|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |
| Phenotypic Information (metabolism pathway, cancer, disease, phenome) |
| |
| |
| Gene-Gene Network Information: Co-Expression Network, Interacting Genes & KEGG |
| |
|
| Gene Summary for MAPK14 |
| Basic gene info. | Gene symbol | MAPK14 |
| Gene name | mitogen-activated protein kinase 14 | |
| Synonyms | CSBP|CSBP1|CSBP2|CSPB1|EXIP|Mxi2|PRKM14|PRKM15|RK|SAPK2A|p38|p38ALPHA | |
| Cytomap | UCSC genome browser: 6p21.3-p21.2 | |
| Genomic location | chr6 :35995453-36079013 | |
| Type of gene | protein-coding | |
| RefGenes | NM_001315.2, NM_139012.2,NM_139013.2,NM_139014.2, | |
| Ensembl id | ENSG00000112062 | |
| Description | CSAID-binding proteinCsaids binding proteinMAP kinase 14MAP kinase Mxi2MAP kinase p38 alphaMAX-interacting protein 2cytokine suppressive anti-inflammatory drug binding proteincytokine suppressive anti-inflammatory drug-binding proteinmitogen-activ | |
| Modification date | 20141222 | |
| dbXrefs | MIM : 600289 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000112062 | ||
| HPRD : 02619 | ||
| Vega : OTTHUMG00000159806 | ||
| Protein | UniProt: Q16539 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_MAPK14 | |
| BioGPS: 1432 | ||
| Gene Expression Atlas: ENSG00000112062 | ||
| The Human Protein Atlas: ENSG00000112062 | ||
| Pathway | NCI Pathway Interaction Database: MAPK14 | |
| KEGG: MAPK14 | ||
| REACTOME: MAPK14 | ||
| ConsensusPathDB | ||
| Pathway Commons: MAPK14 | ||
| Metabolism | MetaCyc: MAPK14 | |
| HUMANCyc: MAPK14 | ||
| Regulation | Ensembl's Regulation: ENSG00000112062 | |
| miRBase: chr6 :35,995,453-36,079,013 | ||
| TargetScan: NM_001315 | ||
| cisRED: ENSG00000112062 | ||
| Context | iHOP: MAPK14 | |
| cancer metabolism search in PubMed: MAPK14 | ||
| UCL Cancer Institute: MAPK14 | ||
| Assigned class in ccmGDB | A - This gene has a literature evidence and it belongs to cancer gene. | |
| References showing role of MAPK14 in cancer cell metabolism | 1. Grossi V, Peserico A, Tezil T, Simone C (2014) p38alpha MAPK pathway: a key factor in colorectal cancer therapy and chemoresistance. World J Gastroenterol 20: 9744-9758. doi: 10.3748/wjg.v20.i29.9744. pmid: 4123363. go to article | |
| Top |
| Phenotypic Information for MAPK14(metabolism pathway, cancer, disease, phenome) |
| Cancer | CGAP: MAPK14 |
| Familial Cancer Database: MAPK14 | |
| * This gene is included in those cancer gene databases. |
|
|
|
|
|
| . | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Oncogene 1 | Significant driver gene in | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| cf) number; DB name 1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 2 Tumor Suppressor gene; https://bioinfo.uth.edu/TSGene/, 3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html, 4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php, 1Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/ |
| REACTOME_METABOLISM_OF_MRNA REACTOME_METABOLISM_OF_RNA | |
| OMIM | 600289; gene. 600289; gene. |
| Orphanet | |
| Disease | KEGG Disease: MAPK14 |
| MedGen: MAPK14 (Human Medical Genetics with Condition) | |
| ClinVar: MAPK14 | |
| Phenotype | MGI: MAPK14 (International Mouse Phenotyping Consortium) |
| PhenomicDB: MAPK14 | |
| Mutations for MAPK14 |
| * Under tables are showing count per each tissue to give us broad intuition about tissue specific mutation patterns.You can go to the detailed page for each mutation database's web site. |
| - Statistics for Tissue and Mutation type | Top |
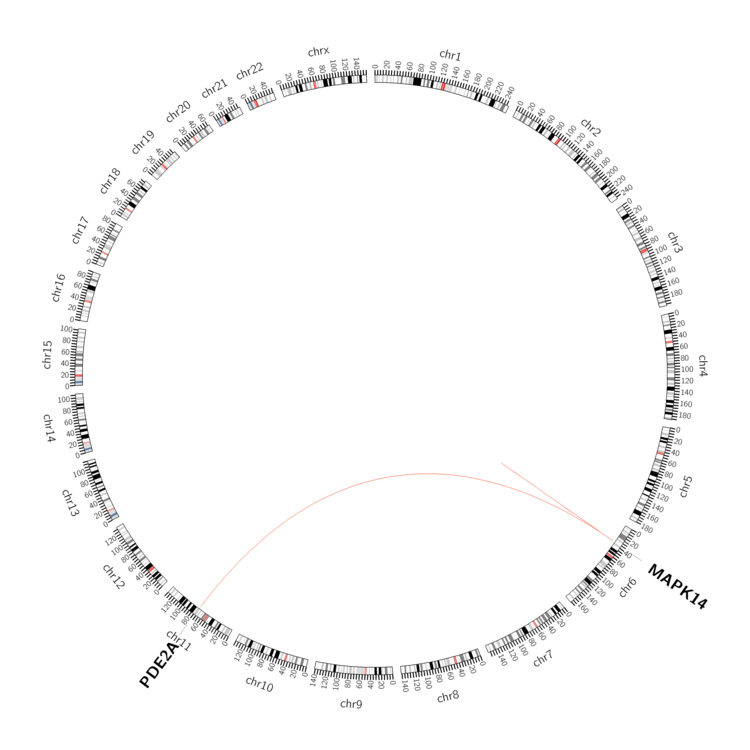 |
| - For Inter-chromosomal Variations |
| * Inter-chromosomal variantions includes 'interchromosomal amplicon to amplicon', 'interchromosomal amplicon to non-amplified dna', 'interchromosomal insertion', 'Interchromosomal unknown type'. |
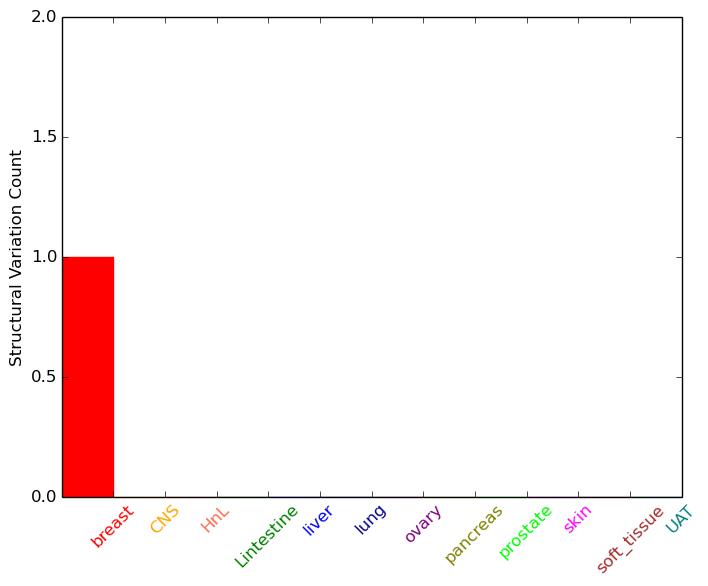 |
| - For Intra-chromosomal Variations |
| * Intra-chromosomal variantions includes 'intrachromosomal amplicon to amplicon', 'intrachromosomal amplicon to non-amplified dna', 'intrachromosomal deletion', 'intrachromosomal fold-back inversion', 'intrachromosomal inversion', 'intrachromosomal tandem duplication', 'Intrachromosomal unknown type', 'intrachromosomal with inverted orientation', 'intrachromosomal with non-inverted orientation'. |
 |
| Sample | Symbol_a | Chr_a | Start_a | End_a | Symbol_b | Chr_b | Start_b | End_b |
| breast | MAPK14 | chr6 | 36060468 | 36060468 | PDE2A | chr11 | 72370761 | 72370761 |
| breast | MAPK14 | chr6 | 36067515 | 36067915 | MAPK14 | chr6 | 36072669 | 36073069 |
| ovary | MAPK14 | chr6 | 36031610 | 36031630 | MAPK14 | chr6 | 36031769 | 36031789 |
| cf) Tissue number; Tissue name (1;Breast, 2;Central_nervous_system, 3;Haematopoietic_and_lymphoid_tissue, 4;Large_intestine, 5;Liver, 6;Lung, 7;Ovary, 8;Pancreas, 9;Prostate, 10;Skin, 11;Soft_tissue, 12;Upper_aerodigestive_tract) |
| * From mRNA Sanger sequences, Chitars2.0 arranged chimeric transcripts. This table shows MAPK14 related fusion information. |
| ID | Head Gene | Tail Gene | Accession | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a |
| AI652202 | TPI1 | 18 | 72 | 12 | 6979938 | 6979992 | MAPK14 | 69 | 205 | 6 | 36076697 | 36076833 | |
| CA429031 | MAPK14 | 18 | 442 | 6 | 36078112 | 36078536 | MEMO1 | 438 | 660 | 2 | 32254755 | 32264853 | |
| BF183021 | MAPK14 | 16 | 74 | 6 | 36075265 | 36075323 | MAPK14 | 72 | 576 | 6 | 36075319 | 36076574 | |
| BM826654 | MAPK14 | 1 | 124 | 6 | 36038712 | 36038835 | POP5 | 107 | 422 | 12 | 121017498 | 121017812 | |
| L35253 | LEMD3 | 11 | 287 | 12 | 65563588 | 65563864 | MAPK14 | 277 | 1539 | 6 | 35995917 | 36076386 | |
| Top |
| Mutation type/ Tissue ID | brca | cns | cerv | endome | haematopo | kidn | Lintest | liver | lung | ns | ovary | pancre | prost | skin | stoma | thyro | urina | |||
| Total # sample | 1 | |||||||||||||||||||
| GAIN (# sample) | 1 | |||||||||||||||||||
| LOSS (# sample) |
| cf) Tissue ID; Tissue type (1; Breast, 2; Central_nervous_system, 3; Cervix, 4; Endometrium, 5; Haematopoietic_and_lymphoid_tissue, 6; Kidney, 7; Large_intestine, 8; Liver, 9; Lung, 10; NS, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary_tract) |
| Top |
|
 |
| Top |
| Stat. for Non-Synonymous SNVs (# total SNVs=26) | (# total SNVs=11) |
 |  |
(# total SNVs=1) | (# total SNVs=1) |
 |  |
| Top |
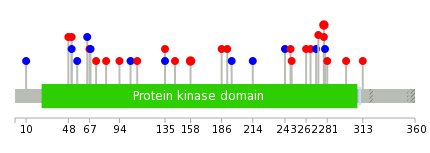
| * When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site,primary_histology,mutation(aa),pubmedID. |
| GRCh37 position | Mutation(aa) | Unique sampleID count |
| chr6:36075327-36075327 | p.D313N | 3 |
| chr6:36020511-36020511 | p.A51V | 3 |
| chr6:36070418-36070418 | p.N278I | 3 |
| chr6:36044336-36044336 | p.C211C | 2 |
| chr6:36041850-36041850 | p.V158M | 2 |
| chr6:36040714-36040714 | p.D124N | 1 |
| chr6:36043701-36043701 | p.P191H | 1 |
| chr6:36020553-36020553 | p.A65V | 1 |
| chr6:36070422-36070422 | p.P279P | 1 |
| chr6:36040747-36040747 | p.L135I | 1 |
| Top |
|
 |
| Point Mutation/ Tissue ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| # sample | 1 | 3 | 10 | 2 | 1 | 3 | 1 | 3 | 1 | 5 | 1 | 5 | ||||||||
| # mutation | 1 | 3 | 9 | 2 | 1 | 3 | 1 | 3 | 1 | 5 | 1 | 5 | ||||||||
| nonsynonymous SNV | 2 | 7 | 1 | 1 | 2 | 1 | 2 | 3 | 1 | 3 | ||||||||||
| synonymous SNV | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 2 |
| cf) Tissue ID; Tissue type (1; BLCA[Bladder Urothelial Carcinoma], 2; BRCA[Breast invasive carcinoma], 3; CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], 4; COAD[Colon adenocarcinoma], 5; GBM[Glioblastoma multiforme], 6; Glioma Low Grade, 7; HNSC[Head and Neck squamous cell carcinoma], 8; KICH[Kidney Chromophobe], 9; KIRC[Kidney renal clear cell carcinoma], 10; KIRP[Kidney renal papillary cell carcinoma], 11; LAML[Acute Myeloid Leukemia], 12; LUAD[Lung adenocarcinoma], 13; LUSC[Lung squamous cell carcinoma], 14; OV[Ovarian serous cystadenocarcinoma ], 15; PAAD[Pancreatic adenocarcinoma], 16; PRAD[Prostate adenocarcinoma], 17; SKCM[Skin Cutaneous Melanoma], 18:STAD[Stomach adenocarcinoma], 19:THCA[Thyroid carcinoma], 20:UCEC[Uterine Corpus Endometrial Carcinoma]) |
| Top |
| * We represented just top 10 SNVs. When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site, primary_histology, mutation(aa), pubmedID. |
| Genomic Position | Mutation(aa) | Unique sampleID count |
| chr6:36070418 | p.N278S,MAPK14 | 3 |
| chr6:36041850 | p.V158M,MAPK14 | 2 |
| chr6:36044345 | p.G110A,MAPK14 | 1 |
| chr6:36075327 | p.A281S,MAPK14 | 1 |
| chr6:36027100 | p.L135I,MAPK14 | 1 |
| chr6:36063825 | p.C262F,MAPK14 | 1 |
| chr6:35995964 | p.L135L,MAPK14 | 1 |
| chr6:36075361 | p.R273W,MAPK14 | 1 |
| chr6:36040654 | p.A144T,MAPK14 | 1 |
| chr6:36063827 | p.D313N,MAPK14 | 1 |
| * Copy number data were extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered on Jan-05-2015. Function ProcessCNAData in TCGA-Assembler package was used to obtain gene-level copy number value which is calculated as the average copy number of the genomic region of a gene. |
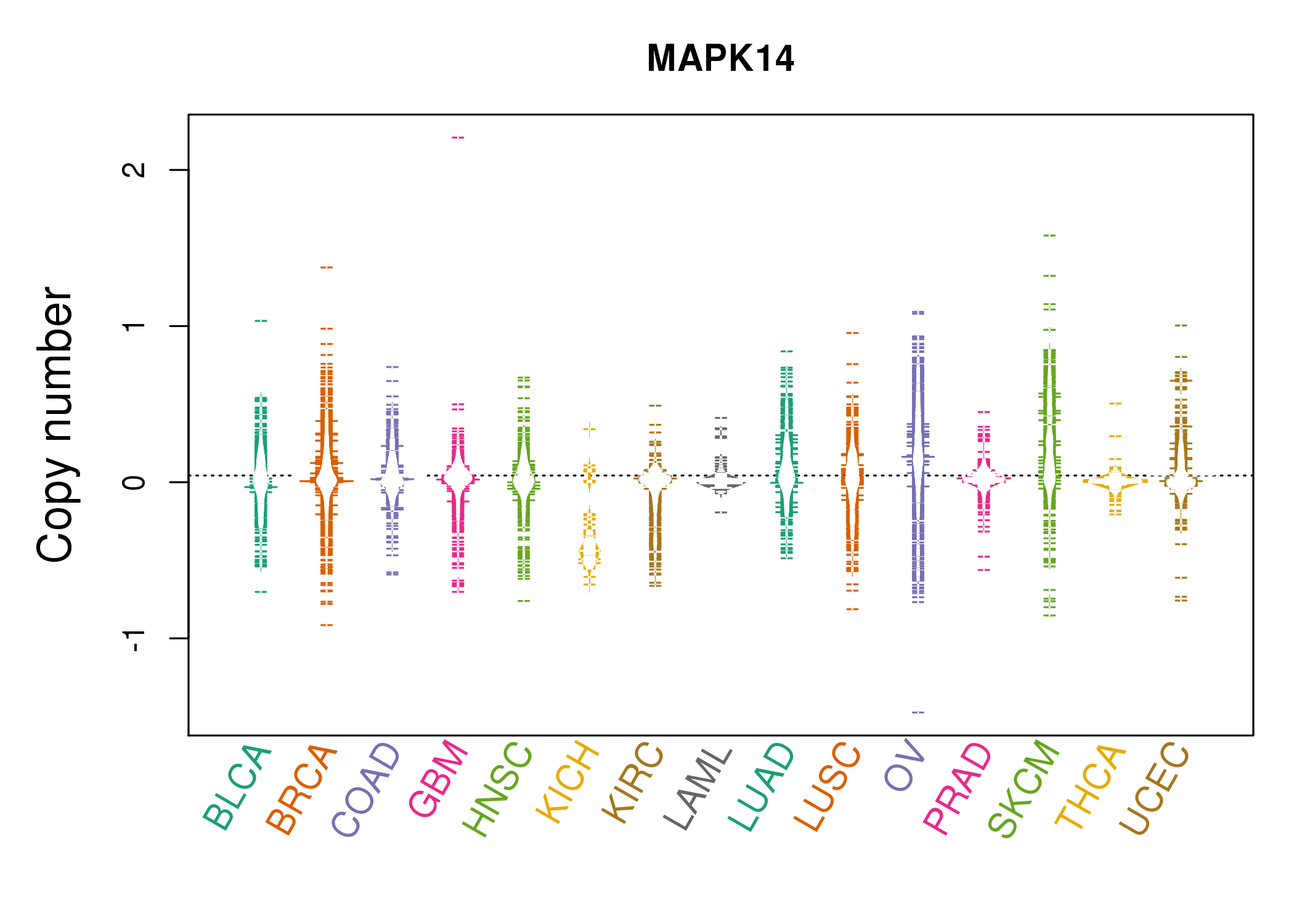 |
| cf) Tissue ID[Tissue type]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma], KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ], PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma] |
| Top |
| Gene Expression for MAPK14 |
| * CCLE gene expression data were extracted from CCLE_Expression_Entrez_2012-10-18.res: Gene-centric RMA-normalized mRNA expression data. |
 |
| Top |
| *RPPA protein expression data were extracted from TCPA (The Cancer Proteome Atlas). Normalized data based on replicated based normalization (RBN) was used to draw following figures. |
 |
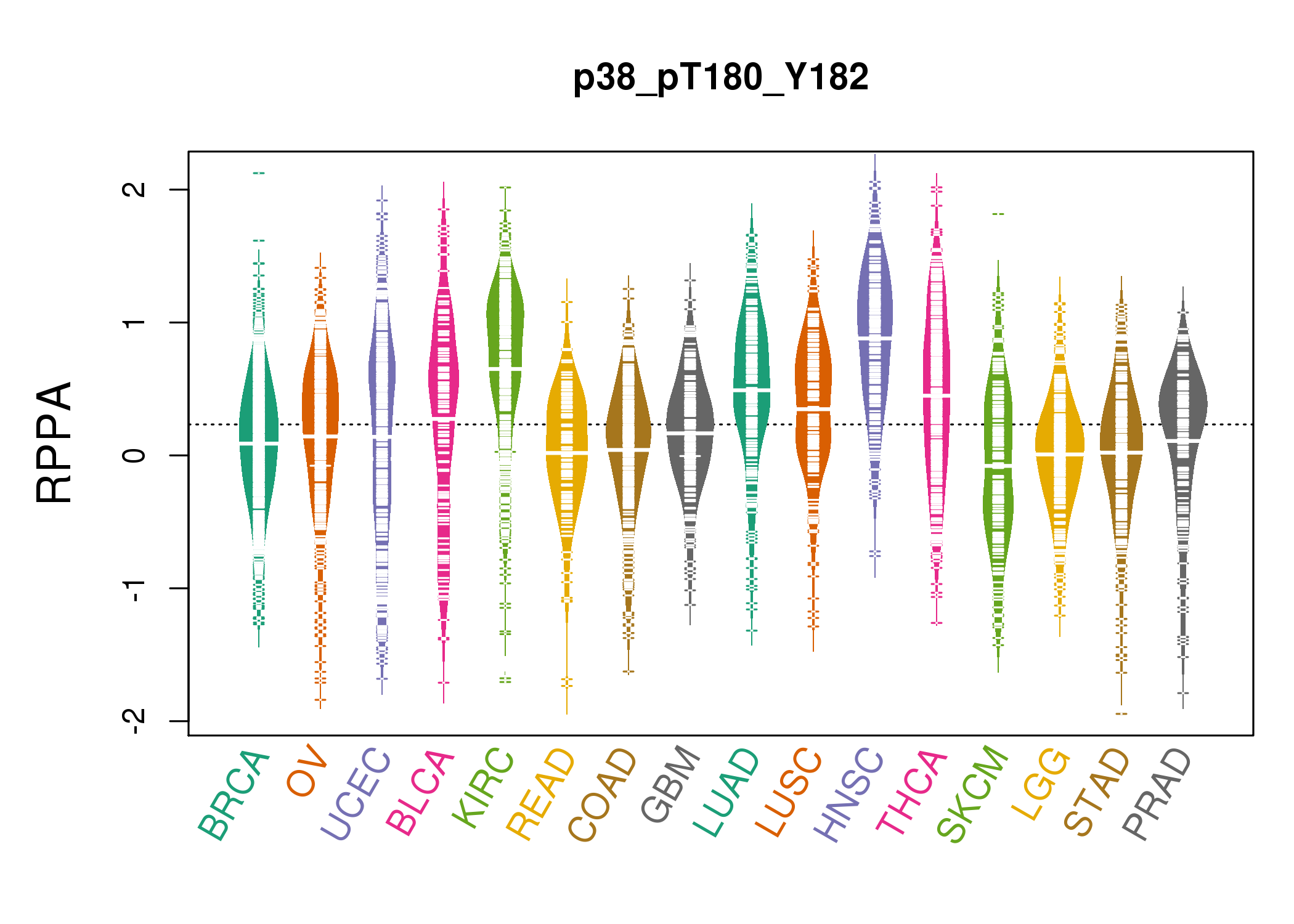 |
| * Normalized gene expression data of RNASeqV2 was extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered at Jan-05-2015. Only eight cancer types have enough normal control samples for differential expression analysis. (t test, adjusted p<0.05 (using Benjamini-Hochberg FDR)) |
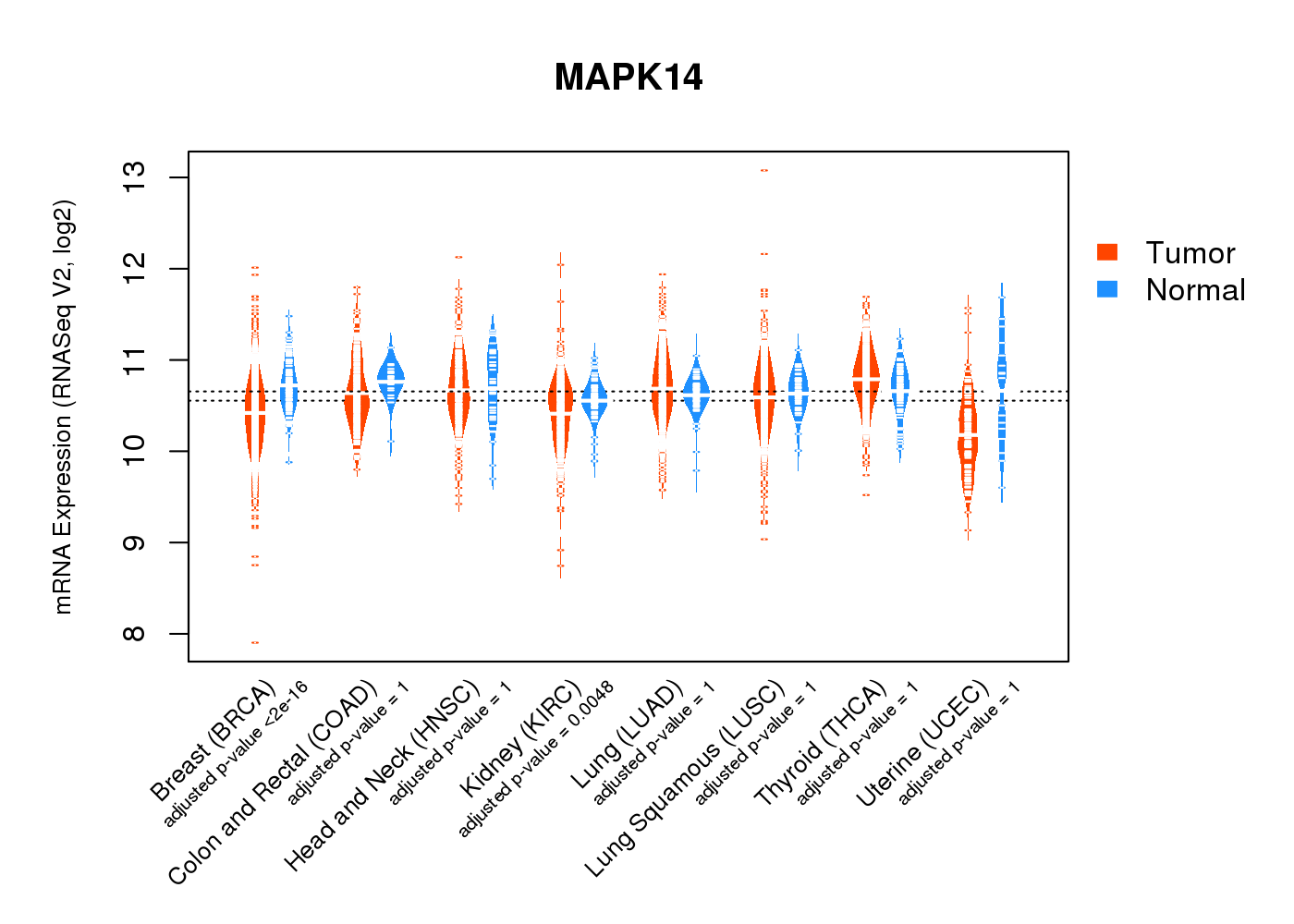 |
| Top |
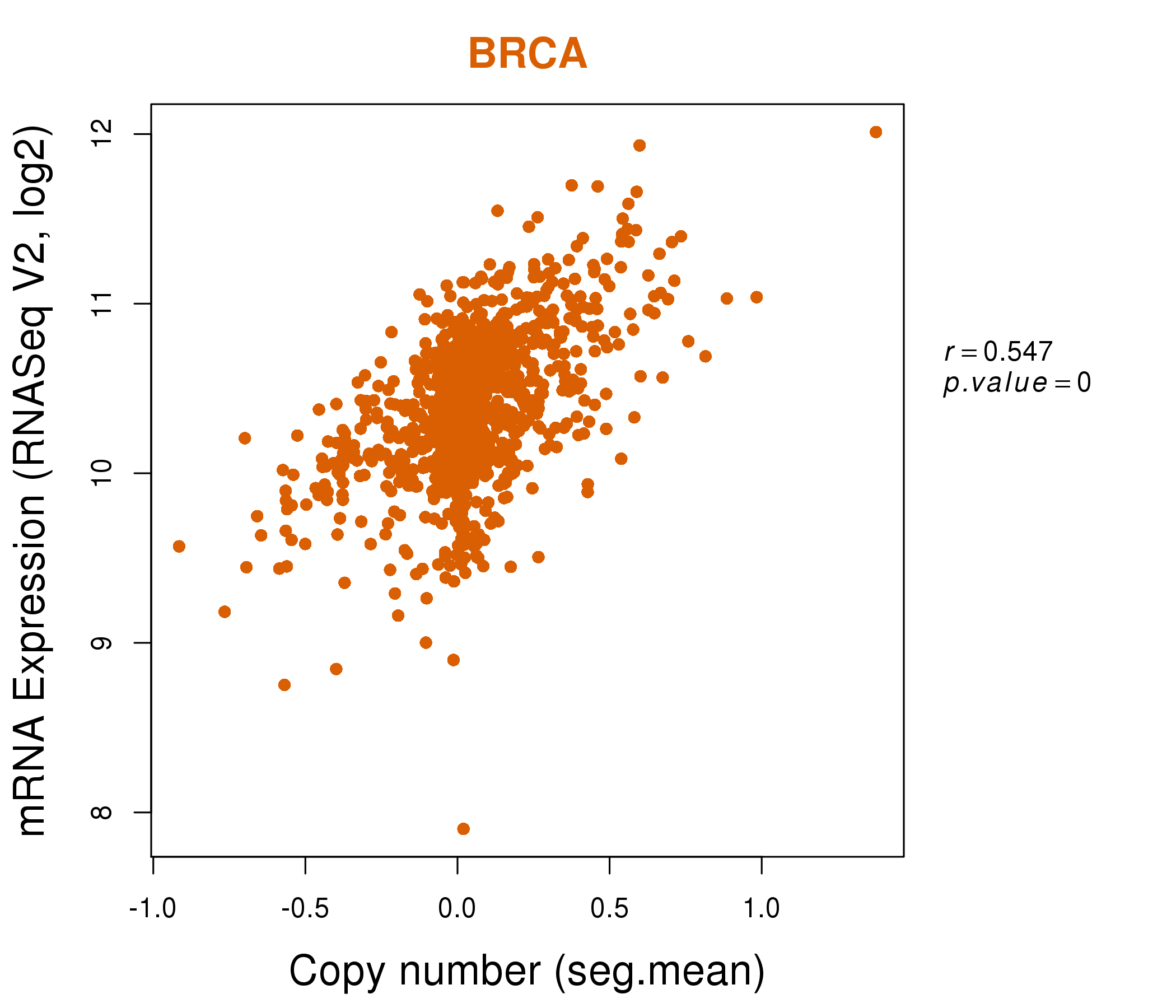
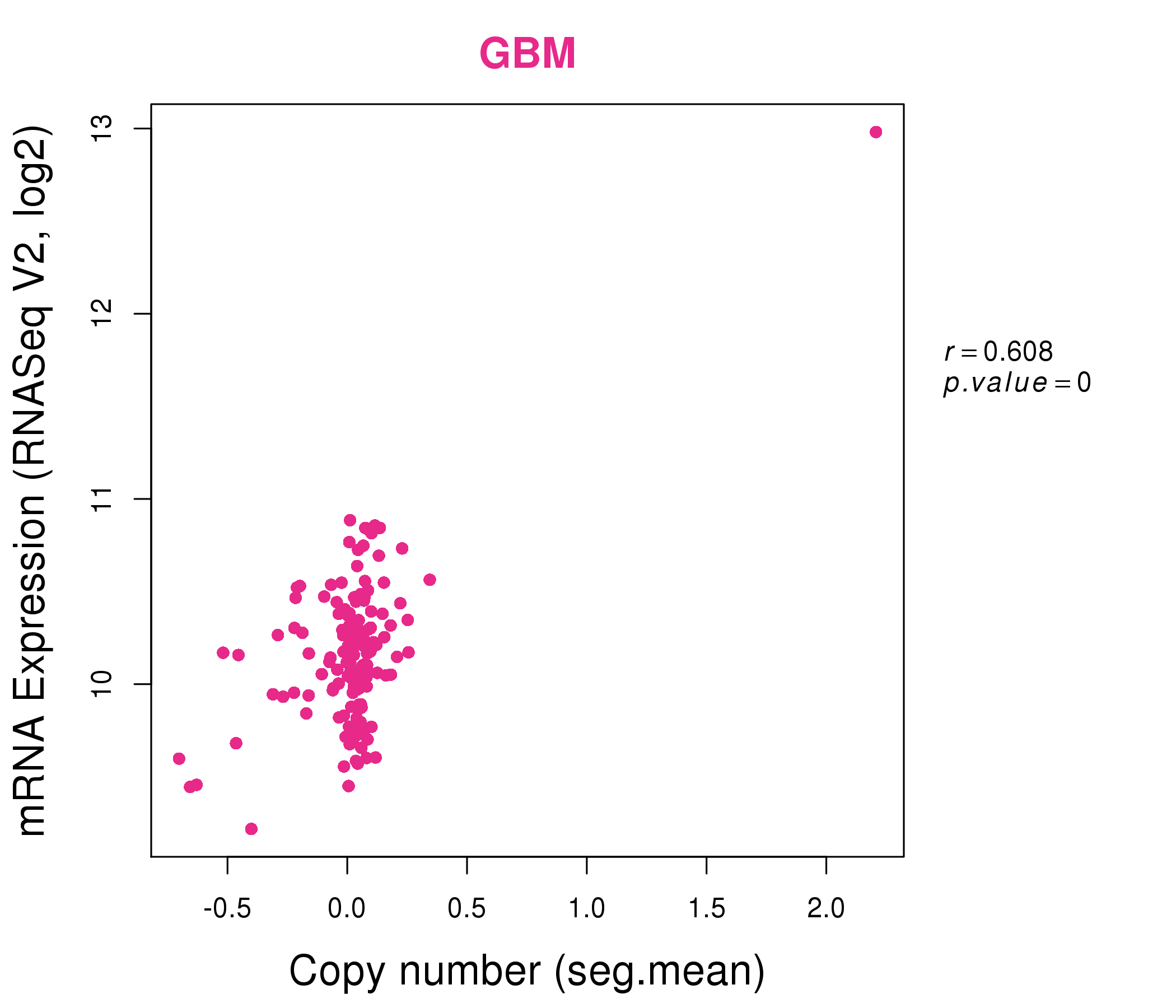
| * This plots show the correlation between CNV and gene expression. |
: Open all plots for all cancer types
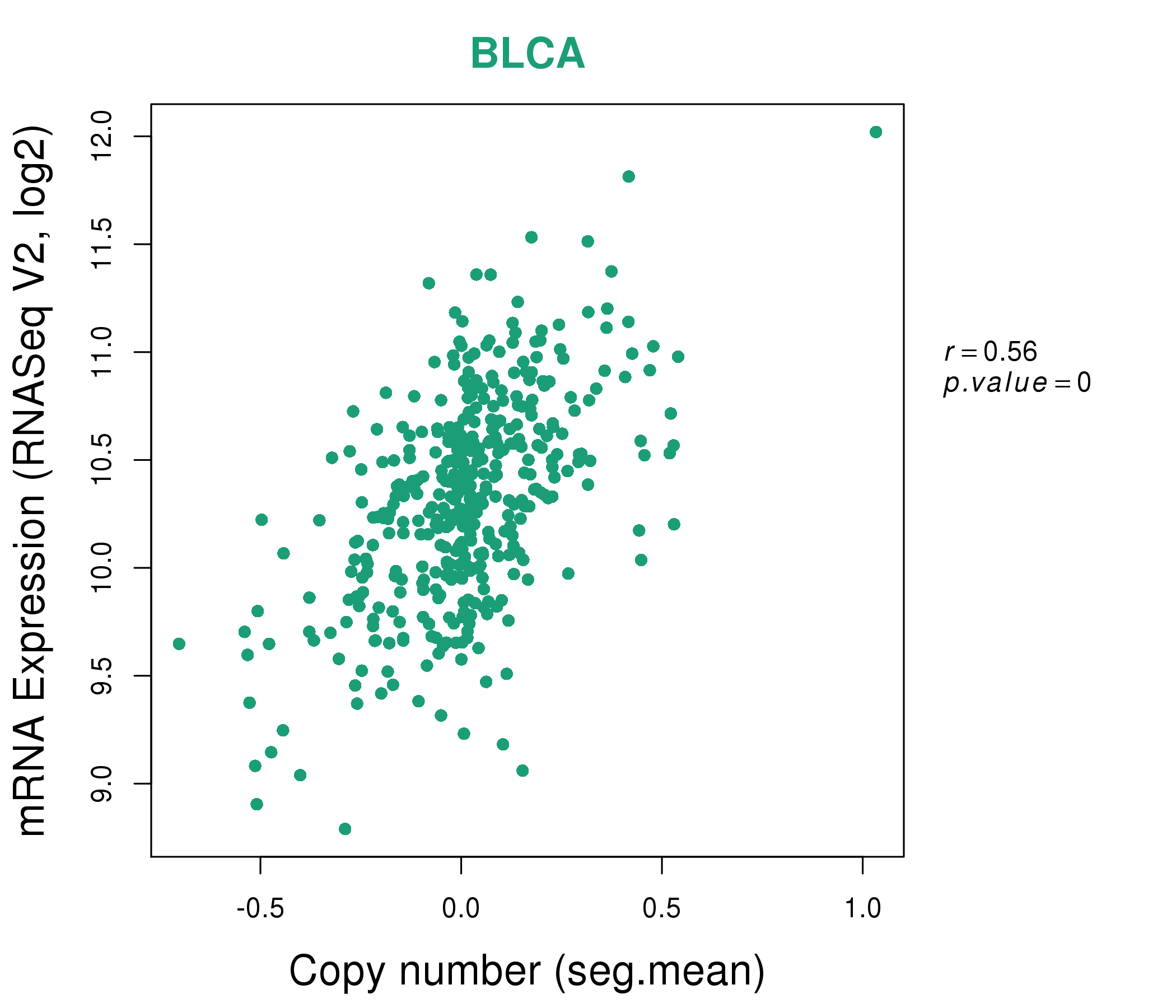 |
|
 |
|
| Top |
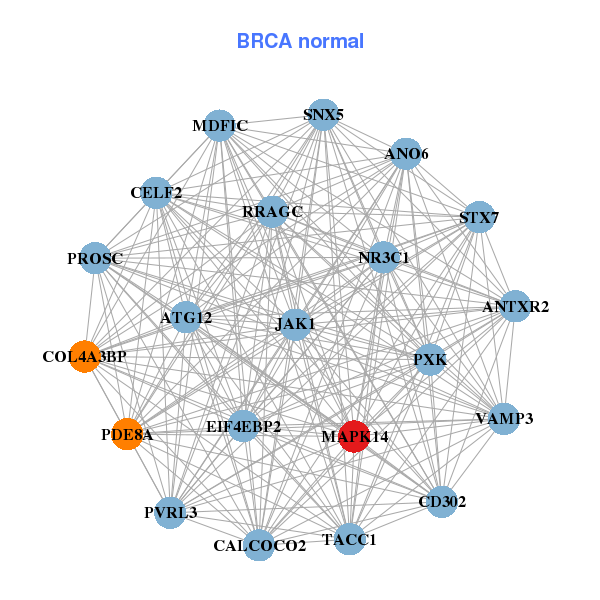
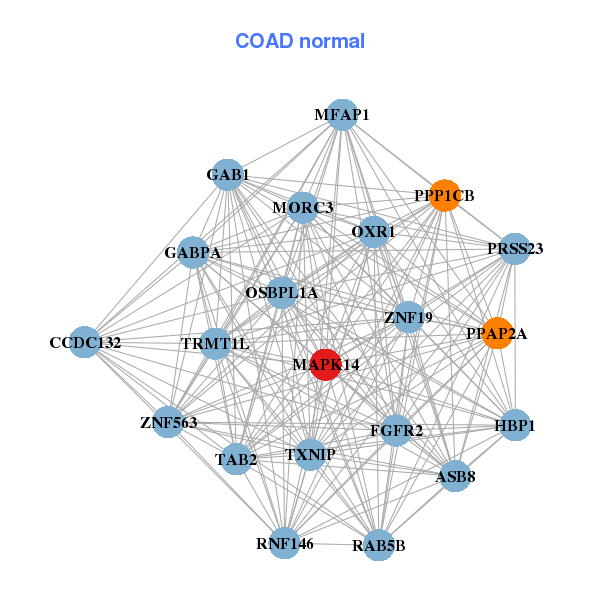
| Gene-Gene Network Information |
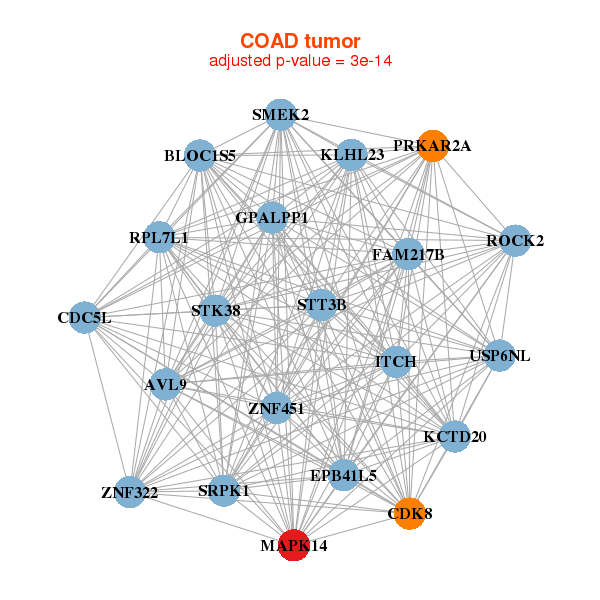
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
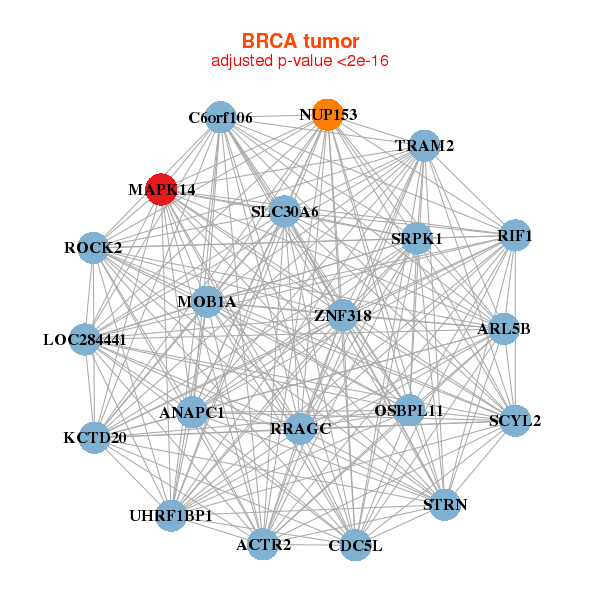 |
| ||||
| ACTR2,ANAPC1,ARL5B,C6orf106,CDC5L,KCTD20,LOC284441, MAPK14,MOB1A,NUP153,OSBPL11,RIF1,ROCK2,RRAGC, SCYL2,SLC30A6,SRPK1,STRN,TRAM2,UHRF1BP1,ZNF318 | ANO6,ANTXR2,ATG12,CALCOCO2,CD302,CELF2,COL4A3BP, EIF4EBP2,JAK1,MAPK14,MDFIC,NR3C1,PDE8A,PROSC, PVRL3,PXK,RRAGC,SNX5,STX7,TACC1,VAMP3 | ||||
 |
| ||||
| AVL9,FAM217B,CDC5L,CDK8,EPB41L5,ITCH,KCTD20, GPALPP1,KLHL23,MAPK14,BLOC1S5,PRKAR2A,ROCK2,RPL7L1, SMEK2,SRPK1,STK38,STT3B,USP6NL,ZNF322,ZNF451 | ASB8,TRMT1L,CCDC132,FGFR2,GAB1,GABPA,HBP1, MAPK14,MFAP1,MORC3,OSBPL1A,OXR1,PPAP2A,PPP1CB, PRSS23,RAB5B,RNF146,TAB2,TXNIP,ZNF19,ZNF563 |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
| Top |
: Open all interacting genes' information including KEGG pathway for all interacting genes from DAVID
| Top |
| Pharmacological Information for MAPK14 |
| DB Category | DB Name | DB's ID and Url link |
| Chemistry | BindingDB | Q16539; -. |
| Chemistry | ChEMBL | CHEMBL2094115; -. |
| Chemistry | GuidetoPHARMACOLOGY | 1499; -. |
| Chemistry | BindingDB | Q16539; -. |
| Chemistry | ChEMBL | CHEMBL2094115; -. |
| Chemistry | GuidetoPHARMACOLOGY | 1499; -. |
| Organism-specific databases | PharmGKB | PA30621; -. |
| Organism-specific databases | PharmGKB | PA30621; -. |
| Organism-specific databases | CTD | 1432; -. |
| Organism-specific databases | CTD | 1432; -. |
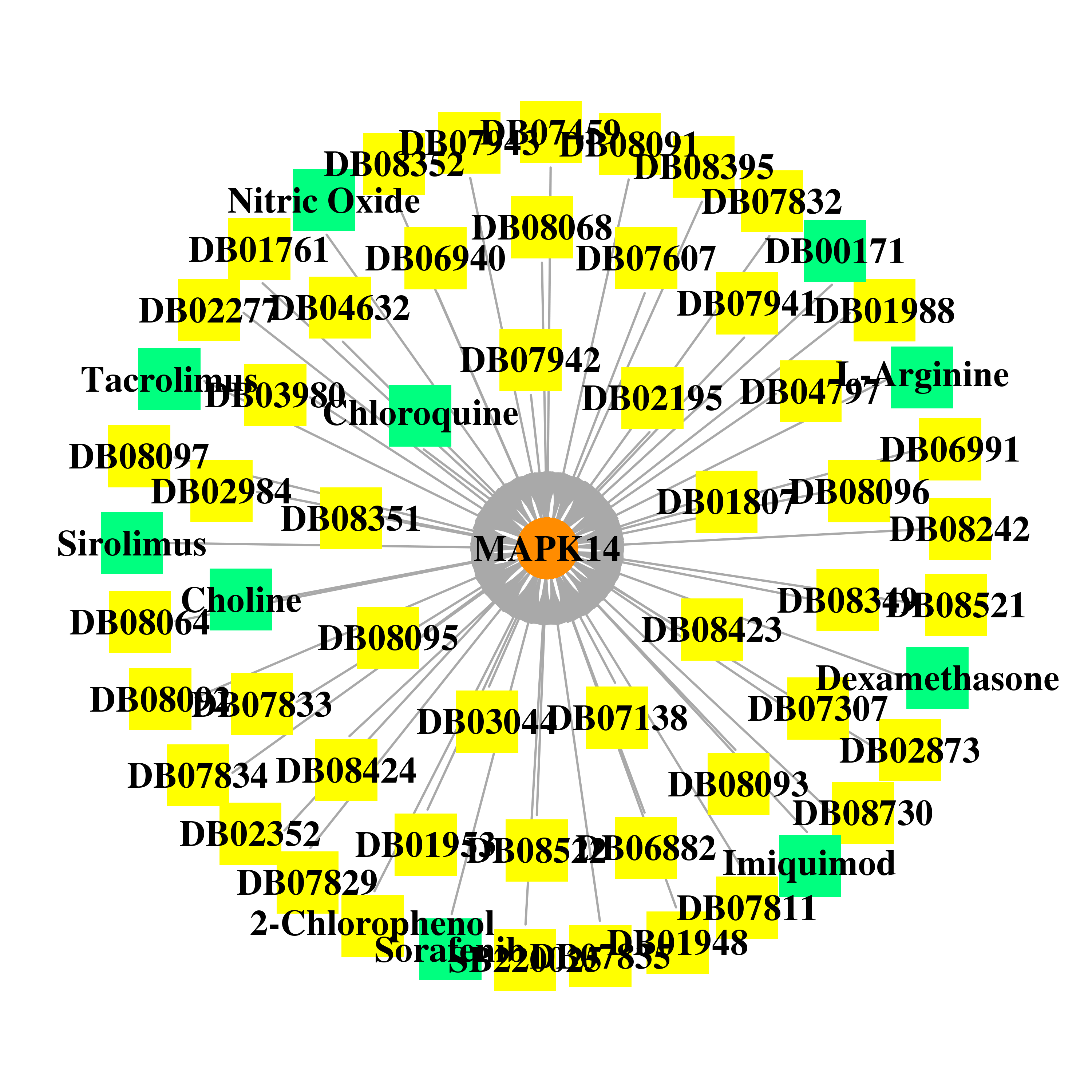
| * Gene Centered Interaction Network. |
 |
| * Drug Centered Interaction Network. |
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB01761 | mitogen-activated protein kinase 14 | experimental | 4-[5-[2-(1-Phenyl-Ethylamino)-Pyrimidin-4-Yl]-1-Methyl-4-(3-Trifluoromethylphenyl)-1h-Imidazol-2-Yl]-Piperidine |  |  |
| DB01807 | mitogen-activated protein kinase 14 | experimental | N-[(3z)-5-Tert-Butyl-2-Phenyl-1,2-Dihydro-3h-Pyrazol-3-Ylidene]-N'-(4-Chlorophenyl)Urea |  |  |
| DB01948 | mitogen-activated protein kinase 14 | experimental | 1-(2,6-Dichlorophenyl)-5-(2,4-Difluorophenyl)-7-Piperidin-4-Yl-3,4-Dihydroquinolin-2(1h)-One |  |  |
| DB01953 | mitogen-activated protein kinase 14 | experimental | Inhibitor of P38 Kinase |  |  |
| DB01988 | mitogen-activated protein kinase 14 | experimental | 6((S)-3-Benzylpiperazin-1-Yl)-3-(Naphthalen-2-Yl)-4-(Pyridin-4-Yl)Pyrazine |  |  |
| DB02195 | mitogen-activated protein kinase 14 | experimental | 3-(4-Fluorophenyl)-1-Hydroxy-2-(Pyridin-4-Yl)-1h-Pyrrolo[3,2-B]Pyridine |  |  |
| DB02277 | mitogen-activated protein kinase 14 | experimental | 1-(5-Tert-Butyl-2-Methyl-2h-Pyrazol-3-Yl)-3-(4-Chloro-Phenyl)-Urea |  |  |
| DB02352 | mitogen-activated protein kinase 14 | experimental | 3-(Benzyloxy)Pyridin-2-Amine |  |  |
| DB02873 | mitogen-activated protein kinase 14 | experimental | 1-(2,6-Dichlorophenyl)-5-(2,4-Difluorophenyl)-7-Piperazin-1-Yl-3,4-Dihydroquinazolin-2(1h)-One |  |  |
| DB02984 | mitogen-activated protein kinase 14 | experimental | 4-[3-Methylsulfanylanilino]-6,7-Dimethoxyquinazoline |  |  |
| DB03044 | mitogen-activated protein kinase 14 | experimental | 1-(5-Tert-Butyl-2-P-Tolyl-2h-Pyrazol-3-Yl)-3-[4-(2-Morpholin-4-Yl-Ethoxy)-Naphthalen-1-Yl]-Urea | 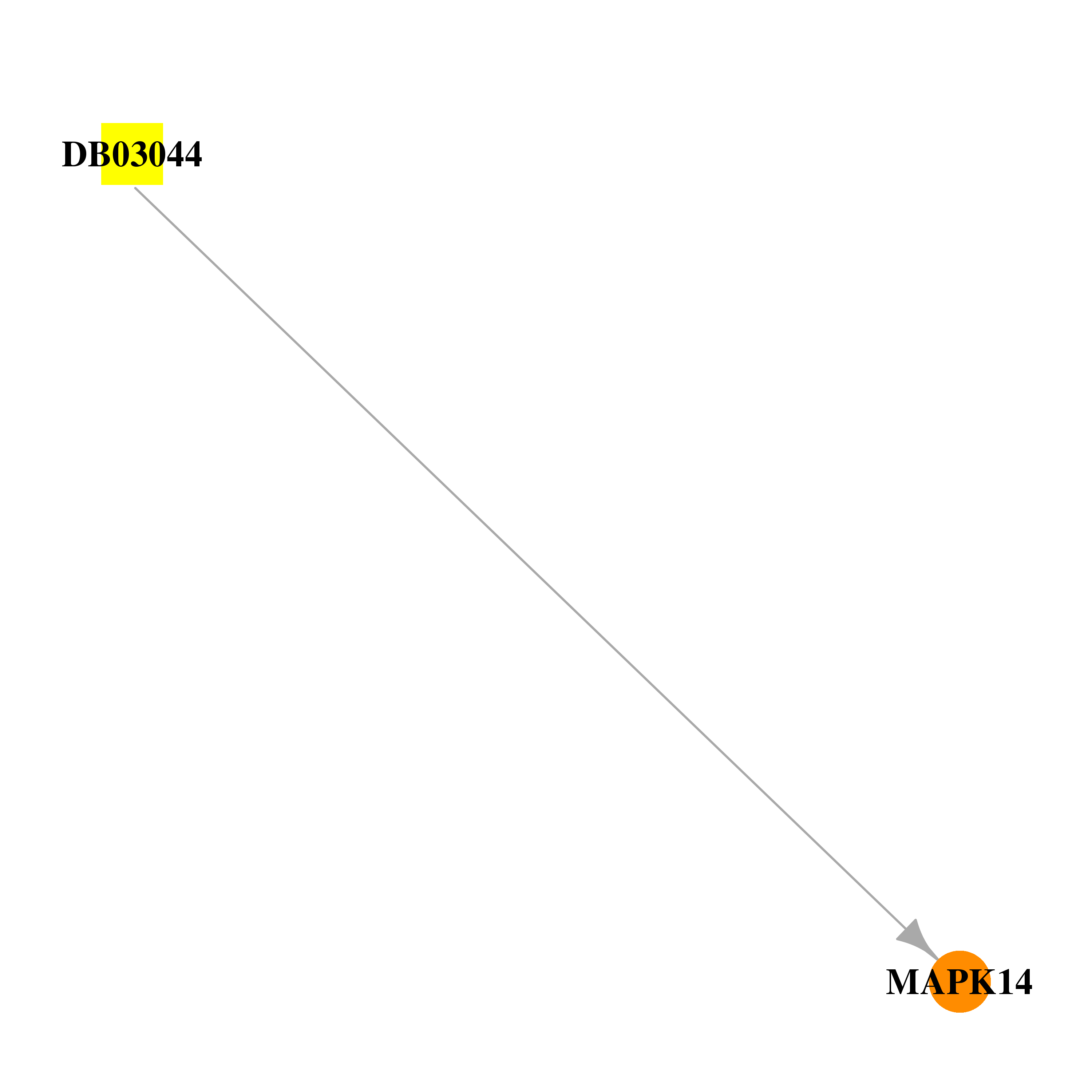 |  |
| DB03110 | mitogen-activated protein kinase 14 | experimental | 2-Chlorophenol |  |  |
| DB03980 | mitogen-activated protein kinase 14 | experimental | 4-(Fluorophenyl)-1-Cyclopropylmethyl-5-(2-Amino-4-Pyrimidinyl)Imidazole |  |  |
| DB04338 | mitogen-activated protein kinase 14 | experimental | SB220025 |  |  |
| DB04632 | mitogen-activated protein kinase 14 | experimental | 4-(2-HYDROXYBENZYLAMINO)-N-(3-(4-FLUOROPHENOXY)PHENYL)PIPERIDINE-1-SULFONAMIDE |  |  |
| DB04797 | mitogen-activated protein kinase 14 | experimental | Triazolopyridine |  |  |
| DB06882 | mitogen-activated protein kinase 14 | experimental | 1-[1-(3-aminophenyl)-3-tert-butyl-1H-pyrazol-5-yl]-3-naphthalen-1-ylurea |  |  |
| DB06940 | mitogen-activated protein kinase 14 | experimental | N-ethyl-4-{[5-(methoxycarbamoyl)-2-methylphenyl]amino}-5-methylpyrrolo[2,1-f][1,2,4]triazine-6-carboxamide |  |  |
| DB06991 | mitogen-activated protein kinase 14 | experimental | N-[2-methyl-5-(methylcarbamoyl)phenyl]-2-{[(1R)-1-methylpropyl]amino}-1,3-thiazole-5-carboxamide |  |  |
| DB07138 | mitogen-activated protein kinase 14 | experimental | 5-(2,6-dichlorophenyl)-2-[(2,4-difluorophenyl)sulfanyl]-6H-pyrimido[1,6-b]pyridazin-6-one |  |  |
| DB07307 | mitogen-activated protein kinase 14 | experimental | N-cyclopropyl-4-methyl-3-[1-(2-methylphenyl)phthalazin-6-yl]benzamide |  |  |
| DB07459 | mitogen-activated protein kinase 14 | experimental | 4-PHENOXY-N-(PYRIDIN-2-YLMETHYL)BENZAMIDE |  |  |
| DB07607 | mitogen-activated protein kinase 14 | experimental | 4-[5-(3-IODO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-1H-IMIDAZOL-4-YL]-PYRIDINE |  |  |
| DB07811 | mitogen-activated protein kinase 14 | experimental | N-cyclopropyl-2',6-dimethyl-4'-(5-methyl-1,3,4-oxadiazol-2-yl)biphenyl-3-carboxamide |  |  |
| DB07829 | mitogen-activated protein kinase 14 | experimental | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE |  |  |
| DB07832 | mitogen-activated protein kinase 14 | experimental | 4-{4-[(5-hydroxy-2-methylphenyl)amino]quinolin-7-yl}-1,3-thiazole-2-carbaldehyde |  |  |
| DB07833 | mitogen-activated protein kinase 14 | experimental | N-(3-cyanophenyl)-2'-methyl-5'-(5-methyl-1,3,4-oxadiazol-2-yl)-4-biphenylcarboxamide |  |  |
| DB07834 | mitogen-activated protein kinase 14 | experimental | N-(cyclopropylmethyl)-2'-methyl-5'-(5-methyl-1,3,4-oxadiazol-2-yl)biphenyl-4-carboxamide |  |  |
| DB07835 | mitogen-activated protein kinase 14 | experimental | N~3~-cyclopropyl-N~4~'-(cyclopropylmethyl)-6-methylbiphenyl-3,4'-dicarboxamide |  |  |
| DB07941 | mitogen-activated protein kinase 14 | experimental | 3-{3-bromo-4-[(2,4-difluorobenzyl)oxy]-6-methyl-2-oxopyridin-1(2H)-yl}-N,4-dimethylbenzamide |  |  |
| DB07942 | mitogen-activated protein kinase 14 | experimental | 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine |  |  |
| DB07943 | mitogen-activated protein kinase 14 | experimental | 2-{4-[5-(4-chlorophenyl)-4-pyrimidin-4-yl-1H-pyrazol-3-yl]piperidin-1-yl}-2-oxoethanol |  |  |
| DB08064 | mitogen-activated protein kinase 14 | experimental | N-(3-TERT-BUTYL-1H-PYRAZOL-5-YL)-N'-{4-CHLORO-3-[(PYRIDIN-3-YLOXY)METHYL]PHENYL}UREA |  | 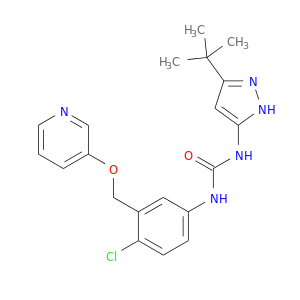 |
| DB08068 | mitogen-activated protein kinase 14 | experimental | N-[4-CHLORO-3-(PYRIDIN-3-YLOXYMETHYL)-PHENYL]-3-FLUORO- |  |  |
| DB08091 | mitogen-activated protein kinase 14 | experimental | 3-FLUORO-5-MORPHOLIN-4-YL-N-[3-(2-PYRIDIN-4-YLETHYL)-1H-INDOL-5-YL]BENZAMIDE |  |  |
| DB08092 | mitogen-activated protein kinase 14 | experimental | 3-FLUORO-N-1H-INDOL-5-YL-5-MORPHOLIN-4-YLBENZAMIDE |  |  |
| DB08093 | mitogen-activated protein kinase 14 | experimental | 3-(1-NAPHTHYLMETHOXY)PYRIDIN-2-AMINE |  |  |
| DB08095 | mitogen-activated protein kinase 14 | experimental | 3-(2-CHLOROPHENYL)-1-(2-{[(1S)-2-HYDROXY-1,2-DIMETHYLPROPYL]AMINO}PYRIMIDIN-4-YL)-1-(4-METHOXYPHENYL)UREA |  |  |
| DB08096 | mitogen-activated protein kinase 14 | experimental | 8-(2-CHLOROPHENYLAMINO)-2-(2,6-DIFLUOROPHENYLAMINO)-9-ETHYL-9H-PURINE-1,7-DIIUM |  |  |
| DB08097 | mitogen-activated protein kinase 14 | experimental | 2-(2,6-DIFLUOROPHENOXY)-N-(2-FLUOROPHENYL)-9-ISOPROPYL-9H-PURIN-8-AMINE |  |  |
| DB08242 | mitogen-activated protein kinase 14 | experimental | N,4-dimethyl-3-[(1-phenyl-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide |  |  |
| DB08349 | mitogen-activated protein kinase 14 | experimental | N-cyclopropyl-3-{[1-(2,4-difluorophenyl)-7-methyl-6-oxo-6,7-dihydro-1H-pyrazolo[3,4-b]pyridin-4-yl]amino}-4-methylbenzamide |  |  |
| DB08351 | mitogen-activated protein kinase 14 | experimental | N-cyclopropyl-4-methyl-3-{2-[(2-morpholin-4-ylethyl)amino]quinazolin-6-yl}benzamide |  |  |
| DB08352 | mitogen-activated protein kinase 14 | experimental | 6-[4-(2-fluorophenyl)-1,3-oxazol-5-yl]-N-(1-methylethyl)-1,3-benzothiazol-2-amine |  |  |
| DB08395 | mitogen-activated protein kinase 14 | experimental | 2-(ETHOXYMETHYL)-4-(4-FLUOROPHENYL)-3-[2-(2-HYDROXYPHENOXY)PYRIMIDIN-4-YL]ISOXAZOL-5(2H)-ONE |  |  |
| DB08423 | mitogen-activated protein kinase 14 | experimental | [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL][3-(PIPERIDIN-4-YLOXY)PHENYL]METHANONE |  |  |
| DB08424 | mitogen-activated protein kinase 14 | experimental | [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE |  |  |
| DB08521 | mitogen-activated protein kinase 14 | experimental | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE |  |  |
| DB08522 | mitogen-activated protein kinase 14 | experimental | 4-(4-FLUOROPHENYL)-1-CYCLOROPROPYLMETHYL-5-(4-PYRIDYL)-IMIDAZOLE |  |  |
| DB08730 | mitogen-activated protein kinase 14 | experimental | 3-FLUORO-5-MORPHOLIN-4-YL-N-[1-(2-PYRIDIN-4-YLETHYL)-1H-INDOL-6-YL]BENZAMIDE |  |  |
| DB00724 | mitogen-activated protein kinase 14 | approved; investigational | Imiquimod |  |  |
| DB00122 | mitogen-activated protein kinase 14 | approved; nutraceutical | Choline |  |  |
| DB00864 | mitogen-activated protein kinase 14 | approved; investigational | Tacrolimus |  |  |
| DB01234 | mitogen-activated protein kinase 14 | approved; investigational | Dexamethasone | 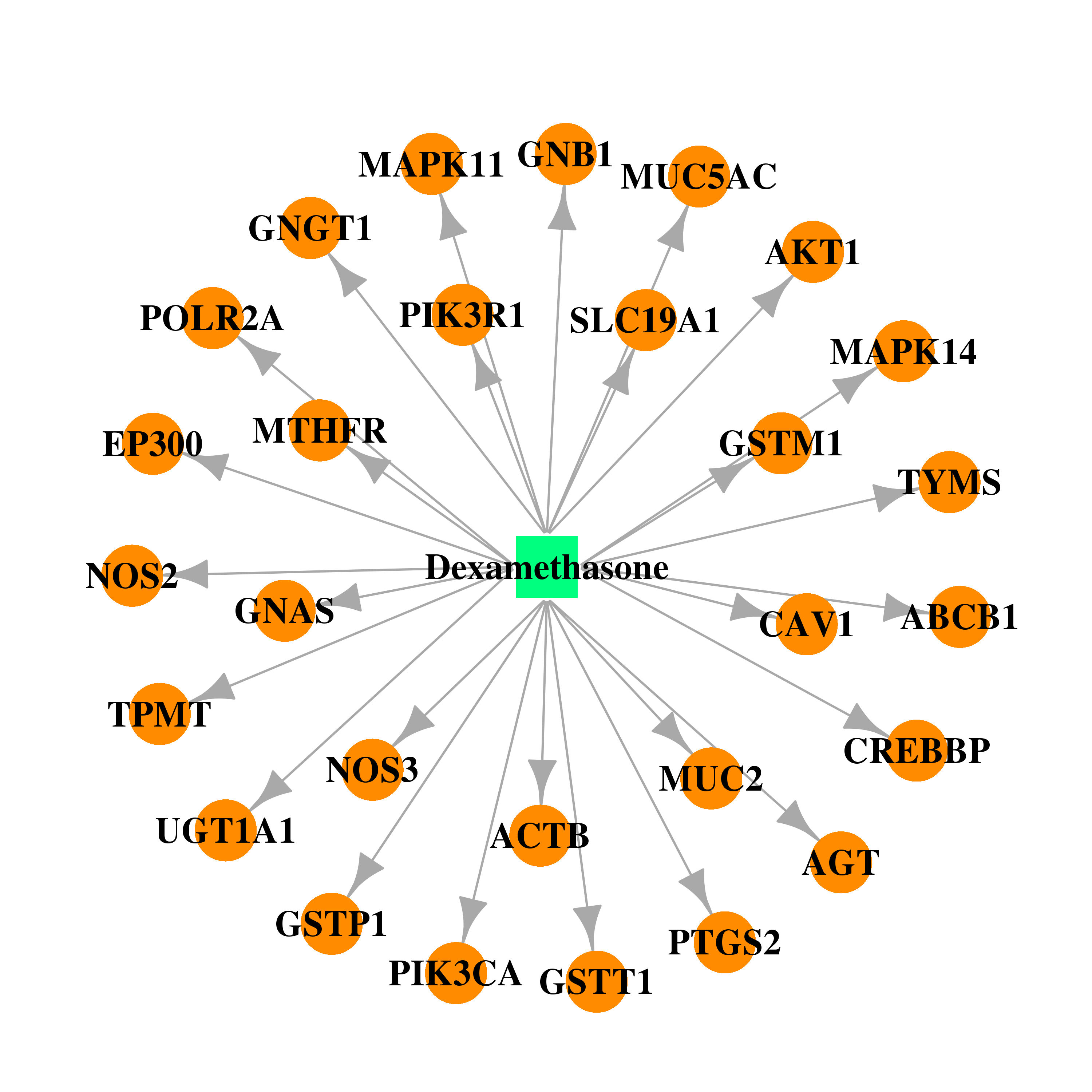 | 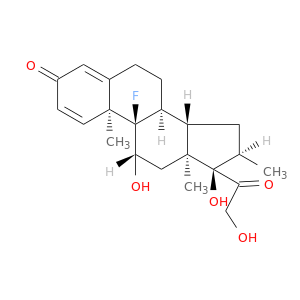 |
| DB00608 | mitogen-activated protein kinase 14 | approved | Chloroquine |  | 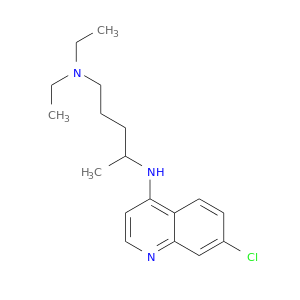 |
| DB00877 | mitogen-activated protein kinase 14 | approved; investigational | Sirolimus | 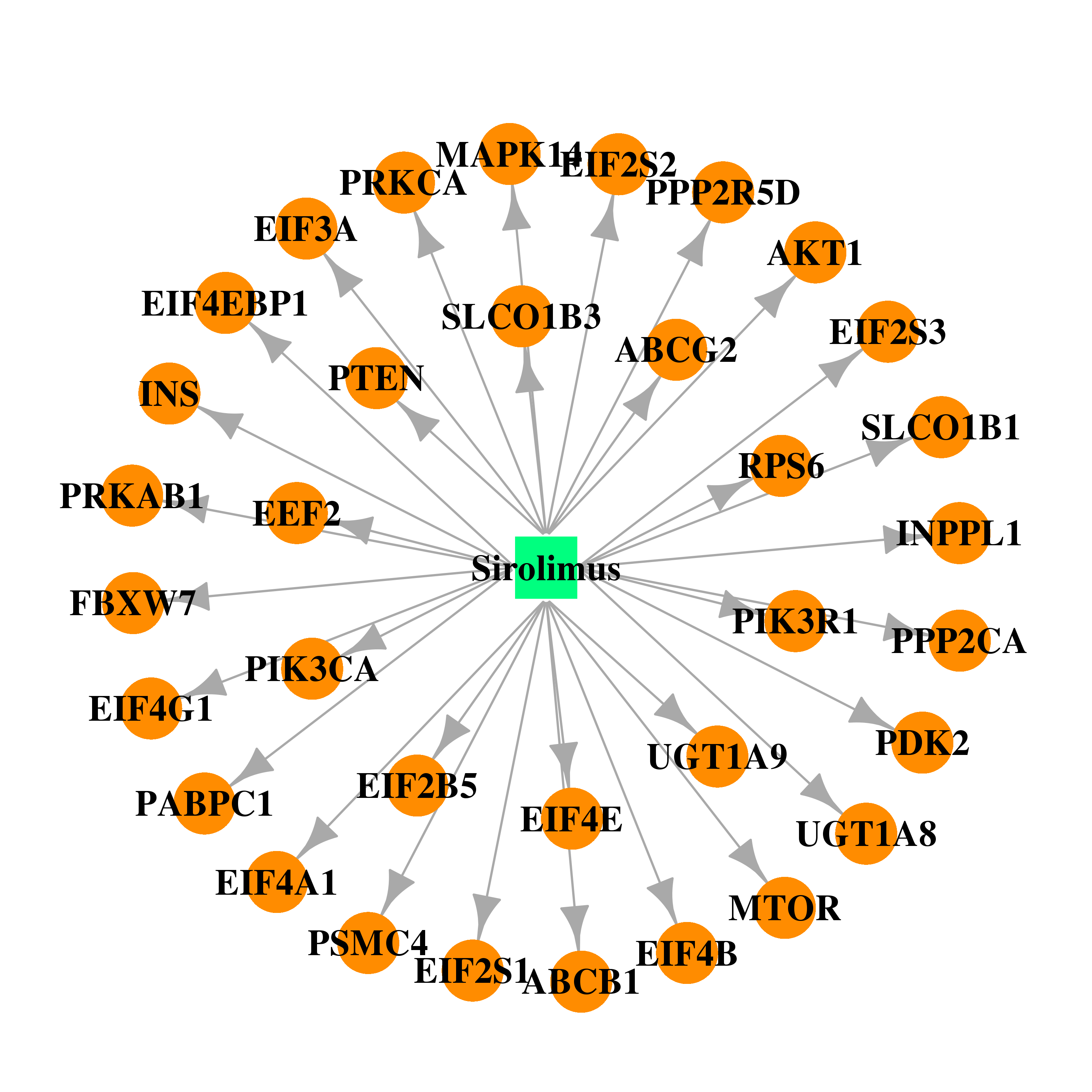 |  |
| DB00171 | mitogen-activated protein kinase 14 | approved; nutraceutical | Adenosine triphosphate |  |  |
| DB00398 | mitogen-activated protein kinase 14 | approved; investigational | Sorafenib | 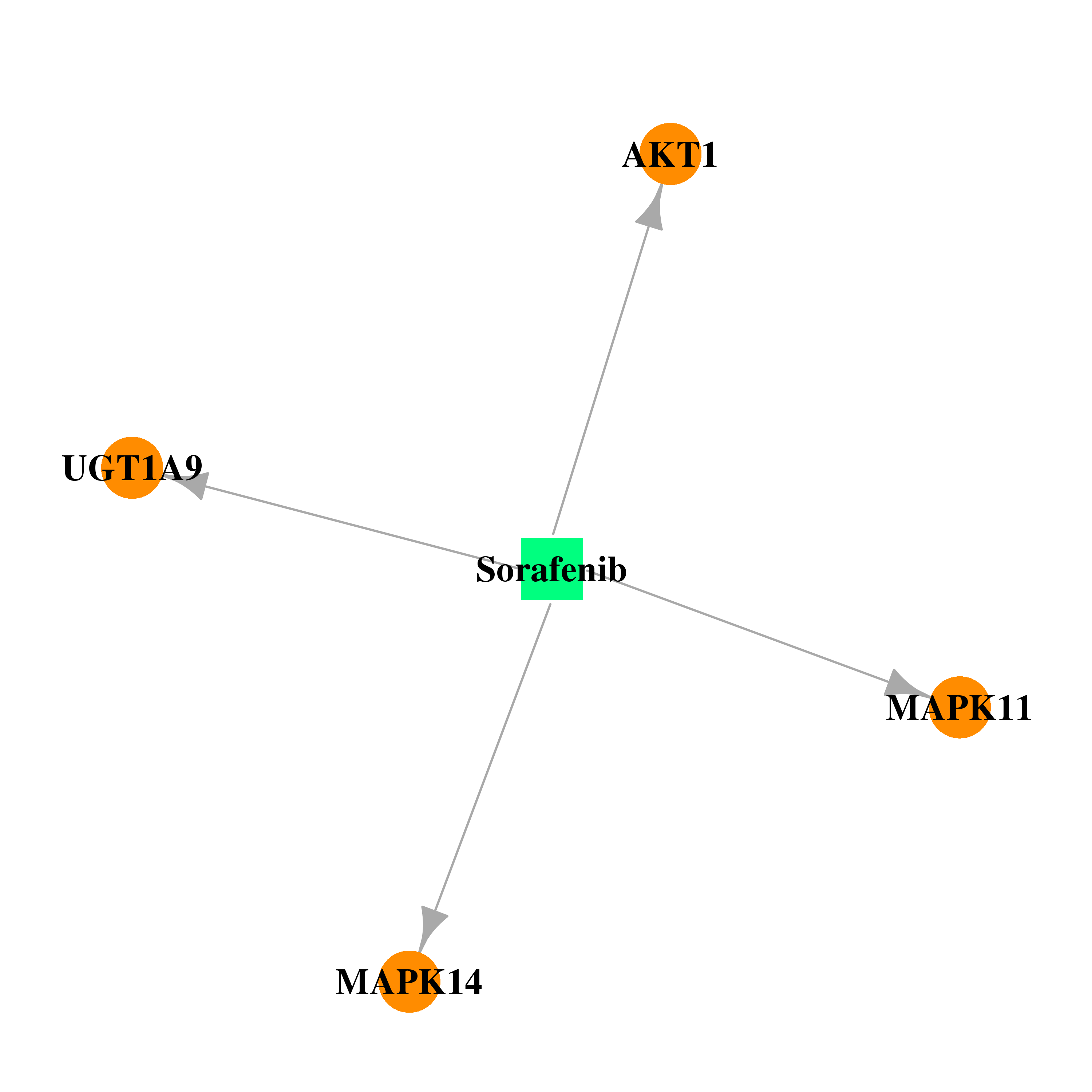 |  |
| DB00125 | mitogen-activated protein kinase 14 | approved; nutraceutical | L-Arginine | 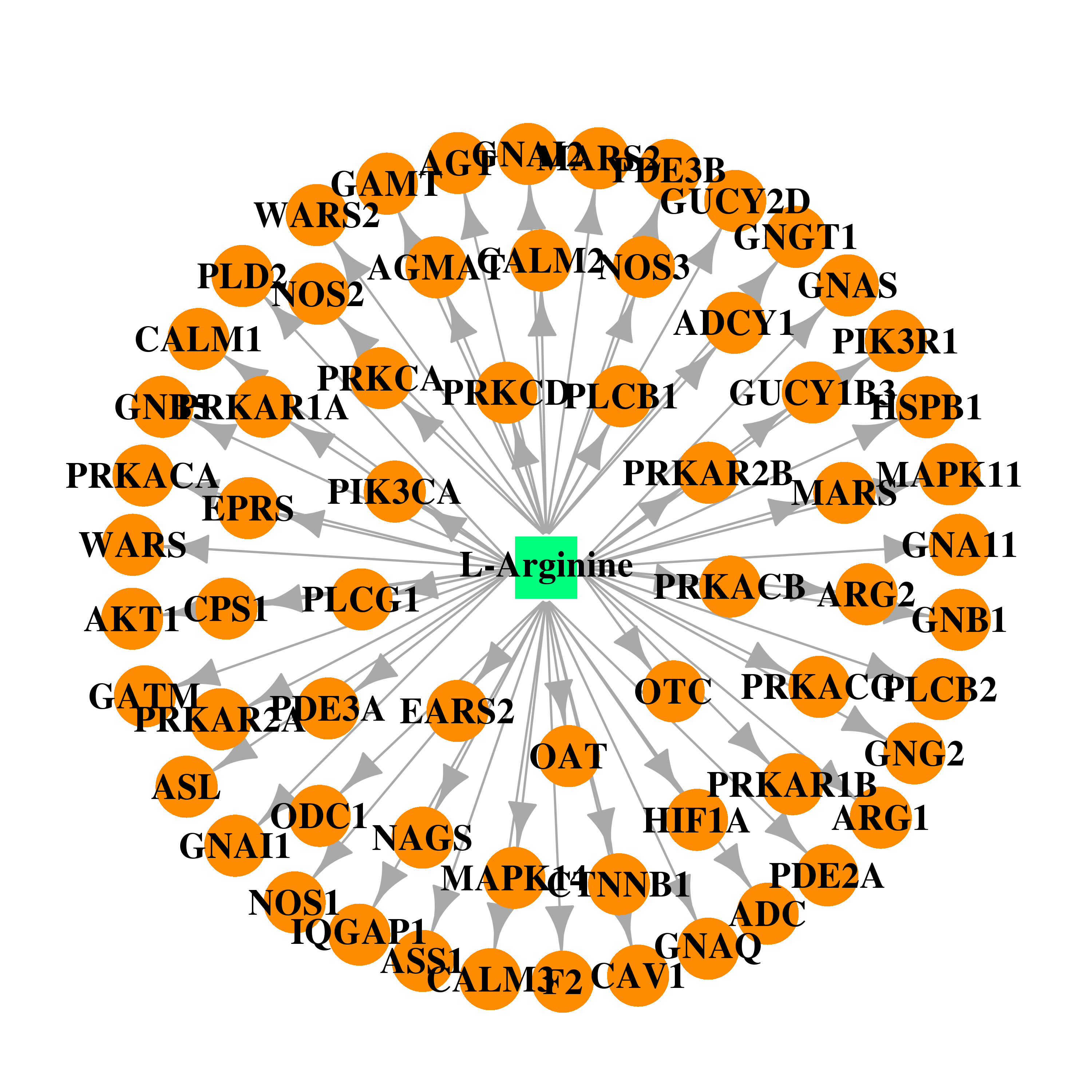 | 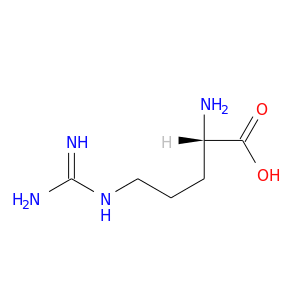 |
| DB00435 | mitogen-activated protein kinase 14 | approved | Nitric Oxide |  |  |
| Top |
| Cross referenced IDs for MAPK14 |
| * We obtained these cross-references from Uniprot database. It covers 150 different DBs, 18 categories. http://www.uniprot.org/help/cross_references_section |
: Open all cross reference information
|
Copyright © 2016-Present - The Univsersity of Texas Health Science Center at Houston @ |