|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |
| Phenotypic Information (metabolism pathway, cancer, disease, phenome) |
| |
| |
| Gene-Gene Network Information: Co-Expression Network, Interacting Genes & KEGG |
| |
|
| Gene Summary for DHFR |
| Basic gene info. | Gene symbol | DHFR |
| Gene name | dihydrofolate reductase | |
| Synonyms | DHFRP1|DYR | |
| Cytomap | UCSC genome browser: 5q11.2-q13.2 | |
| Genomic location | chr5 :79922044-79950800 | |
| Type of gene | protein-coding | |
| RefGenes | NM_000791.3, NM_001290354.1,NM_001290357.1,NR_110936.1, | |
| Ensembl id | ENSG00000228716 | |
| Description | - | |
| Modification date | 20141207 | |
| dbXrefs | MIM : 126060 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000228716 | ||
| HPRD : 00519 | ||
| Vega : OTTHUMG00000162529 | ||
| Protein | UniProt: go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_DHFR | |
| BioGPS: 1719 | ||
| Gene Expression Atlas: ENSG00000228716 | ||
| The Human Protein Atlas: ENSG00000228716 | ||
| Pathway | NCI Pathway Interaction Database: DHFR | |
| KEGG: DHFR | ||
| REACTOME: DHFR | ||
| ConsensusPathDB | ||
| Pathway Commons: DHFR | ||
| Metabolism | MetaCyc: DHFR | |
| HUMANCyc: DHFR | ||
| Regulation | Ensembl's Regulation: ENSG00000228716 | |
| miRBase: chr5 :79,922,044-79,950,800 | ||
| TargetScan: NM_000791 | ||
| cisRED: ENSG00000228716 | ||
| Context | iHOP: DHFR | |
| cancer metabolism search in PubMed: DHFR | ||
| UCL Cancer Institute: DHFR | ||
| Assigned class in ccmGDB | C | |
| Top |
| Phenotypic Information for DHFR(metabolism pathway, cancer, disease, phenome) |
| Cancer | CGAP: DHFR |
| Familial Cancer Database: DHFR | |
| * This gene is included in those cancer gene databases. |
|
|
|
|
|
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Oncogene 1 | Significant driver gene in | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| cf) number; DB name 1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 2 Tumor Suppressor gene; https://bioinfo.uth.edu/TSGene/, 3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html, 4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php, 1Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/ |
| REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | |
| OMIM | |
| Orphanet | |
| Disease | KEGG Disease: DHFR |
| MedGen: DHFR (Human Medical Genetics with Condition) | |
| ClinVar: DHFR | |
| Phenotype | MGI: DHFR (International Mouse Phenotyping Consortium) |
| PhenomicDB: DHFR | |
| Mutations for DHFR |
| * Under tables are showing count per each tissue to give us broad intuition about tissue specific mutation patterns.You can go to the detailed page for each mutation database's web site. |
| There's no structural variation information in COSMIC data for this gene. |
| * From mRNA Sanger sequences, Chitars2.0 arranged chimeric transcripts. This table shows DHFR related fusion information. |
| ID | Head Gene | Tail Gene | Accession | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a |
| M33552 | DHFR | 1 | 46 | 5 | 79946335 | 79946450 | LSP1 | 41 | 1631 | 11 | 1874307 | 1913492 | |
| BC068072 | ZMIZ1 | 1 | 1267 | 10 | 81070711 | 81073434 | DHFR | 1262 | 1315 | 5 | 79947669 | 79947722 | |
| DB727835 | DHFR | 2 | 66 | 5 | 79947600 | 79947664 | TM9SF3 | 62 | 675 | 10 | 98321775 | 98346664 | |
| DA541950 | DHFR | 1 | 107 | 5 | 79947318 | 79947424 | PSMD2 | 108 | 556 | 3 | 184019408 | 184020492 | |
| L42531 | GSS | 1 | 1690 | 20 | 33516405 | 33543560 | DHFR | 1691 | 1811 | 5 | 79946695 | 79946815 | |
| AK123238 | ZNF483 | 2 | 2294 | 9 | 114309598 | 114311891 | DHFR | 2285 | 2366 | 5 | 79946785 | 79946866 | |
| BQ328186 | DHFR | 181 | 259 | 5 | 79945841 | 79945919 | SPPL3 | 250 | 586 | 12 | 121222150 | 121229364 | |
| Top |
| Mutation type/ Tissue ID | brca | cns | cerv | endome | haematopo | kidn | Lintest | liver | lung | ns | ovary | pancre | prost | skin | stoma | thyro | urina | |||
| Total # sample | 1 | 1 | ||||||||||||||||||
| GAIN (# sample) | ||||||||||||||||||||
| LOSS (# sample) | 1 | 1 |
| cf) Tissue ID; Tissue type (1; Breast, 2; Central_nervous_system, 3; Cervix, 4; Endometrium, 5; Haematopoietic_and_lymphoid_tissue, 6; Kidney, 7; Large_intestine, 8; Liver, 9; Lung, 10; NS, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary_tract) |
| Top |
|
 |
| Top |
| Stat. for Non-Synonymous SNVs (# total SNVs=11) | (# total SNVs=2) |
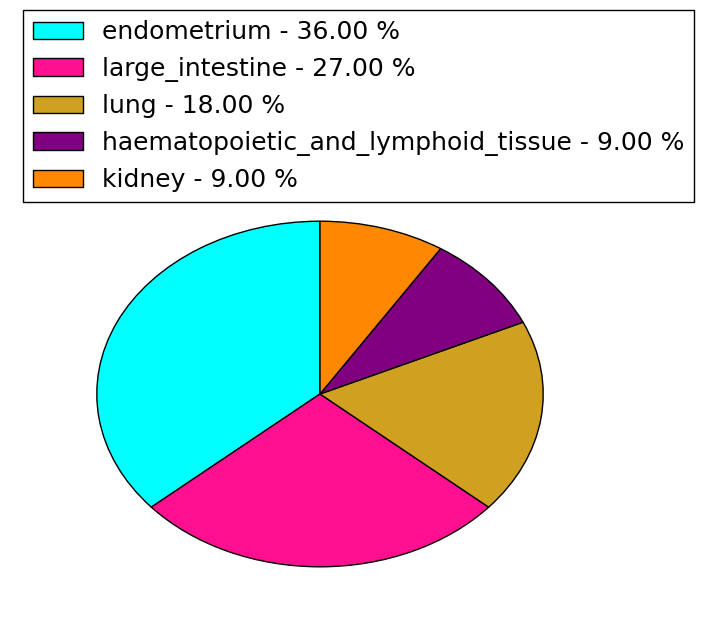 |  |
(# total SNVs=0) | (# total SNVs=0) |
| Top |
| * When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site,primary_histology,mutation(aa),pubmedID. |
| GRCh37 position | Mutation(aa) | Unique sampleID count |
| chr5:79945224-79945224 | p.L76I | 2 |
| chr5:79945254-79945254 | p.R66* | 2 |
| chr5:79945226-79945226 | p.V75A | 1 |
| chr5:79924930-79924930 | p.F180F | 1 |
| chr5:79945268-79945268 | p.I61T | 1 |
| chr5:79924953-79924953 | p.E173K | 1 |
| chr5:79949865-79949865 | p.R33T | 1 |
| chr5:79924971-79924971 | p.L167I | 1 |
| chr5:79924972-79924972 | p.V166V | 1 |
| chr5:79933712-79933712 | p.S120Y | 1 |
| Top |
|
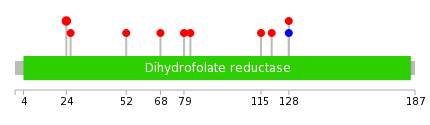 |
| Point Mutation/ Tissue ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| # sample | 1 | 3 | 1 | 1 | 4 | |||||||||||||||
| # mutation | 1 | 3 | 1 | 1 | 5 | |||||||||||||||
| nonsynonymous SNV | 1 | 3 | 1 | 1 | 4 | |||||||||||||||
| synonymous SNV | 1 |
| cf) Tissue ID; Tissue type (1; BLCA[Bladder Urothelial Carcinoma], 2; BRCA[Breast invasive carcinoma], 3; CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], 4; COAD[Colon adenocarcinoma], 5; GBM[Glioblastoma multiforme], 6; Glioma Low Grade, 7; HNSC[Head and Neck squamous cell carcinoma], 8; KICH[Kidney Chromophobe], 9; KIRC[Kidney renal clear cell carcinoma], 10; KIRP[Kidney renal papillary cell carcinoma], 11; LAML[Acute Myeloid Leukemia], 12; LUAD[Lung adenocarcinoma], 13; LUSC[Lung squamous cell carcinoma], 14; OV[Ovarian serous cystadenocarcinoma ], 15; PAAD[Pancreatic adenocarcinoma], 16; PRAD[Prostate adenocarcinoma], 17; SKCM[Skin Cutaneous Melanoma], 18:STAD[Stomach adenocarcinoma], 19:THCA[Thyroid carcinoma], 20:UCEC[Uterine Corpus Endometrial Carcinoma]) |
| Top |
| * We represented just top 10 SNVs. When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site, primary_histology, mutation(aa), pubmedID. |
| Genomic Position | Mutation(aa) | Unique sampleID count |
| chr5:79945224 | p.L24I,DHFR | 2 |
| chr5:79945217 | p.F128F,DHFR | 1 |
| chr5:79924930 | p.E120D,DHFR | 1 |
| chr5:79924954 | p.L115I,DHFR | 1 |
| chr5:79924971 | p.S128A | 1 |
| chr5:79924972 | p.L82I,DHFR | 1 |
| chr5:79929781 | p.H79Y,DHFR | 1 |
| chr5:79929790 | p.S68Y,DHFR | 1 |
| chr5:79933712 | p.P52Q,DHFR | 1 |
| chr5:79933760 | p.R26K,DHFR | 1 |
| * Copy number data were extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered on Jan-05-2015. Function ProcessCNAData in TCGA-Assembler package was used to obtain gene-level copy number value which is calculated as the average copy number of the genomic region of a gene. |
 |
| cf) Tissue ID[Tissue type]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma], KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ], PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma] |
| Top |
| Gene Expression for DHFR |
| * CCLE gene expression data were extracted from CCLE_Expression_Entrez_2012-10-18.res: Gene-centric RMA-normalized mRNA expression data. |
 |
| * Normalized gene expression data of RNASeqV2 was extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered at Jan-05-2015. Only eight cancer types have enough normal control samples for differential expression analysis. (t test, adjusted p<0.05 (using Benjamini-Hochberg FDR)) |
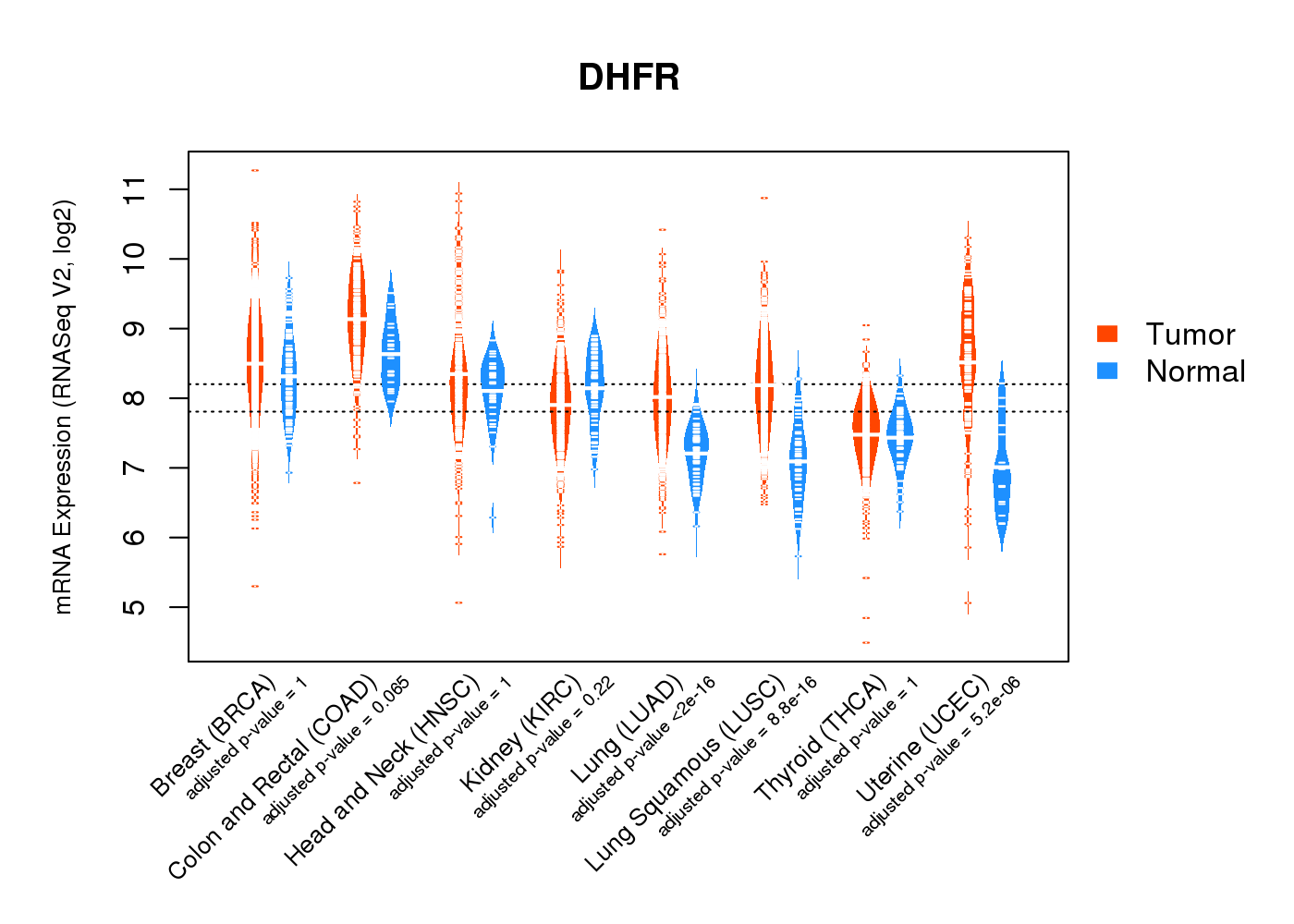 |
| Top |
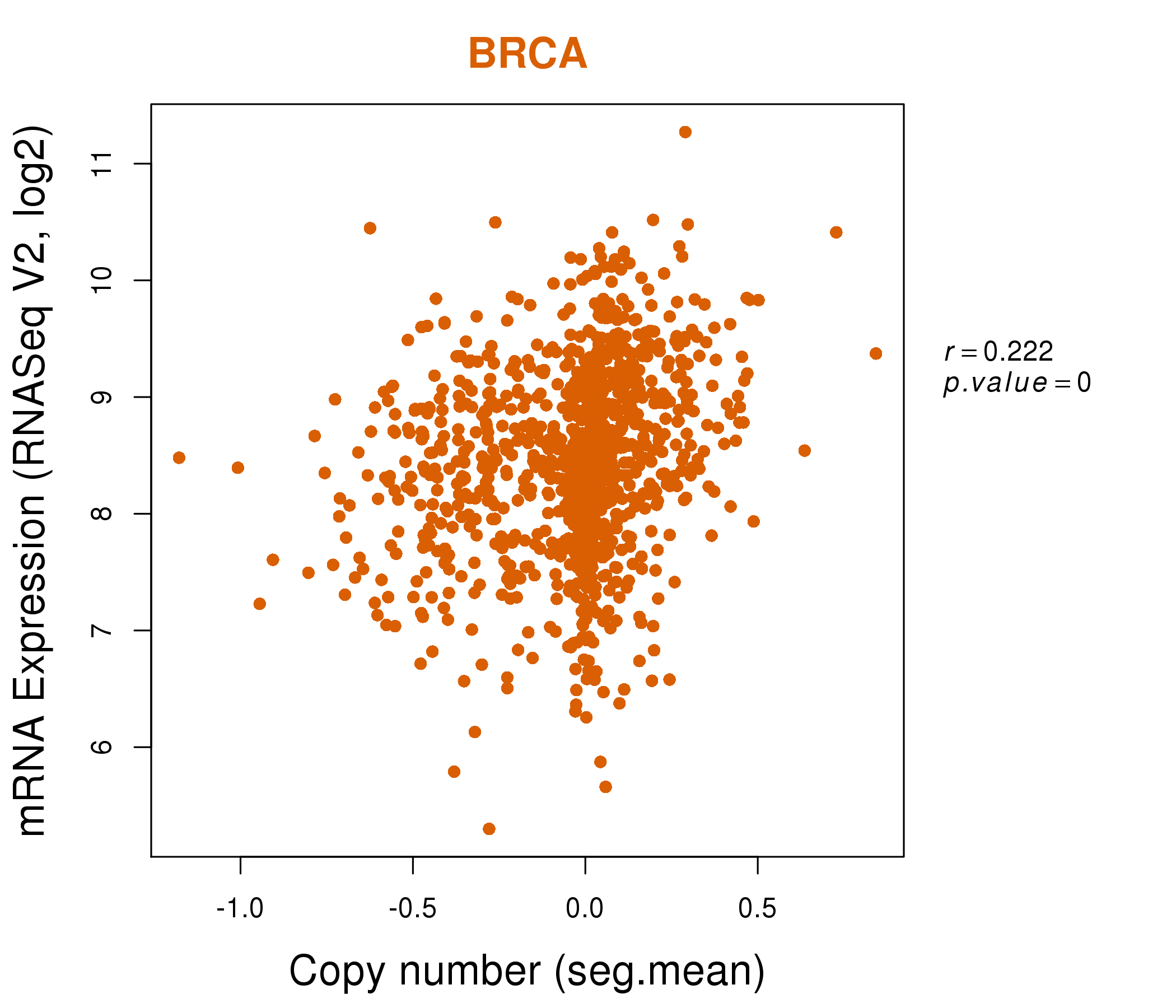
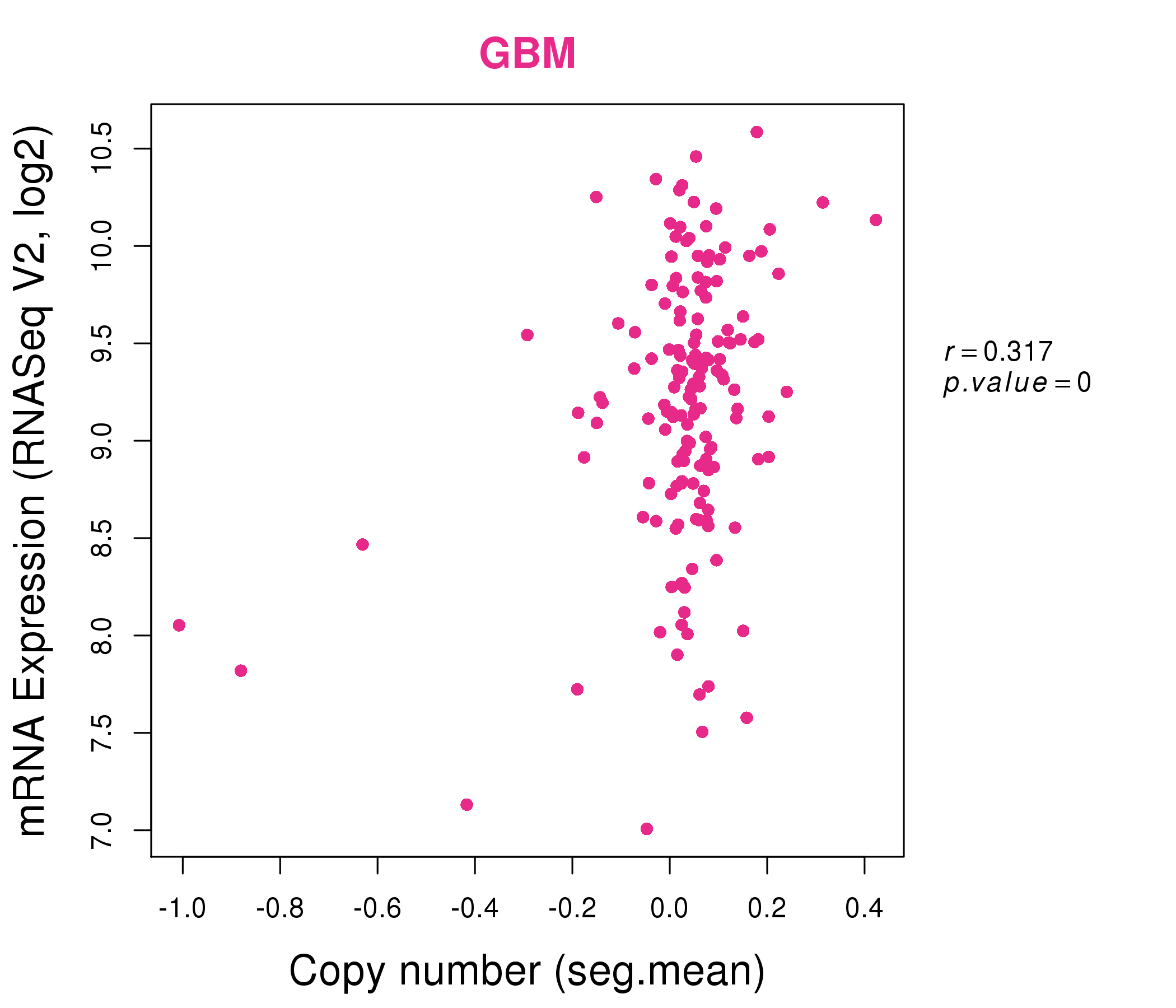
| * This plots show the correlation between CNV and gene expression. |
: Open all plots for all cancer types
 |
|
 |
|
| Top |
| Gene-Gene Network Information |
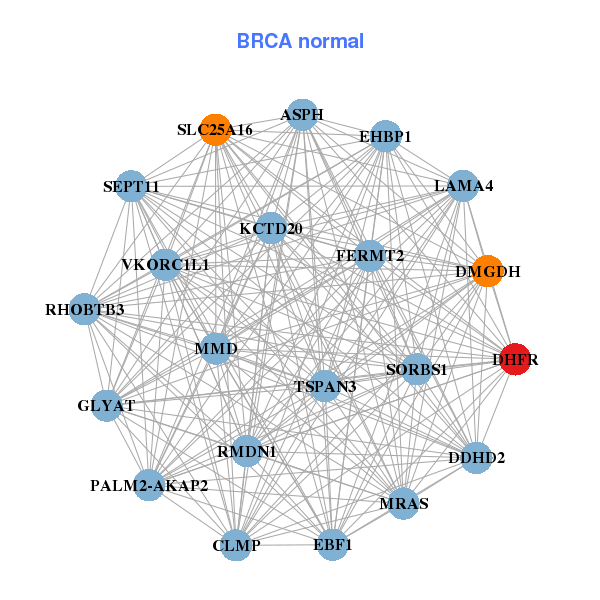
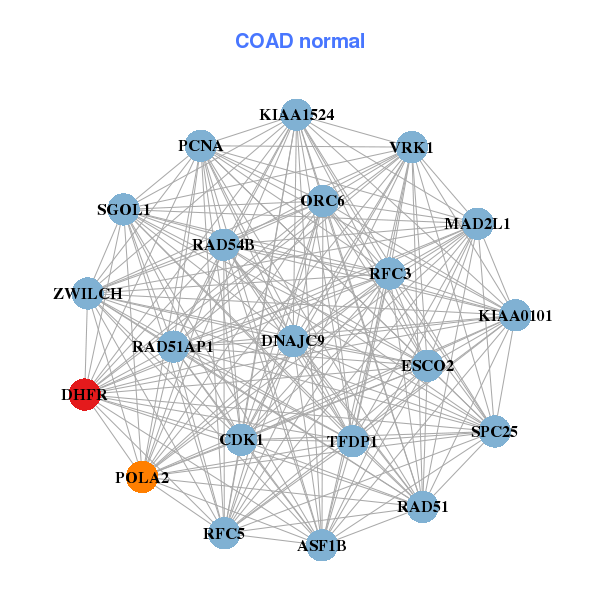
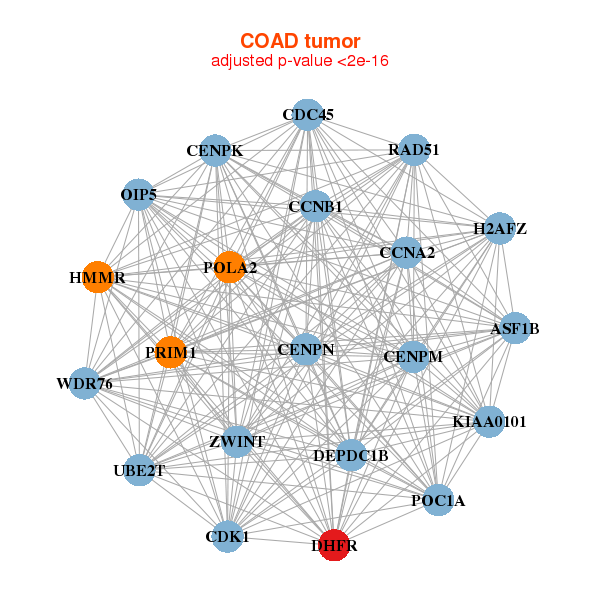
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
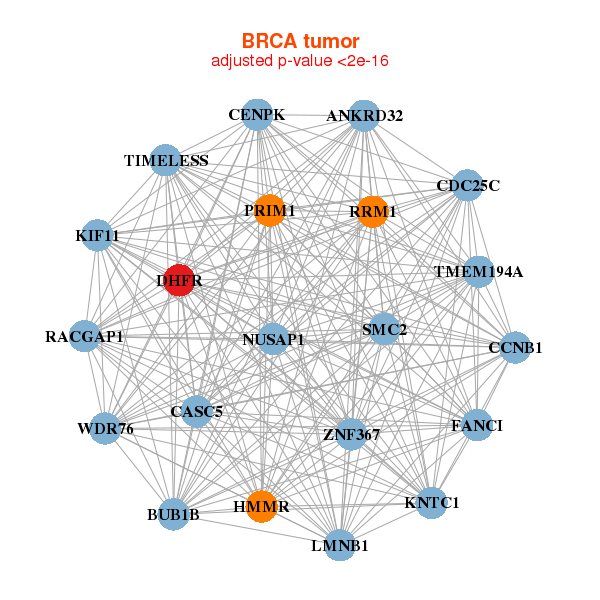 |
| ||||
| ANKRD32,BUB1B,CASC5,CCNB1,CDC25C,CENPK,DHFR, FANCI,HMMR,KIF11,KNTC1,LMNB1,NUSAP1,PRIM1, RACGAP1,RRM1,SMC2,TIMELESS,TMEM194A,WDR76,ZNF367 | CLMP,ASPH,DDHD2,DHFR,DMGDH,EBF1,EHBP1, RMDN1,FERMT2,GLYAT,KCTD20,LAMA4,MMD,MRAS, PALM2-AKAP2,RHOBTB3,SEPT11,SLC25A16,SORBS1,TSPAN3,VKORC1L1 | ||||
 |
| ||||
| ASF1B,CCNA2,CCNB1,CDC45,CDK1,CENPK,CENPM, CENPN,DEPDC1B,DHFR,H2AFZ,HMMR,KIAA0101,OIP5, POC1A,POLA2,PRIM1,RAD51,UBE2T,WDR76,ZWINT | ASF1B,CDK1,DHFR,DNAJC9,ESCO2,KIAA0101,KIAA1524, MAD2L1,ORC6,PCNA,POLA2,RAD51,RAD51AP1,RAD54B, RFC3,RFC5,SGOL1,SPC25,TFDP1,VRK1,ZWILCH |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
| Top |
: Open all interacting genes' information including KEGG pathway for all interacting genes from DAVID
| Top |
| Pharmacological Information for DHFR |
| DB Category | DB Name | DB's ID and Url link |
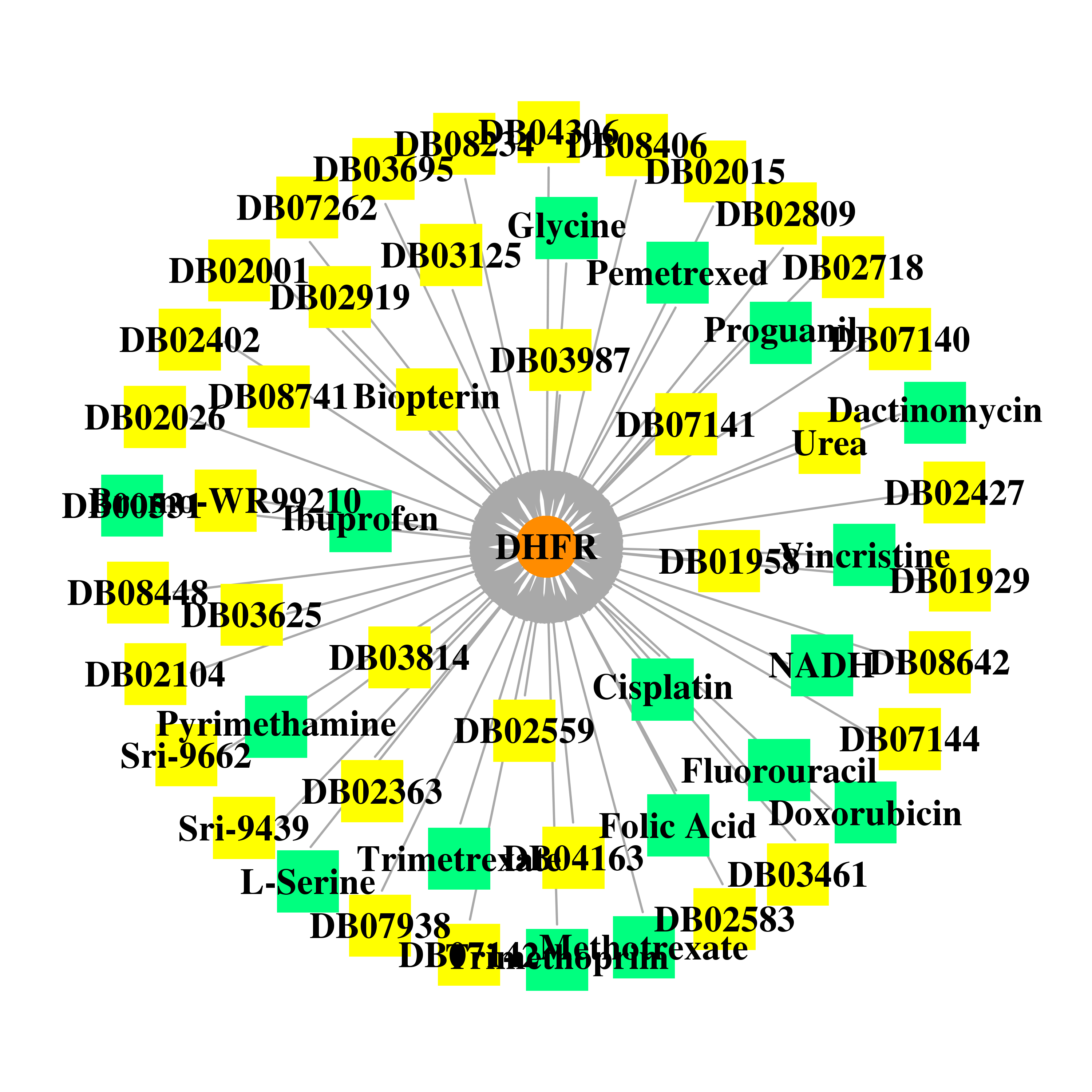
| * Gene Centered Interaction Network. |
 |
| * Drug Centered Interaction Network. |
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB00157 | dihydrofolate reductase | approved; nutraceutical | NADH |  |  |
| DB00205 | dihydrofolate reductase | approved | Pyrimethamine | 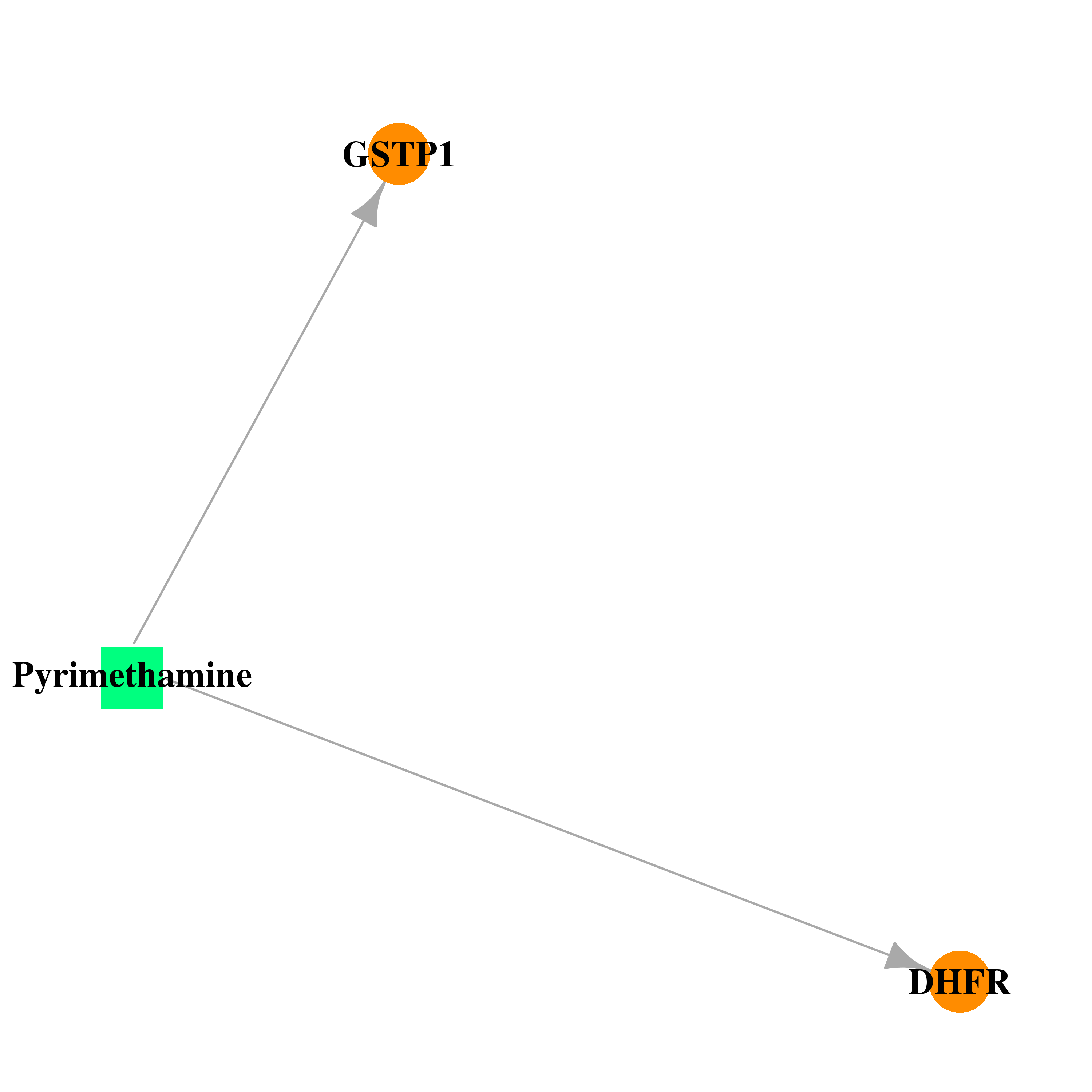 |  |
| DB00440 | dihydrofolate reductase | approved | Trimethoprim | 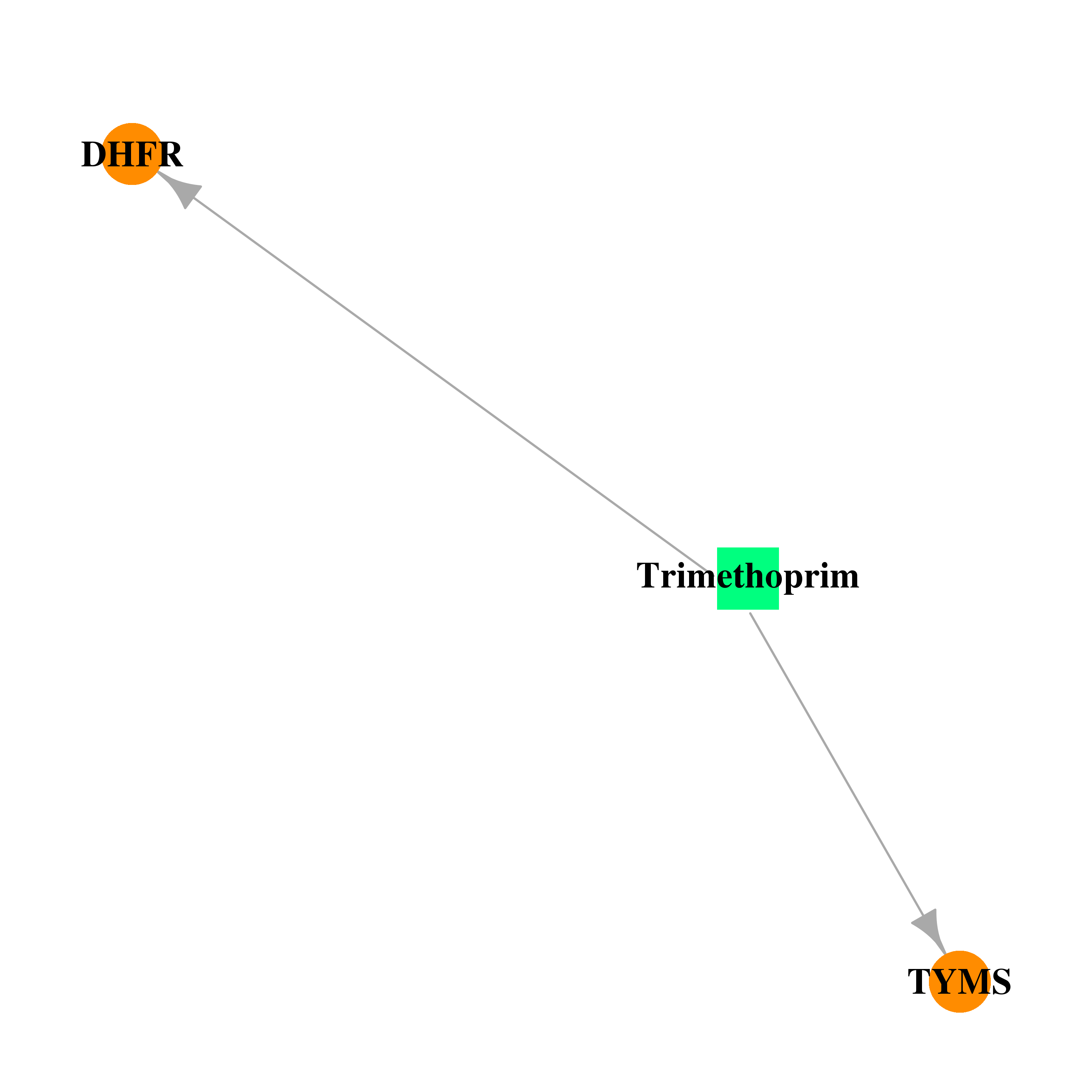 |  |
| DB00563 | dihydrofolate reductase | approved | Methotrexate |  |  |
| DB00642 | dihydrofolate reductase | approved; investigational | Pemetrexed |  |  |
| DB01131 | dihydrofolate reductase | approved | Proguanil |  | 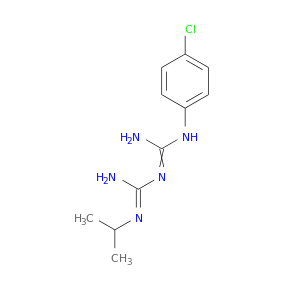 |
| DB01157 | dihydrofolate reductase | approved | Trimetrexate |  | 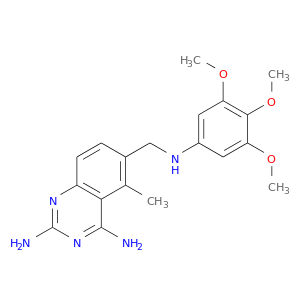 |
| DB02104 | dihydrofolate reductase | experimental | 2,4-Diamino-5-Methyl-6-[(3,4,5-Trimethoxy-N-Methylanilino)Methyl]Pyrido[2,3-D]Pyrimidine |  | 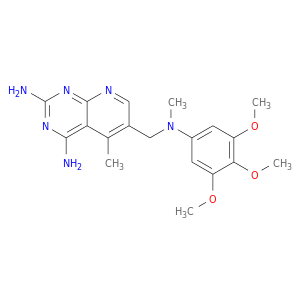 |
| DB02427 | dihydrofolate reductase | experimental | 2,4-Diamino-6-[N-(2',5'-Dimethoxybenzyl)-N-Methylamino]Quinazoline |  | 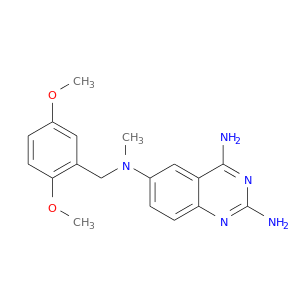 |
| DB02559 | dihydrofolate reductase | experimental | 6-(Octahydro-1h-Indol-1-Ylmethyl)Decahydroquinazoline-2,4-Diamine |  |  |
| DB02583 | dihydrofolate reductase | experimental | N6-(2,5-Dimethoxy-Benzyl)-N6-Methyl-Pyrido[2,3-D]Pyrimidine-2,4,6-Triamine |  |  |
| DB02919 | dihydrofolate reductase | experimental | 2,4-Diamino-6-[N-(3',4',5'-Trimethoxybenzyl)-N-Methylamino]Pyrido[2,3-D]Pyrimidine |  |  |
| DB03060 | dihydrofolate reductase | experimental | Sri-9662 |  |  |
| DB03125 | dihydrofolate reductase | experimental | 2,4-Diamino-5-(3,4,5-Trimethoxy-Benzyl)-Pyrimidin-1-Ium |  |  |
| DB03351 | dihydrofolate reductase | experimental | Sri-9439 |  |  |
| DB03461 | dihydrofolate reductase | experimental | 2'-Monophosphoadenosine 5'-Diphosphoribose | 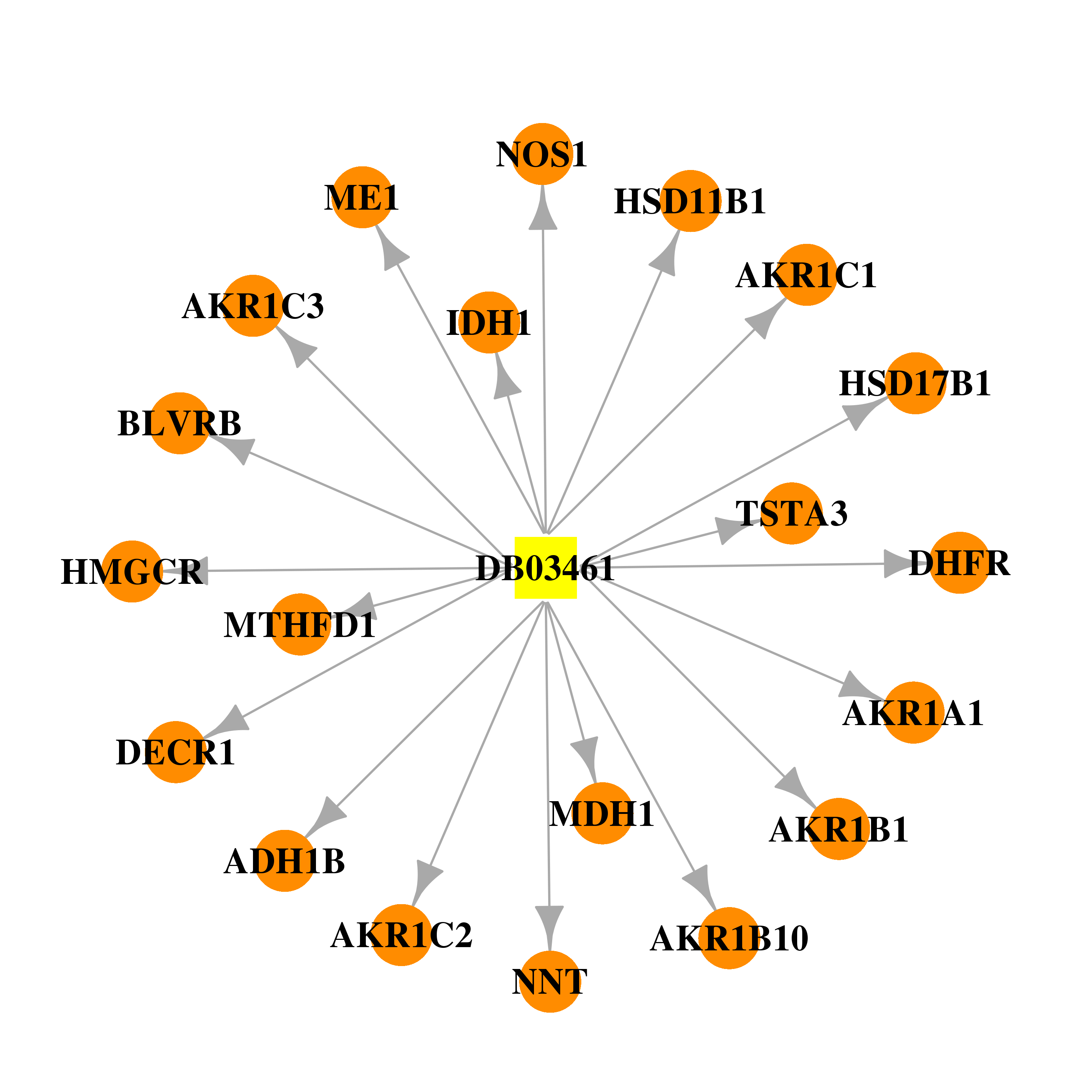 |  |
| DB03695 | dihydrofolate reductase | experimental | 6-(2,5-Dimethoxy-Benzyl)-5-Methyl-Pyrido[2,3-D]Pyrimidine-2,4-Diamine | 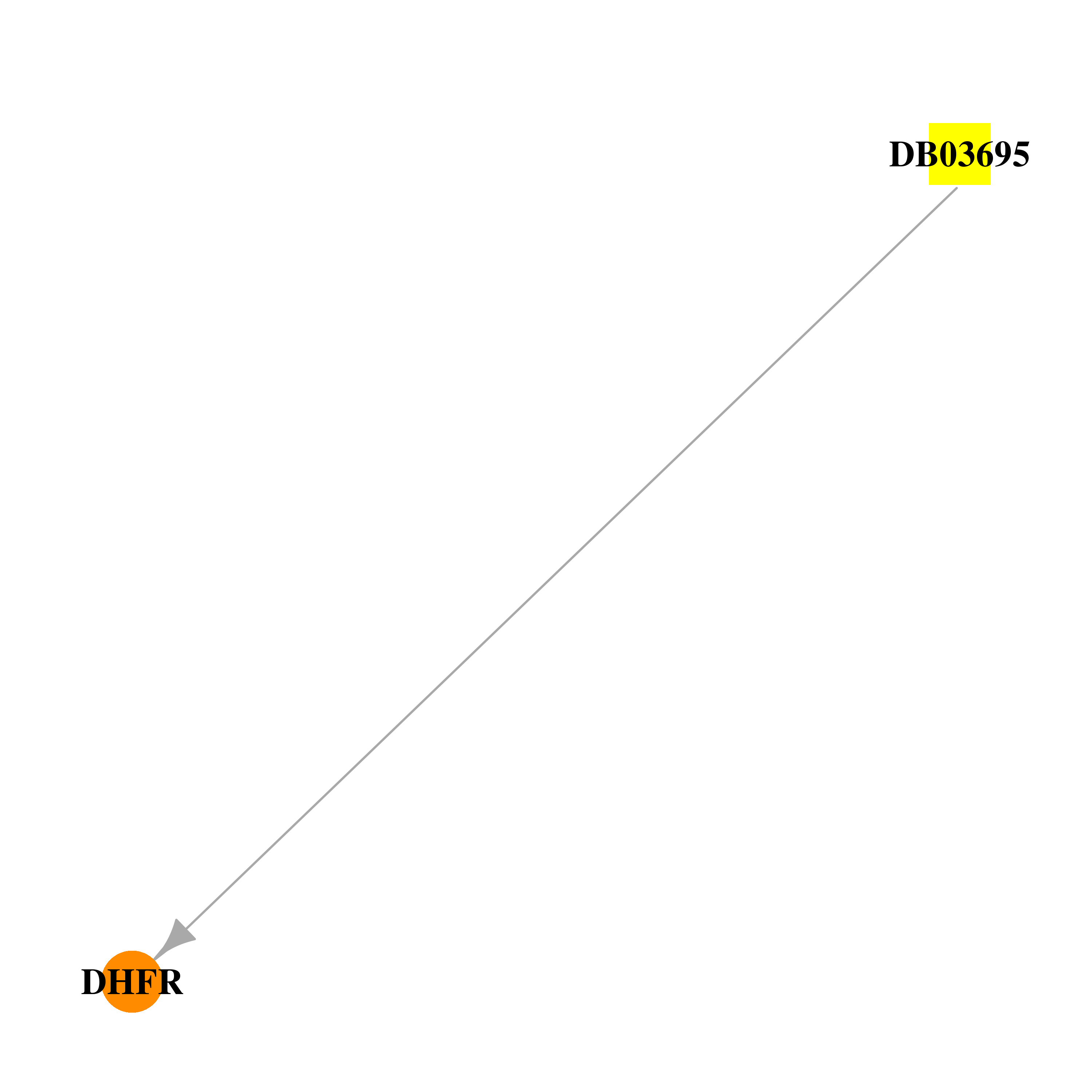 | 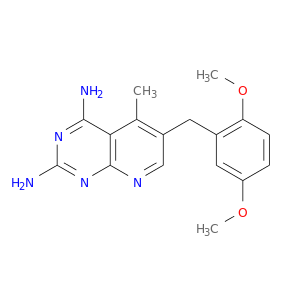 |
| DB03886 | dihydrofolate reductase | experimental | Biopterin | 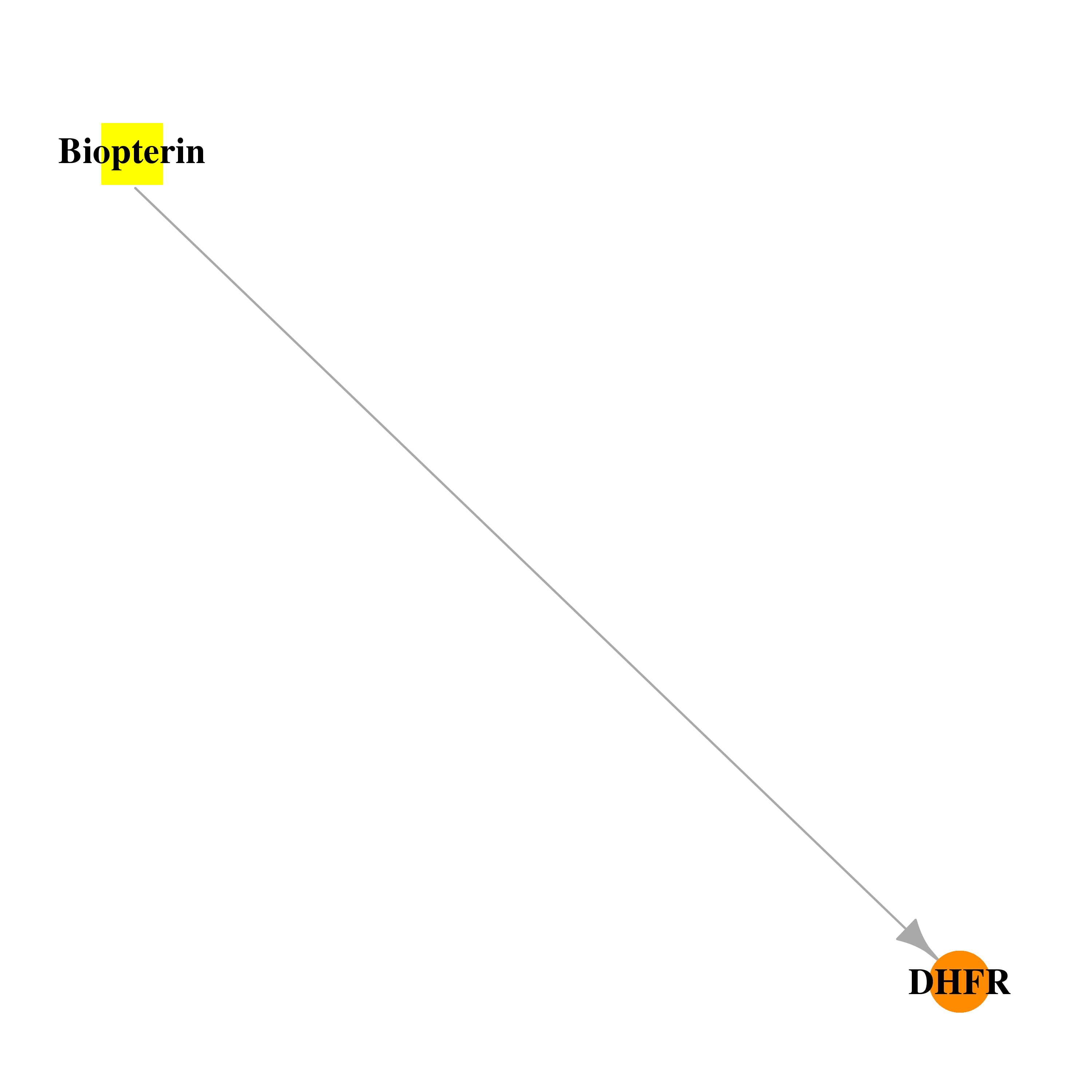 |  |
| DB03987 | dihydrofolate reductase | experimental | 2,4-Diamino-6-[N-(3',5'-Dimethoxybenzyl)-N-Methylamino]Pyrido[2,3-D]Pyrimidine |  |  |
| DB07140 | dihydrofolate reductase | experimental | 5-[(3R)-3-(5-methoxybiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine |  | 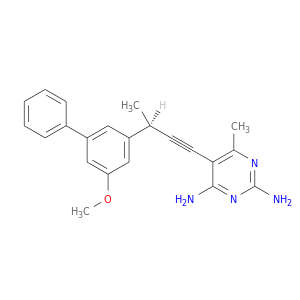 |
| DB07141 | dihydrofolate reductase | experimental | 5-[(3R)-3-(5-methoxy-4'-methylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine |  | 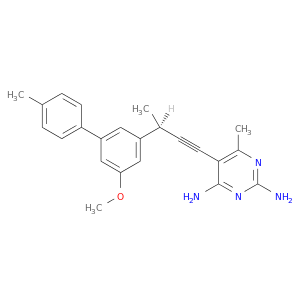 |
| DB07142 | dihydrofolate reductase | experimental | 5-[(3R)-3-(5-methoxy-3',5'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine |  | 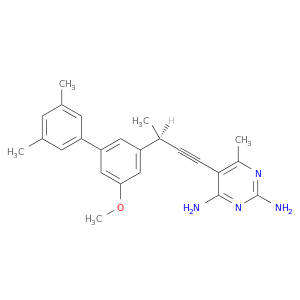 |
| DB07144 | dihydrofolate reductase | experimental | 5-[(3R)-3-(5-methoxy-2',6'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine |  |  |
| DB08234 | dihydrofolate reductase | experimental | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine |  | 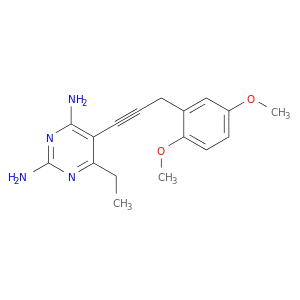 |
| DB08406 | dihydrofolate reductase | experimental | [N-(2,4-DIAMINOPTERIDIN-6-YL)-METHYL]-DIBENZ[B,F]AZEPINE |  |  |
| DB08448 | dihydrofolate reductase | experimental | (4aS)-5-[(2,4-diaminopteridin-6-yl)methyl]-4a,5-dihydro-2H-dibenzo[b,f]azepin-8-ol |  |  |
| DB08642 | dihydrofolate reductase | experimental | (2R,6S)-6-{[methyl(3,4,5-trimethoxyphenyl)amino]methyl}-1,2,5,6,7,8-hexahydroquinazoline-2,4-diamine |  |  |
| DB01929 | dihydrofolate reductase | experimental | 5-Chloryl-2,4,6-Quinazolinetriamine |  |  |
| DB01958 | dihydrofolate reductase | experimental | 5-[4-Tert-Butylphenylsulfanyl]-2,4-Quinazolinediamine |  |  |
| DB02001 | dihydrofolate reductase | experimental | 5-(4-Morpholin-4-Yl-Phenylsulfanyl)-2,4-Quinazolinediamine |  |  |
| DB02402 | dihydrofolate reductase | experimental | 5-(4-Methoxyphenoxy)-2,4-Quinazolinediamine |  |  |
| DB03814 | dihydrofolate reductase | experimental | 2-(N-Morpholino)-Ethanesulfonic Acid | 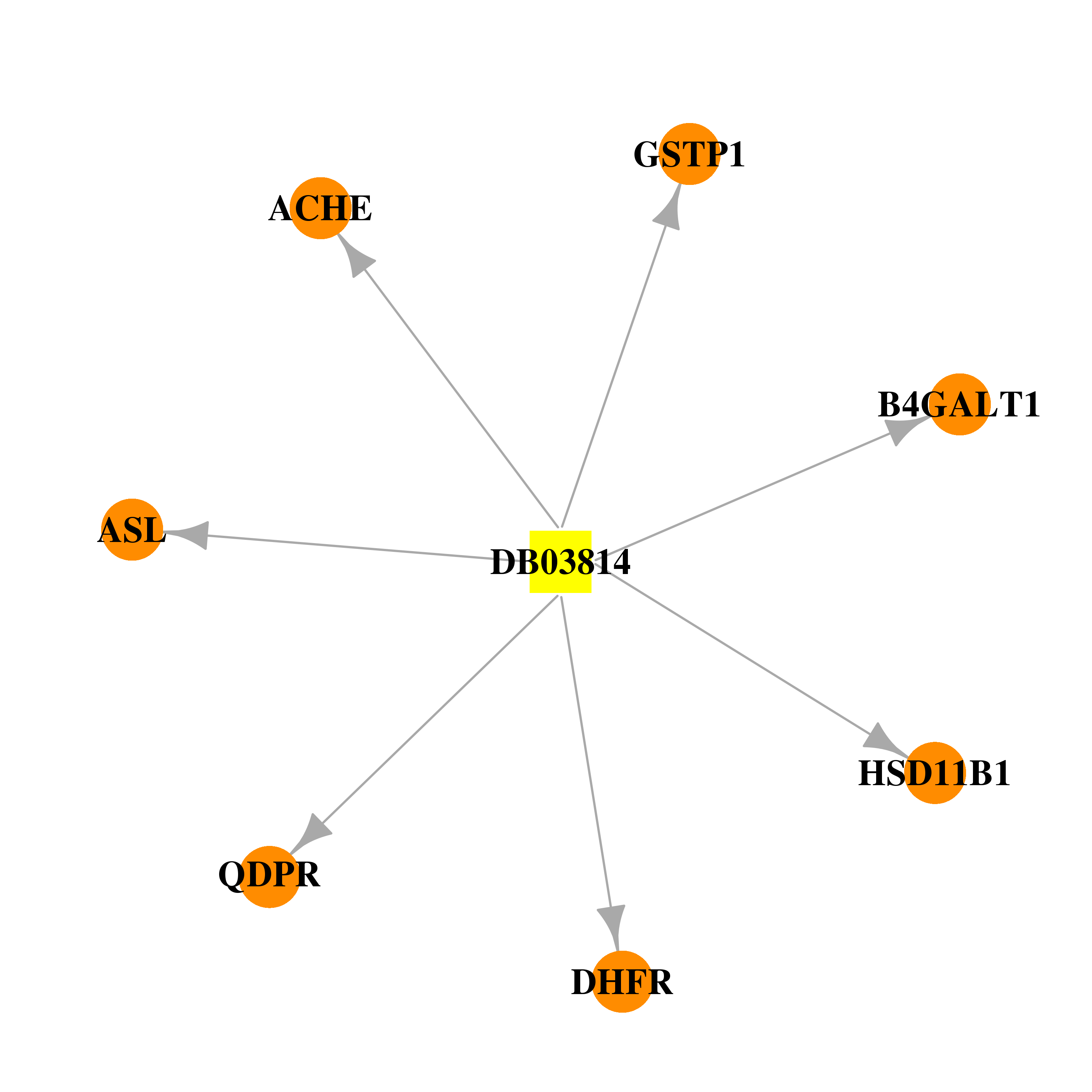 |  |
| DB04163 | dihydrofolate reductase | experimental | 5-Phenylsulfanyl-2,4-Quinazolinediamine |  |  |
| DB04306 | dihydrofolate reductase | experimental | 5-[(4-Methylphenyl)Sulfanyl]-2,4-Quinazolinediamine |  |  |
| DB02026 | dihydrofolate reductase | experimental | Furo[2,3d]Pyrimidine Antifolate |  | 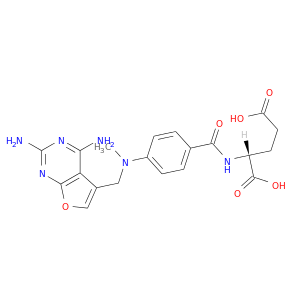 |
| DB02809 | dihydrofolate reductase | experimental | Brodimoprim-4,6-Dicarboxylate |  | 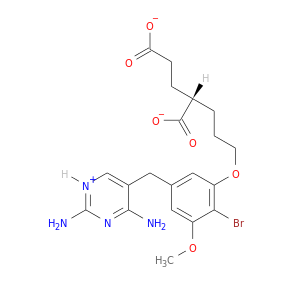 |
| DB04007 | dihydrofolate reductase | experimental | Bromo-WR99210 |  | 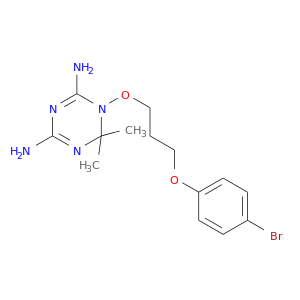 |
| DB02015 | dihydrofolate reductase | experimental | Dihydrofolic Acid |  |  |
| DB02363 | dihydrofolate reductase | experimental | 2'-Monophosphoadenosine-5'-Diphosphate |  |  |
| DB02718 | dihydrofolate reductase | experimental | 5-Formyl-6-Hydrofolic Acid |  |  |
| DB03625 | dihydrofolate reductase | experimental | 5,10-Dideazatetrahydrofolic Acid |  | 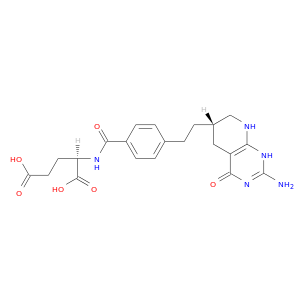 |
| DB03904 | dihydrofolate reductase | experimental | Urea |  | 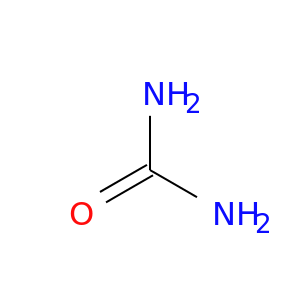 |
| DB07262 | dihydrofolate reductase | experimental | 1-{[N-(1-IMINO-GUANIDINO-METHYL)]SULFANYLMETHYL}-3-TRIFLUOROMETHYL-BENZENE |  | 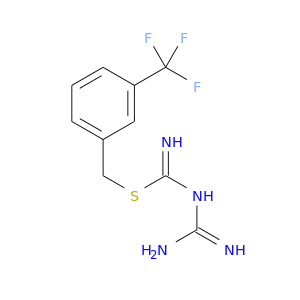 |
| DB07938 | dihydrofolate reductase | experimental | 5-[[(2R)-2-cyclopropyl-7,8-dimethoxy-chroman-5-yl]methyl]pyrimidine-2,4-diamine |  |  |
| DB08741 | dihydrofolate reductase | experimental | 5-[[(2R)-2-cyclopropyl-7,8-dimethoxy-2H-chromen-5-yl]methyl]pyrimidine-2,4-diamine |  | 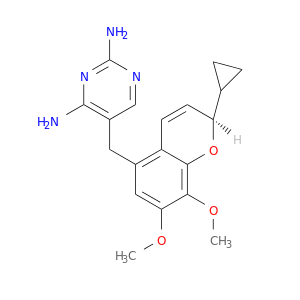 |
| DB00515 | dihydrofolate reductase | approved | Cisplatin |  | 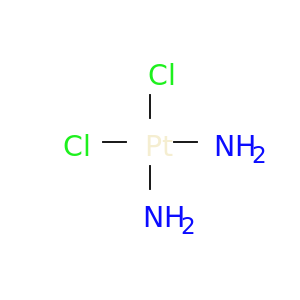 |
| DB00531 | dihydrofolate reductase | approved; investigational | Cyclophosphamide | 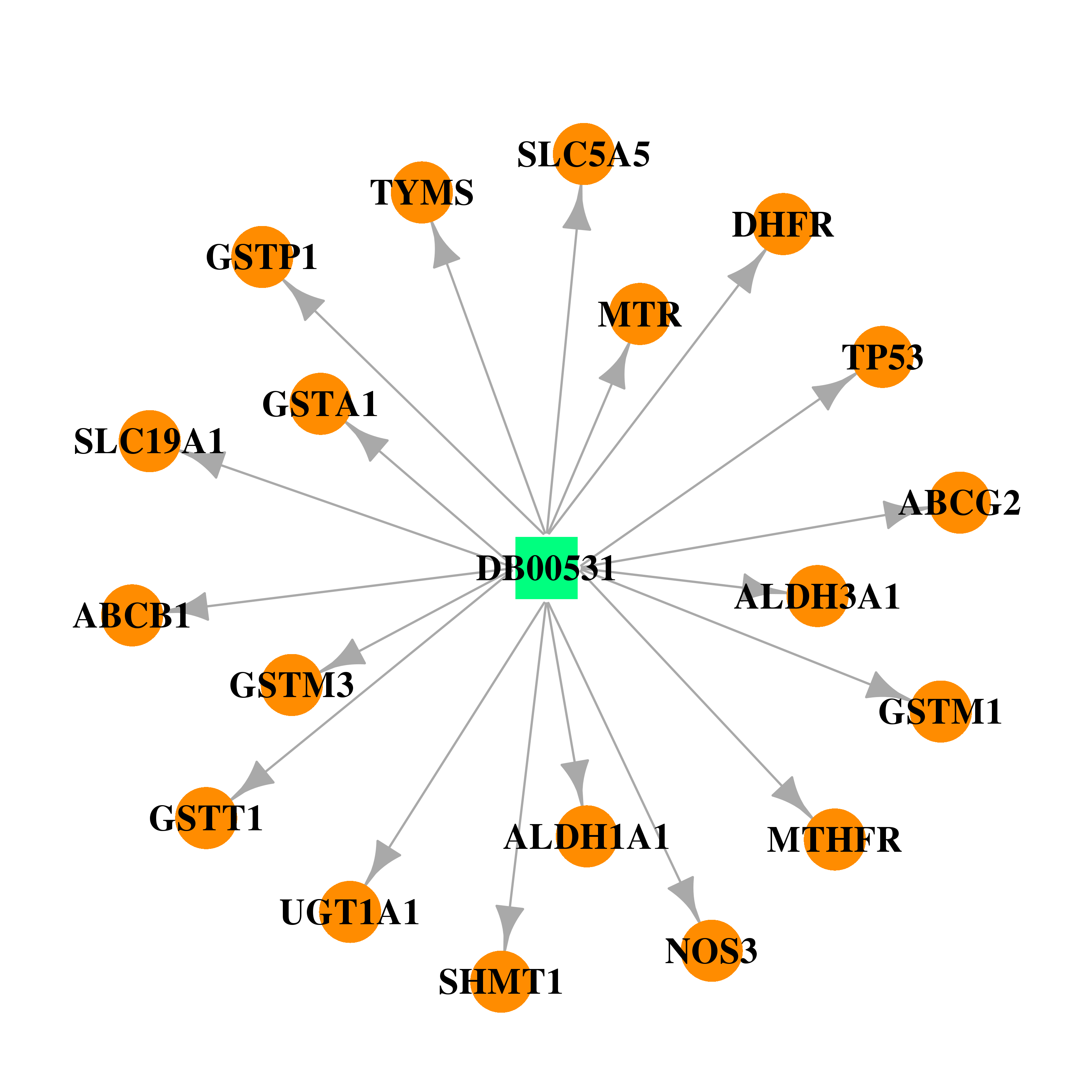 | 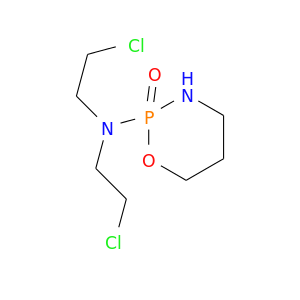 |
| DB00970 | dihydrofolate reductase | approved | Dactinomycin | 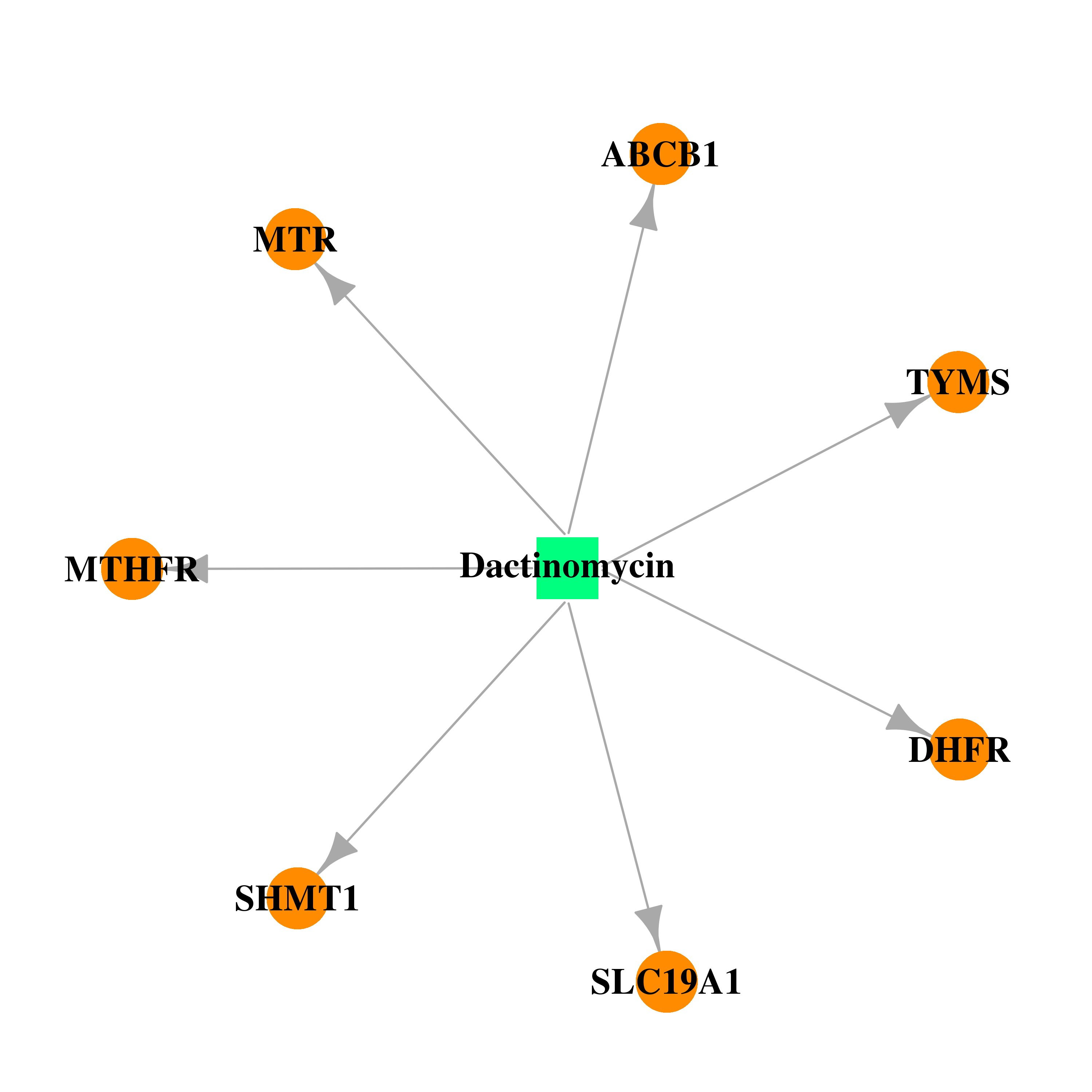 | 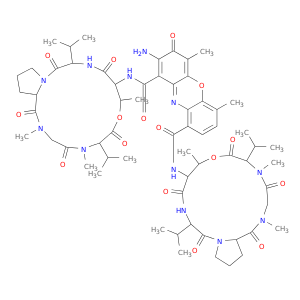 |
| DB00997 | dihydrofolate reductase | approved; investigational | Doxorubicin |  |  |
| DB00541 | dihydrofolate reductase | approved; investigational | Vincristine |  |  |
| DB00145 | dihydrofolate reductase | approved; nutraceutical | Glycine | 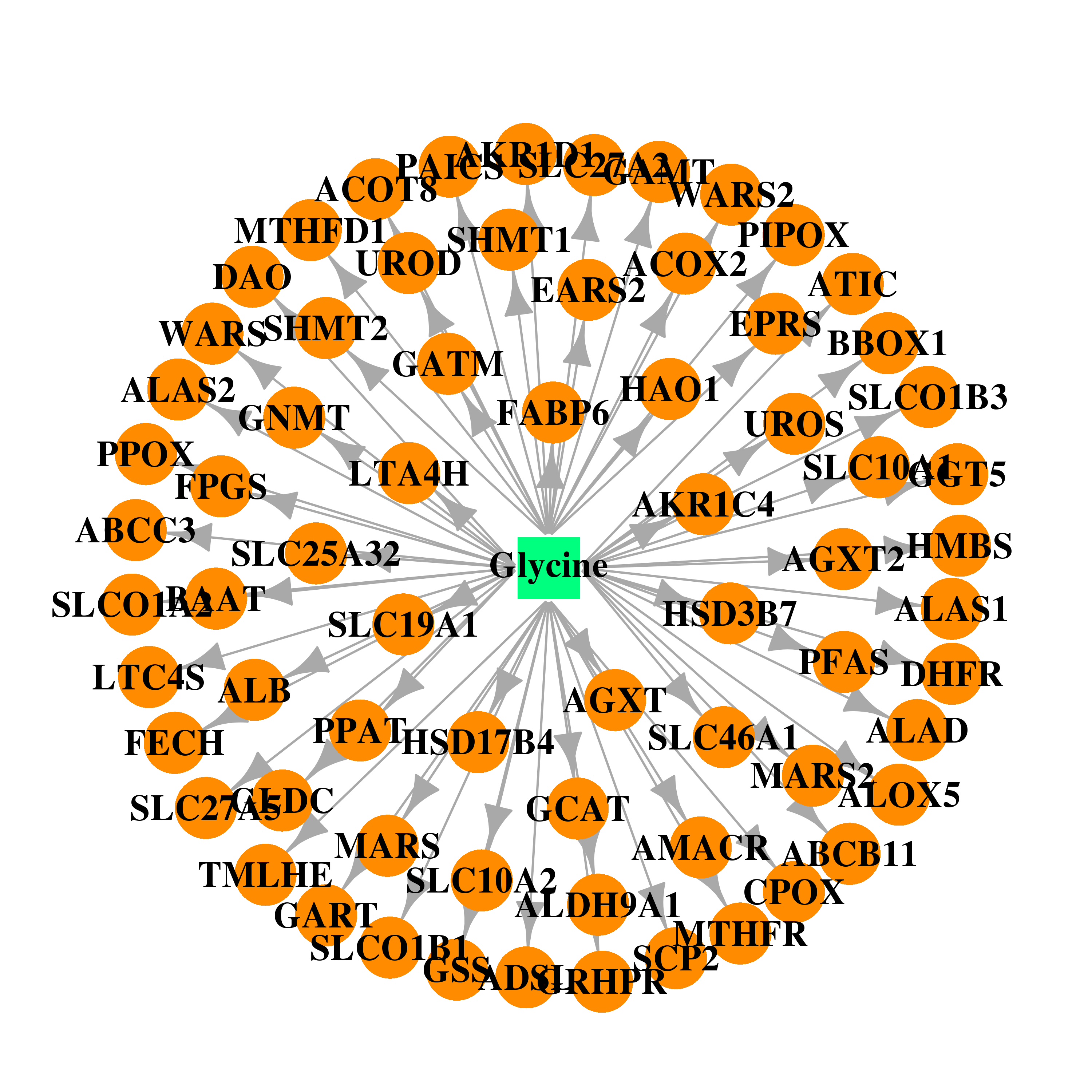 | 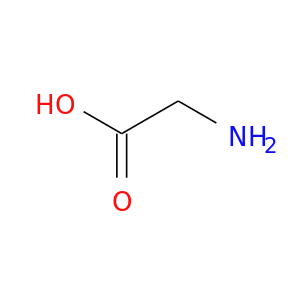 |
| DB00133 | dihydrofolate reductase | approved; nutraceutical | L-Serine | 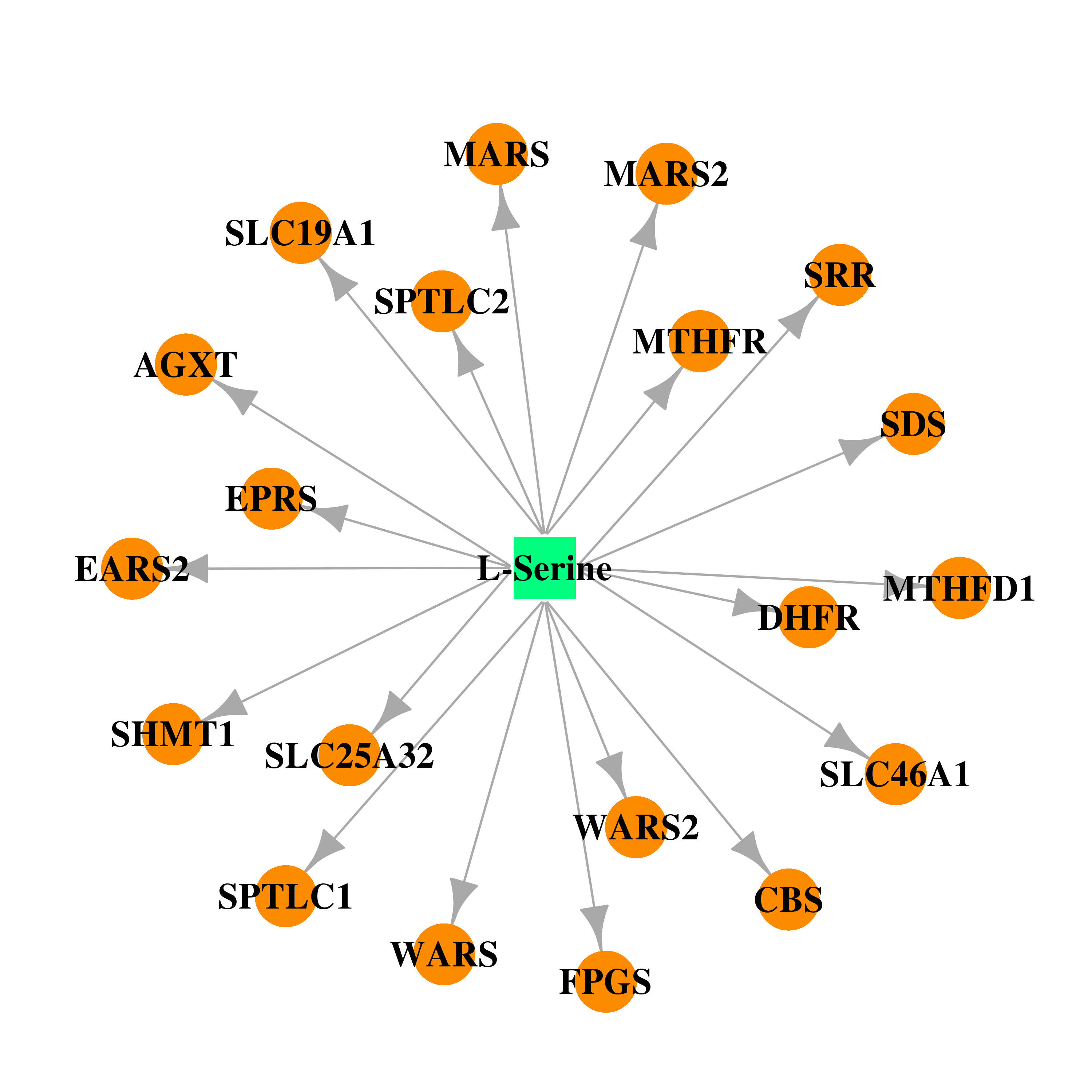 | 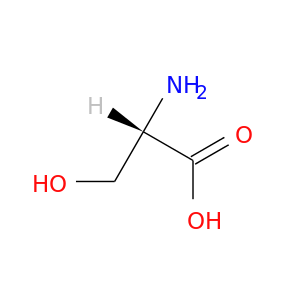 |
| DB00158 | dihydrofolate reductase | approved; nutraceutical | Folic Acid | 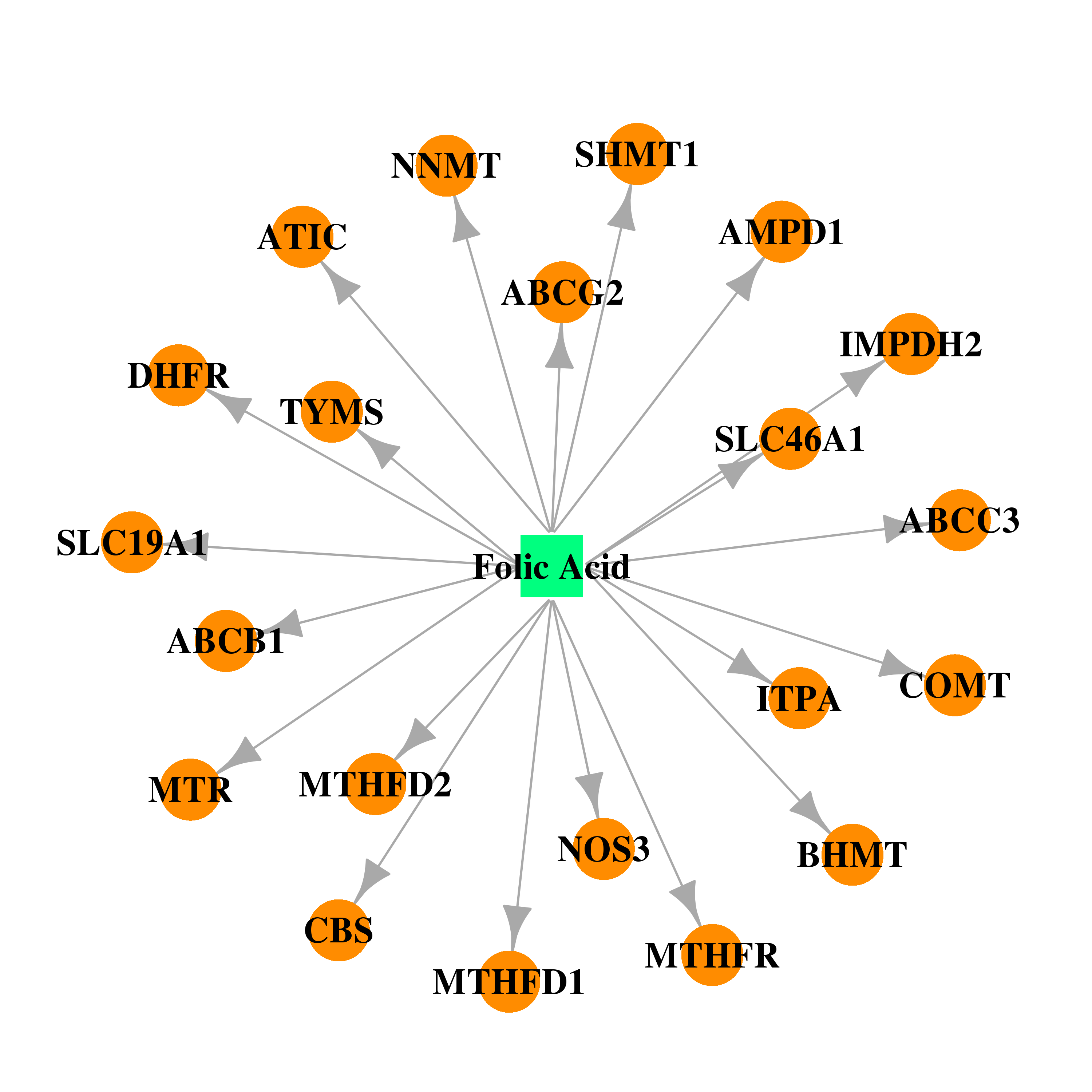 |  |
| DB00544 | dihydrofolate reductase | approved | Fluorouracil |  |  |
| DB01050 | dihydrofolate reductase | approved | Ibuprofen | 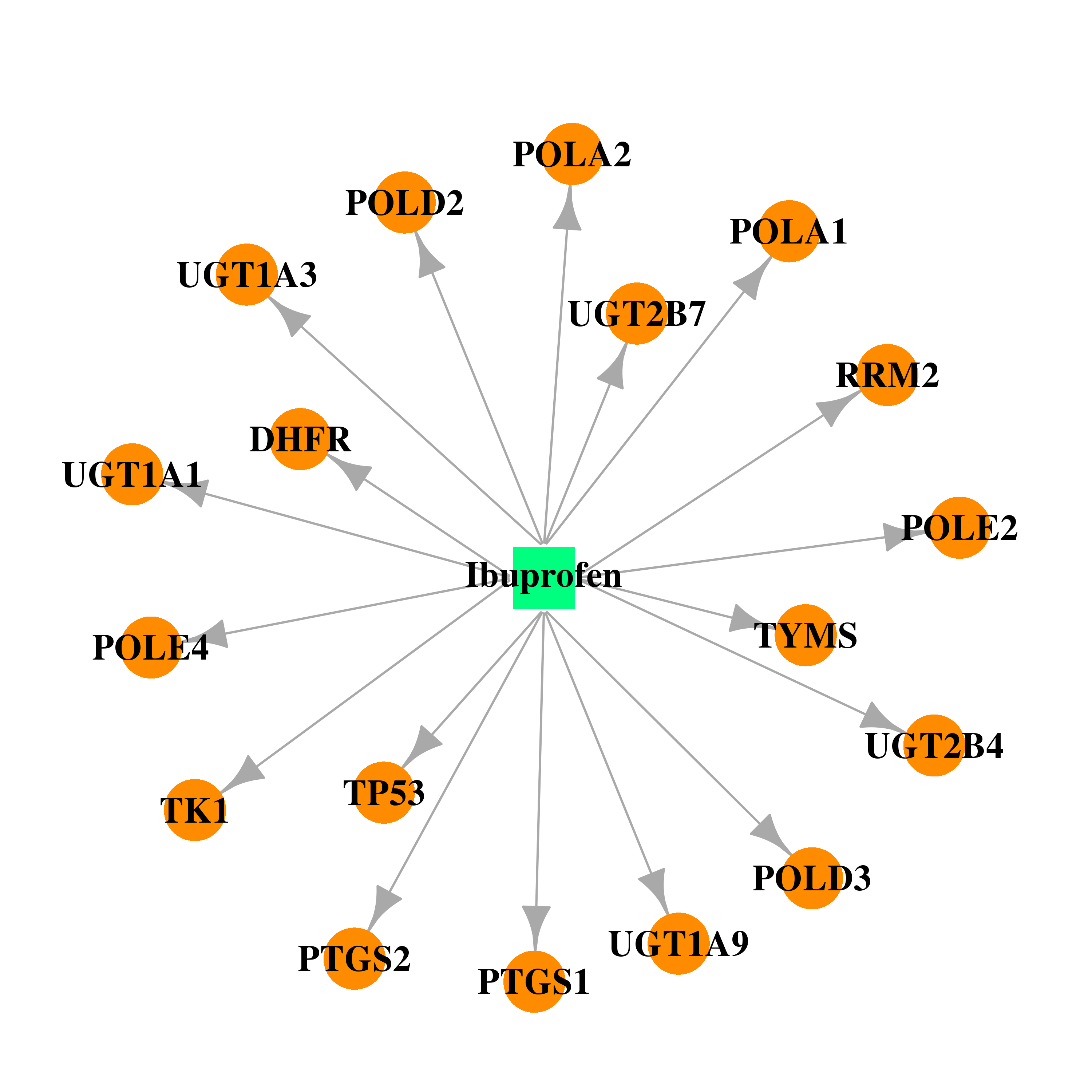 |  |
| Top |
| Cross referenced IDs for DHFR |
| * We obtained these cross-references from Uniprot database. It covers 150 different DBs, 18 categories. http://www.uniprot.org/help/cross_references_section |
: Open all cross reference information
|
Copyright © 2016-Present - The Univsersity of Texas Health Science Center at Houston @ |