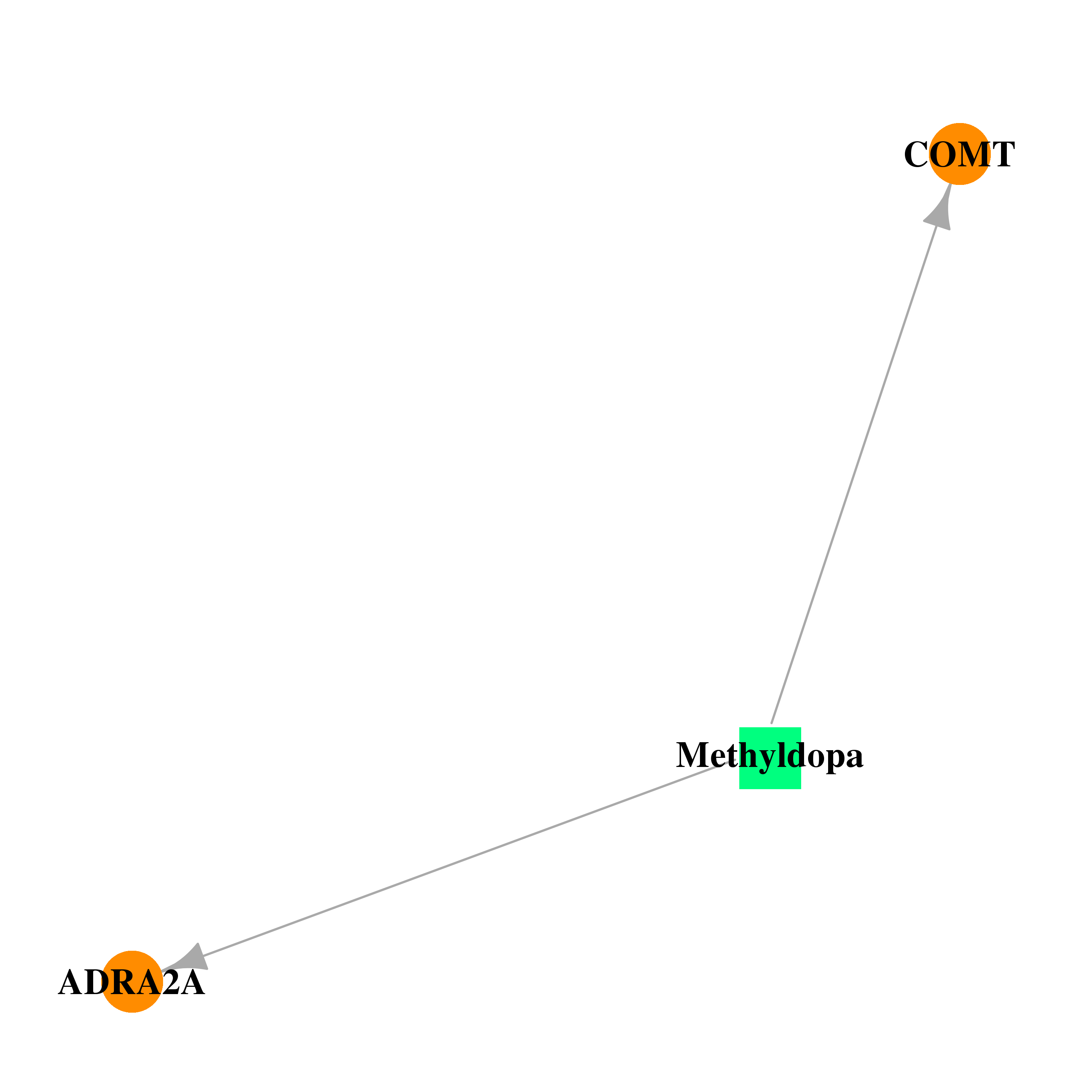 | 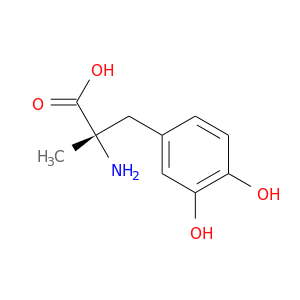 |
| DB00184 | catechol-O-methyltransferase | approved | Nicotine |  | 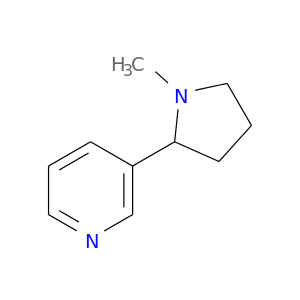 |
| DB00515 | catechol-O-methyltransferase | approved | Cisplatin |  | 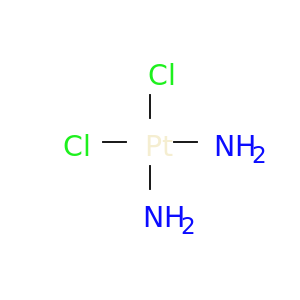 |
| DB00422 | catechol-O-methyltransferase | approved; investigational | Methylphenidate |  |  |
| Top |
| Cross referenced IDs for COMT |
| * We obtained these cross-references from Uniprot database. It covers 150 different DBs, 18 categories. http://www.uniprot.org/help/cross_references_section |
: Open all cross reference information
|
Copyright © 2016-Present - The Univsersity of Texas Health Science Center at Houston @ |