|
Cancer Cell Metabolism Gene DB |
![]() ccmGDB includes..
ccmGDB includes..
![]() cell metabolism pathway genes
cell metabolism pathway genes
- 922 KEGG cell metabolic pathway genes
- 1,597 REACTOME cell metabolic pathway genes
![]() 7 Cancer Gene Source
7 Cancer Gene Source
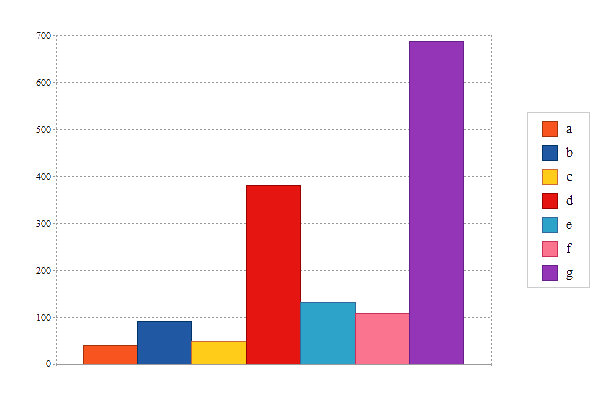
cf) a; Oncogenes, b; TSGene, c; Cancer Gene Census,d; Cancer Genes, e; Network of Cancer Gene,
f; Siginificantly mutated driver genes, g; Therapeutic Vulnerabilities in Cancer
![]() Somatic alterations and genetic features for 2,071 cell metabolism genes with efficent visualization
Somatic alterations and genetic features for 2,071 cell metabolism genes with efficent visualization
- 151238 Somatic nucleotide variants (SNVs) data for 41 primary sites (62,599 samples) from COSMIC,
102399 SNVs for 20 cancer types (5,186 samples) from TCGA
- 6269 Copy number variation (CNV; 4504 copy number
gain, 1784 copy number loss) data
for 17 primary sites (3,062 samples) from COSMIC
- 1971 Structural variation (SV) data for 13 primary sites (412 samples) from COSMIC
- Chitars2.0 - 4,729 chimeric transcripts
![]() Systematic assessment of transcriptome, protein expression and co-expressed gene network
Systematic assessment of transcriptome, protein expression and co-expressed gene network
over 2,000 cell metabolism genes
- Differential gene expression (DEG) plots for 8 TCGA cancer types
- Gene expression in 24 CCLE cancer types
- Correlation between gene expression and CNV for 15 TCGA cancer types
- Co-expressed protein interaction network for 8 cancer TCGA types
![]() Drug-target interaction networks for cell metabolism genes
Drug-target interaction networks for cell metabolism genes
- 4,059 Drugs
- Drug-gene interaction networks
![]() ccmGDB's finding..
ccmGDB's finding..
# DEGs regardless of directionl
|
|
BRCA |
COAD |
HNSC |
KIRC |
LUAD |
LUSC |
THCA |
UCEC |
|
ccmGenes |
||||||||
|
cmGenes |
|
ccmGenes |
BRCA |
COAD |
HNSC |
KIRC |
LUAD |
LUSC |
THCA |
UCEC |
|
Up |
180 |
147 |
133 |
|||||
|
Down |
207 |
141 |
113 |
|
cmGenes |
BRCA |
COAD |
HNSC |
KIRC |
LUAD |
LUSC |
THCA |
UCEC |
|
Up |
||||||||
|
Down |


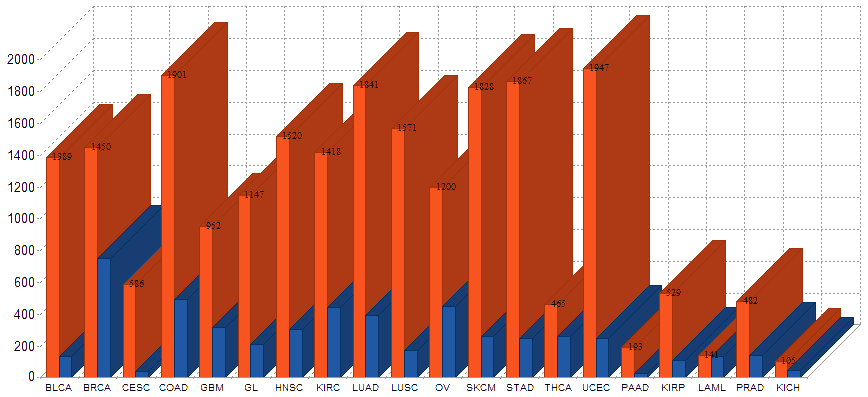
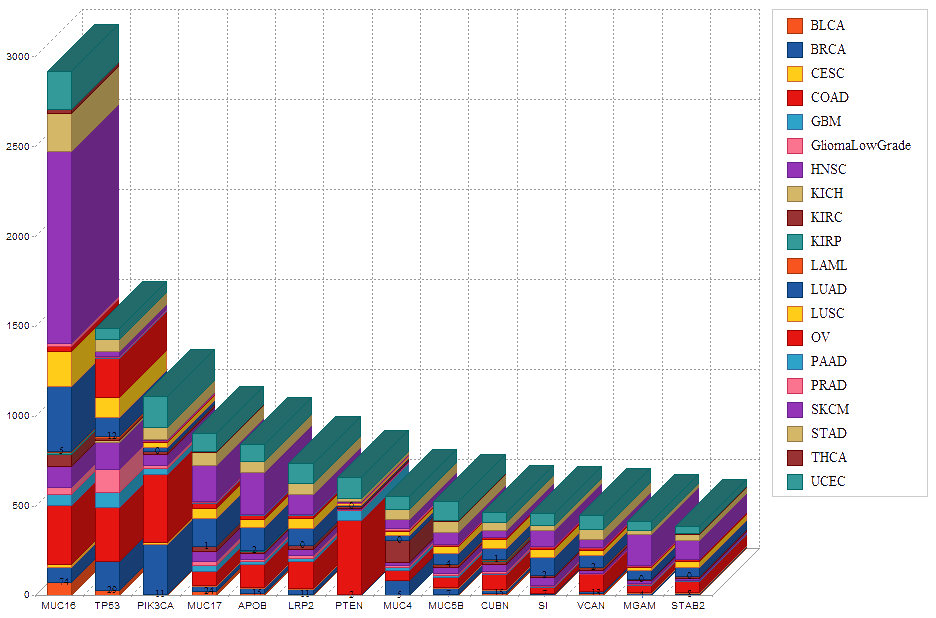
cf) Tissue Abbreviation[Tissue Name]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma],
KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ],
PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma]
![]() References for Databases
References for Databases
1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long
2 Tumor Suppressor gene; http://bioinfo.uth.edu/TSGene/
3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html
4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long
5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php
6 BLCA http://www.nature.com/nature/journal/vaop/ncurrent/full/nature12965.html
BRCA http://www.nature.com/nature/journal/v490/n7418/full/nature11412.html
CLL http://www.nature.com/nature/journal/v505/n7484/full/nature12912.html
COAD http://www.nature.com/nature/journal/v487/n7407/full/nature11252.html
CRC http://www.nature.com/nature/journal/v505/n7484/full/nature12912.html
DLBCL http://www.nature.com/nature/journal/v505/n7484/full/nature12912.html
ESO http://www.nature.com/nature/journal/v505/n7484/full/nature12912.html
GBM http://www.sciencedirect.com/science/article/pii/S0092867413012087
HNSC https://www.sciencemag.org/content/333/6046/1157
KIRC http://www.nature.com/nature/journal/v499/n7456/full/nature12222.html
LAML http://www.nejm.org/doi/full/10.1056/NEJMoa1301689
LUAD http://www.sciencedirect.com/science/article/pii/S0092867412010227
LUSC http://www.nature.com/nature/journal/v489/n7417/full/nature11404.html
MEL http://www.nature.com/nature/journal/v505/n7484/full/nature12912.html
MM http://www.nature.com/nature/journal/v505/n7484/full/nature12912.html
OV http://www.nature.com/nature/journal/v474/n7353/full/nature10166.html
PRAD http://www.nature.com/nature/journal/v505/n7484/full/nature12912.html
UCEC http://www.nature.com/nature/journal/v497/n7447/full/nature12113.html
7 Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/
Catalogue of Cancer Genes; http://www.ncbi.nlm.nih.gov/pubmed/24881052
