|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |
| Phenotypic Information (metabolism pathway, cancer, disease, phenome) |
| |
| |
| Gene-Gene Network Information: Co-Expression Network, Interacting Genes & KEGG |
| |
|
| Gene Summary for DPP4 |
| Basic gene info. | Gene symbol | DPP4 |
| Gene name | dipeptidyl-peptidase 4 | |
| Synonyms | ADABP|ADCP2|CD26|DPPIV|TP103 | |
| Cytomap | UCSC genome browser: 2q24.3 | |
| Genomic location | chr2 :162848754-162931052 | |
| Type of gene | protein-coding | |
| RefGenes | NM_001935.3, | |
| Ensembl id | ENSG00000197635 | |
| Description | ADCP-2DPP IVT-cell activation antigen CD26adenosine deaminase complexing protein 2dipeptidyl peptidase 4dipeptidyl peptidase IVdipeptidylpeptidase 4dipeptidylpeptidase IV (CD26, adenosine deaminase complexing protein 2) | |
| Modification date | 20141222 | |
| dbXrefs | MIM : 102720 | |
| HGNC : HGNC | ||
| Ensembl : ENSG00000197635 | ||
| HPRD : 02187 | ||
| Vega : OTTHUMG00000132056 | ||
| Protein | UniProt: P27487 go to UniProt's Cross Reference DB Table | |
| Expression | CleanEX: HS_DPP4 | |
| BioGPS: 1803 | ||
| Gene Expression Atlas: ENSG00000197635 | ||
| The Human Protein Atlas: ENSG00000197635 | ||
| Pathway | NCI Pathway Interaction Database: DPP4 | |
| KEGG: DPP4 | ||
| REACTOME: DPP4 | ||
| ConsensusPathDB | ||
| Pathway Commons: DPP4 | ||
| Metabolism | MetaCyc: DPP4 | |
| HUMANCyc: DPP4 | ||
| Regulation | Ensembl's Regulation: ENSG00000197635 | |
| miRBase: chr2 :162,848,754-162,931,052 | ||
| TargetScan: NM_001935 | ||
| cisRED: ENSG00000197635 | ||
| Context | iHOP: DPP4 | |
| cancer metabolism search in PubMed: DPP4 | ||
| UCL Cancer Institute: DPP4 | ||
| Assigned class in ccmGDB | A - This gene has a literature evidence and it belongs to cancer gene. | |
| References showing role of DPP4 in cancer cell metabolism | 1. Tilan JU, Krailo M, Barkauskas DA, Galli S, Mtaweh H, et al. (2015) Systemic levels of neuropeptide Y and dipeptidyl peptidase activity in patients with Ewing sarcoma--associations with tumor phenotype and survival. Cancer 121: 697-707. doi: 10.1002/cncr.29090. pmid: 4339539. go to article 2. Femia AP, Raimondi L, Maglieri G, Lodovici M, Mannucci E, et al. (2013) Long-term treatment with Sitagliptin, a dipeptidyl peptidase-4 inhibitor, reduces colon carcinogenesis and reactive oxygen species in 1,2-dimethylhydrazine-induced rats. Int J Cancer 133: 2498-2503. doi: 10.1002/ijc.28260. go to article 3. Milani C, Katayama ML, de Lyra EC, Welsh J, Campos LT, et al. (2013) Transcriptional effects of 1,25 dihydroxyvitamin D(3) physiological and supra-physiological concentrations in breast cancer organotypic culture. BMC Cancer 13: 119. doi: 10.1186/1471-2407-13-119. pmid: 3637238. go to article | |
| Top |
| Phenotypic Information for DPP4(metabolism pathway, cancer, disease, phenome) |
| Cancer | CGAP: DPP4 |
| Familial Cancer Database: DPP4 | |
| * This gene is included in those cancer gene databases. |
|
|
|
|
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Oncogene 1 | Significant driver gene in | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| cf) number; DB name 1 Oncogene; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 2 Tumor Suppressor gene; https://bioinfo.uth.edu/TSGene/, 3 Cancer Gene Census; http://www.nature.com/nrc/journal/v4/n3/abs/nrc1299.html, 4 CancerGenes; http://nar.oxfordjournals.org/content/35/suppl_1/D721.long, 5 Network of Cancer Gene; http://ncg.kcl.ac.uk/index.php, 1Therapeutic Vulnerabilities in Cancer; http://cbio.mskcc.org/cancergenomics/statius/ |
| REACTOME_INTEGRATION_OF_ENERGY_METABOLISM | |
| OMIM | 102720; gene. |
| Orphanet | |
| Disease | KEGG Disease: DPP4 |
| MedGen: DPP4 (Human Medical Genetics with Condition) | |
| ClinVar: DPP4 | |
| Phenotype | MGI: DPP4 (International Mouse Phenotyping Consortium) |
| PhenomicDB: DPP4 | |
| Mutations for DPP4 |
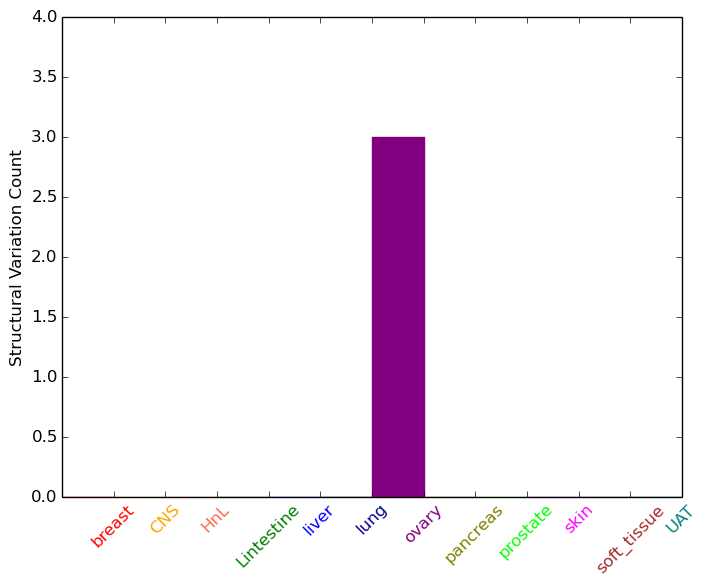
| * Under tables are showing count per each tissue to give us broad intuition about tissue specific mutation patterns.You can go to the detailed page for each mutation database's web site. |
| - Statistics for Tissue and Mutation type | Top |
 |
| - For Inter-chromosomal Variations |
| There's no inter-chromosomal structural variation. |
| - For Intra-chromosomal Variations |
| * Intra-chromosomal variantions includes 'intrachromosomal amplicon to amplicon', 'intrachromosomal amplicon to non-amplified dna', 'intrachromosomal deletion', 'intrachromosomal fold-back inversion', 'intrachromosomal inversion', 'intrachromosomal tandem duplication', 'Intrachromosomal unknown type', 'intrachromosomal with inverted orientation', 'intrachromosomal with non-inverted orientation'. |
 |
| Sample | Symbol_a | Chr_a | Start_a | End_a | Symbol_b | Chr_b | Start_b | End_b |
| ovary | DPP4 | chr2 | 162855084 | 162855104 | SLC4A10 | chr2 | 162822274 | 162822294 |
| ovary | DPP4 | chr2 | 162901614 | 162901634 | DPP4 | chr2 | 162901692 | 162901712 |
| cf) Tissue number; Tissue name (1;Breast, 2;Central_nervous_system, 3;Haematopoietic_and_lymphoid_tissue, 4;Large_intestine, 5;Liver, 6;Lung, 7;Ovary, 8;Pancreas, 9;Prostate, 10;Skin, 11;Soft_tissue, 12;Upper_aerodigestive_tract) |
| * From mRNA Sanger sequences, Chitars2.0 arranged chimeric transcripts. This table shows DPP4 related fusion information. |
| ID | Head Gene | Tail Gene | Accession | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a | Gene_a | qStart_a | qEnd_a | Chromosome_a | tStart_a | tEnd_a |
| BQ348968 | PABPC1 | 170 | 232 | 8 | 101721898 | 101721960 | DPP4 | 223 | 512 | 2 | 162849781 | 162862283 | |
| BG231728 | DPP4 | 4 | 24 | 2 | 162916114 | 162916134 | CD109 | 12 | 32 | 6 | 74457249 | 74457269 | |
| Top |
| Mutation type/ Tissue ID | brca | cns | cerv | endome | haematopo | kidn | Lintest | liver | lung | ns | ovary | pancre | prost | skin | stoma | thyro | urina | |||
| Total # sample | 1 | 1 | ||||||||||||||||||
| GAIN (# sample) | 1 | |||||||||||||||||||
| LOSS (# sample) | 1 |
| cf) Tissue ID; Tissue type (1; Breast, 2; Central_nervous_system, 3; Cervix, 4; Endometrium, 5; Haematopoietic_and_lymphoid_tissue, 6; Kidney, 7; Large_intestine, 8; Liver, 9; Lung, 10; NS, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary_tract) |
| Top |
|
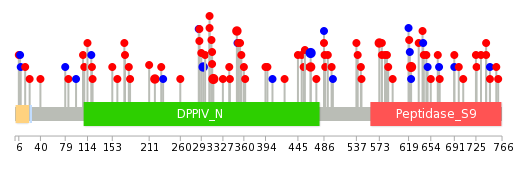 |
| Top |
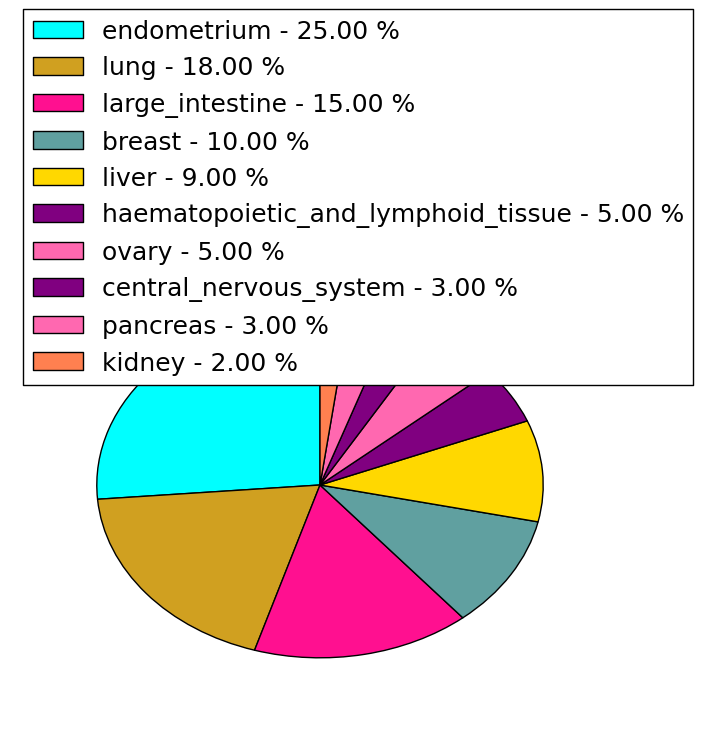
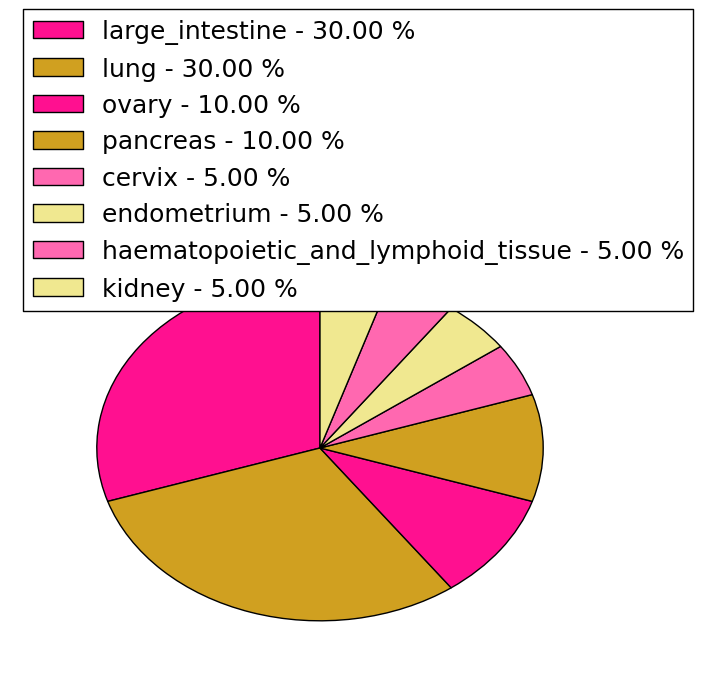
| Stat. for Non-Synonymous SNVs (# total SNVs=77) | (# total SNVs=20) |
 |  |
(# total SNVs=1) | (# total SNVs=0) |
 |
| Top |
| * When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site,primary_histology,mutation(aa),pubmedID. |
| GRCh37 position | Mutation(aa) | Unique sampleID count |
| chr2:162875264-162875264 | p.A465A | 3 |
| chr2:162865112-162865112 | p.C649* | 3 |
| chr2:162881402-162881402 | p.S312Y | 3 |
| chr2:162879274-162879274 | p.W353* | 3 |
| chr2:162865770-162865770 | p.R623Q | 3 |
| chr2:162879287-162879287 | p.S349N | 2 |
| chr2:162902048-162902048 | p.Y120Y | 2 |
| chr2:162891790-162891790 | p.G220S | 2 |
| chr2:162875265-162875265 | p.A465V | 2 |
| chr2:162881449-162881449 | p.G296G | 2 |
| Top |
|
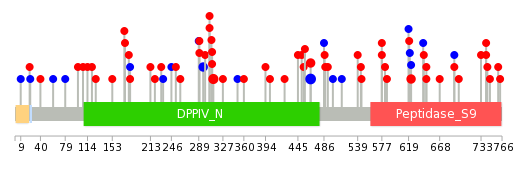 |
| Point Mutation/ Tissue ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| # sample | 1 | 4 | 1 | 11 | 5 | 3 | 10 | 7 | 4 | 9 | 6 | 14 | ||||||||
| # mutation | 1 | 4 | 1 | 10 | 5 | 3 | 12 | 8 | 4 | 11 | 6 | 18 | ||||||||
| nonsynonymous SNV | 1 | 4 | 7 | 5 | 2 | 9 | 3 | 3 | 7 | 5 | 17 | |||||||||
| synonymous SNV | 1 | 3 | 1 | 3 | 5 | 1 | 4 | 1 | 1 |
| cf) Tissue ID; Tissue type (1; BLCA[Bladder Urothelial Carcinoma], 2; BRCA[Breast invasive carcinoma], 3; CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], 4; COAD[Colon adenocarcinoma], 5; GBM[Glioblastoma multiforme], 6; Glioma Low Grade, 7; HNSC[Head and Neck squamous cell carcinoma], 8; KICH[Kidney Chromophobe], 9; KIRC[Kidney renal clear cell carcinoma], 10; KIRP[Kidney renal papillary cell carcinoma], 11; LAML[Acute Myeloid Leukemia], 12; LUAD[Lung adenocarcinoma], 13; LUSC[Lung squamous cell carcinoma], 14; OV[Ovarian serous cystadenocarcinoma ], 15; PAAD[Pancreatic adenocarcinoma], 16; PRAD[Prostate adenocarcinoma], 17; SKCM[Skin Cutaneous Melanoma], 18:STAD[Stomach adenocarcinoma], 19:THCA[Thyroid carcinoma], 20:UCEC[Uterine Corpus Endometrial Carcinoma]) |
| Top |
| * We represented just top 10 SNVs. When you move the cursor on each content, you can see more deailed mutation information on the Tooltip. Those are primary_site, primary_histology, mutation(aa), pubmedID. |
| Genomic Position | Mutation(aa) | Unique sampleID count |
| chr2:162875264 | p.A465A | 4 |
| chr2:162881402 | p.S312Y | 3 |
| chr2:162881449 | p.R623Q | 2 |
| chr2:162875265 | p.A465V | 2 |
| chr2:162865770 | p.G296G | 2 |
| chr2:162902047 | p.S360P | 1 |
| chr2:162851471 | p.L246L | 1 |
| chr2:162875326 | p.F79F | 1 |
| chr2:162881420 | p.E668K | 1 |
| chr2:162865775 | p.K539N | 1 |
| * Copy number data were extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered on Jan-05-2015. Function ProcessCNAData in TCGA-Assembler package was used to obtain gene-level copy number value which is calculated as the average copy number of the genomic region of a gene. |
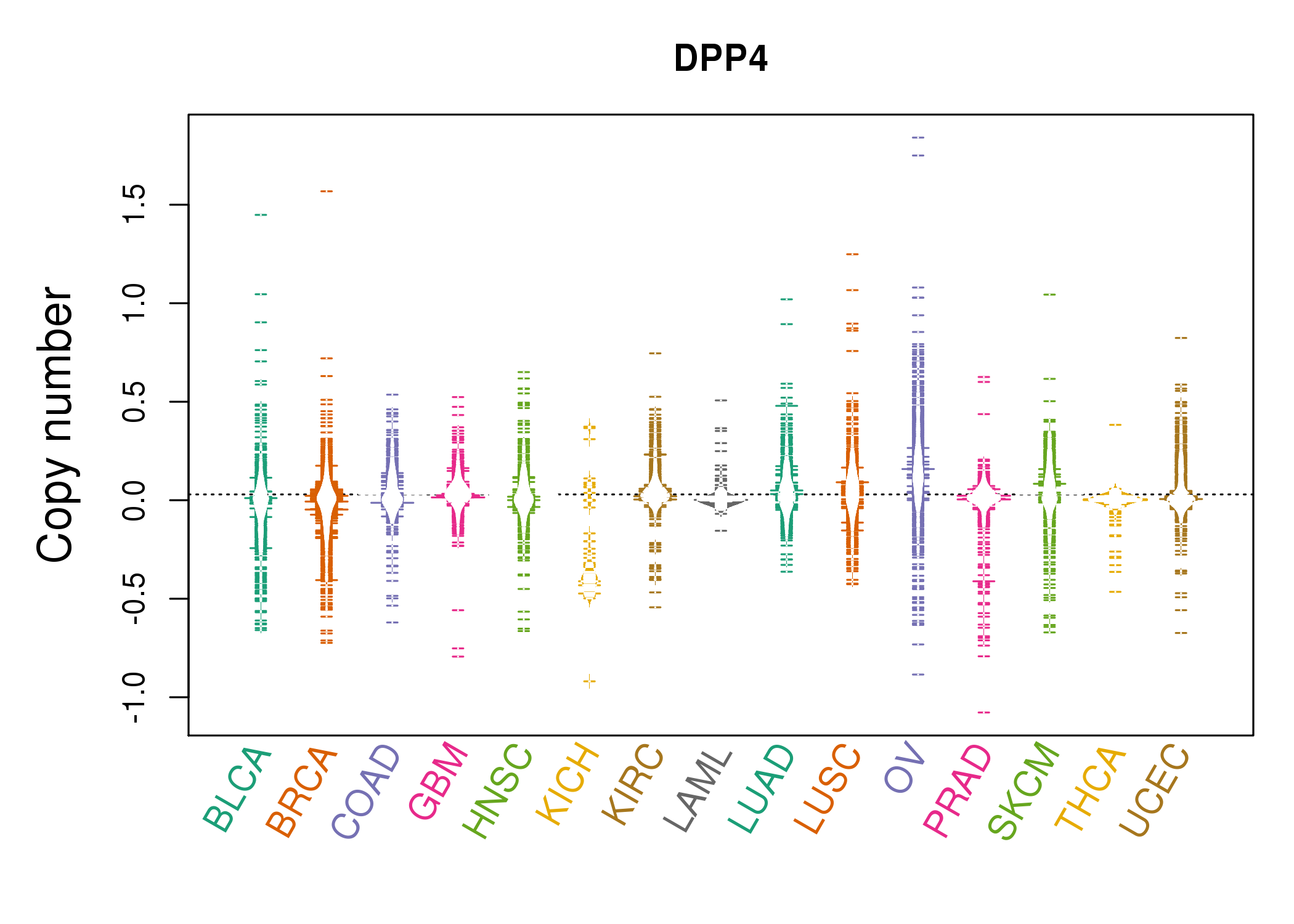 |
| cf) Tissue ID[Tissue type]: BLCA[Bladder Urothelial Carcinoma], BRCA[Breast invasive carcinoma], CESC[Cervical squamous cell carcinoma and endocervical adenocarcinoma], COAD[Colon adenocarcinoma], GBM[Glioblastoma multiforme], Glioma Low Grade, HNSC[Head and Neck squamous cell carcinoma], KICH[Kidney Chromophobe], KIRC[Kidney renal clear cell carcinoma], KIRP[Kidney renal papillary cell carcinoma], LAML[Acute Myeloid Leukemia], LUAD[Lung adenocarcinoma], LUSC[Lung squamous cell carcinoma], OV[Ovarian serous cystadenocarcinoma ], PAAD[Pancreatic adenocarcinoma], PRAD[Prostate adenocarcinoma], SKCM[Skin Cutaneous Melanoma], STAD[Stomach adenocarcinoma], THCA[Thyroid carcinoma], UCEC[Uterine Corpus Endometrial Carcinoma] |
| Top |
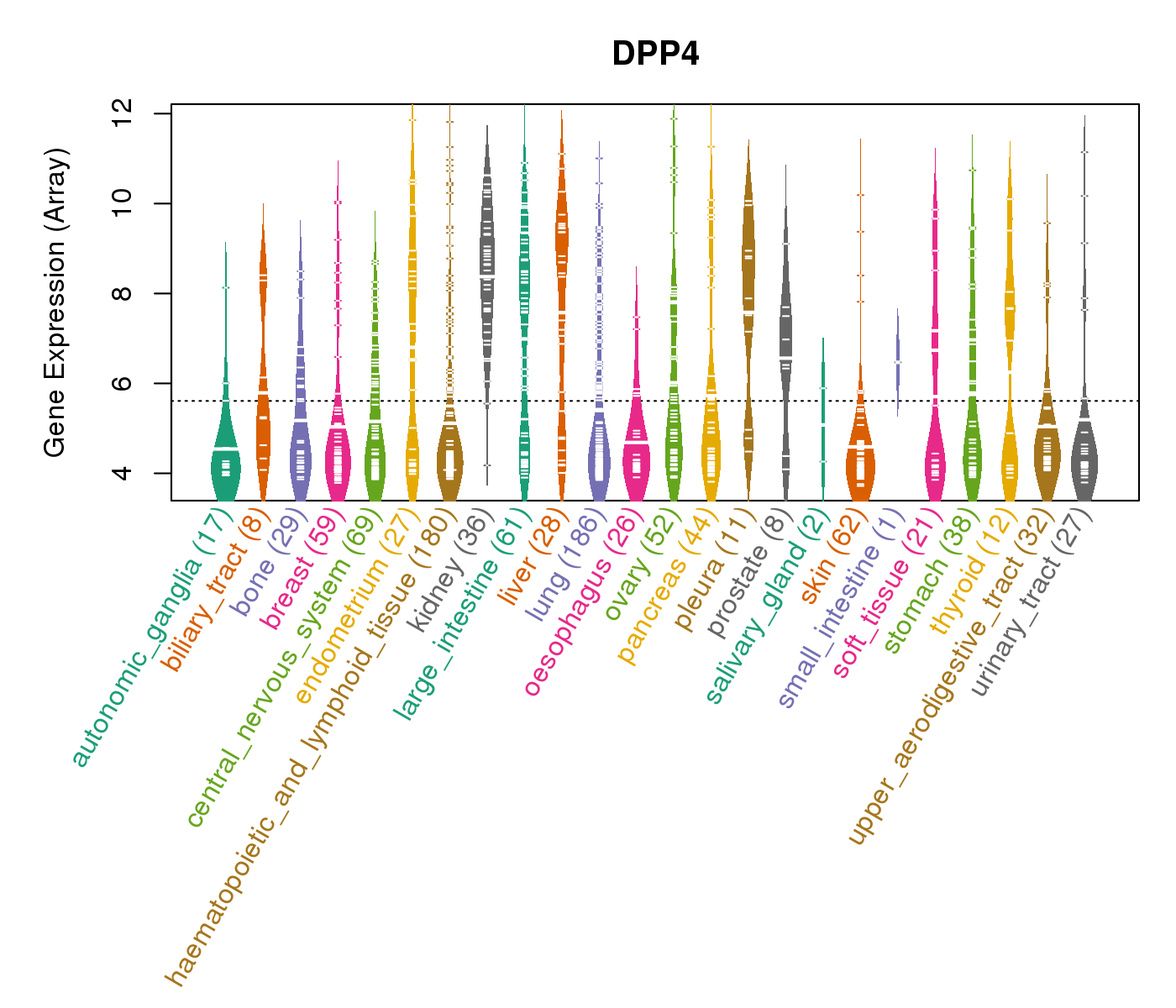
| Gene Expression for DPP4 |
| * CCLE gene expression data were extracted from CCLE_Expression_Entrez_2012-10-18.res: Gene-centric RMA-normalized mRNA expression data. |
 |
| * Normalized gene expression data of RNASeqV2 was extracted from TCGA using R package TCGA-Assembler. The URLs of all public data files on TCGA DCC data server were gathered at Jan-05-2015. Only eight cancer types have enough normal control samples for differential expression analysis. (t test, adjusted p<0.05 (using Benjamini-Hochberg FDR)) |
 |
| Top |
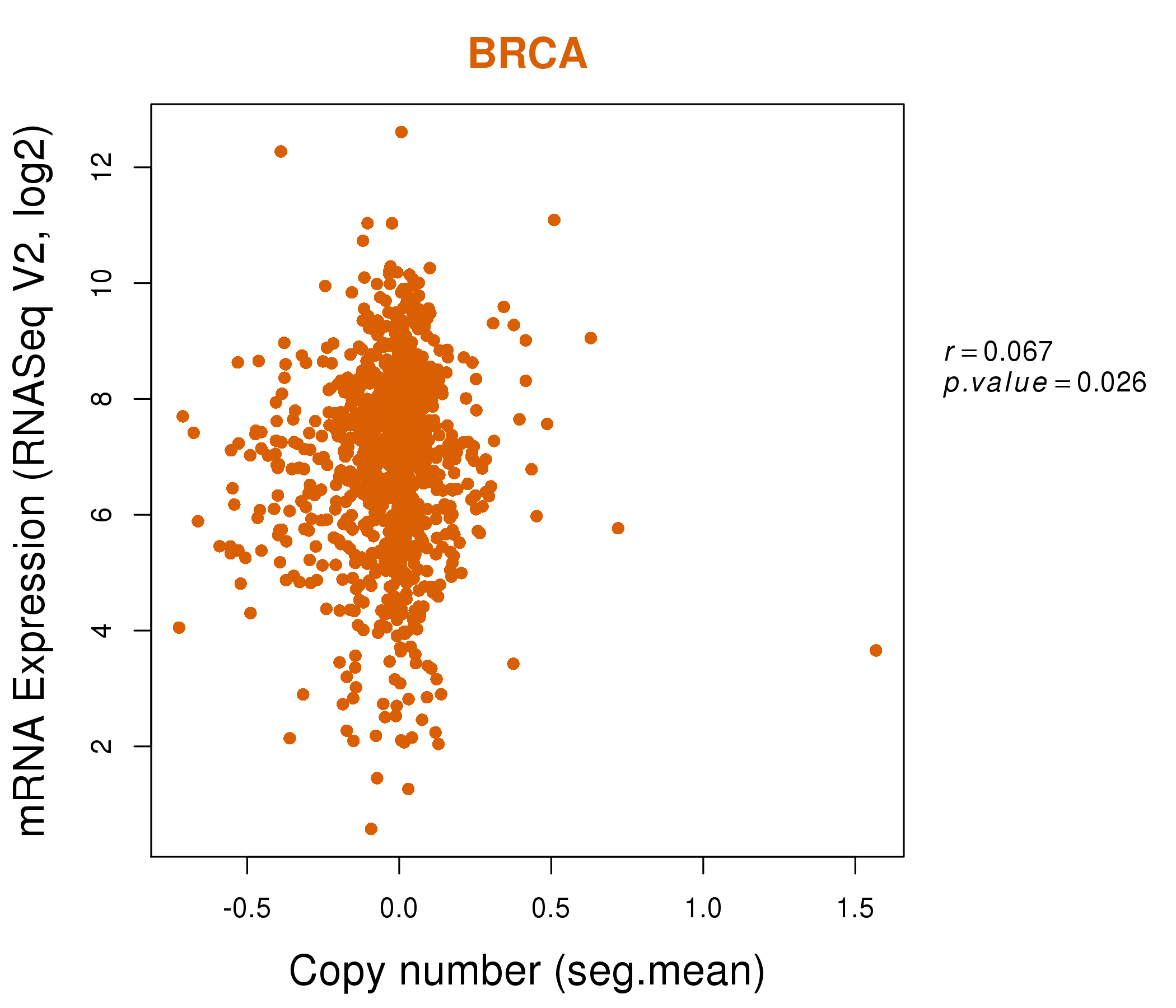
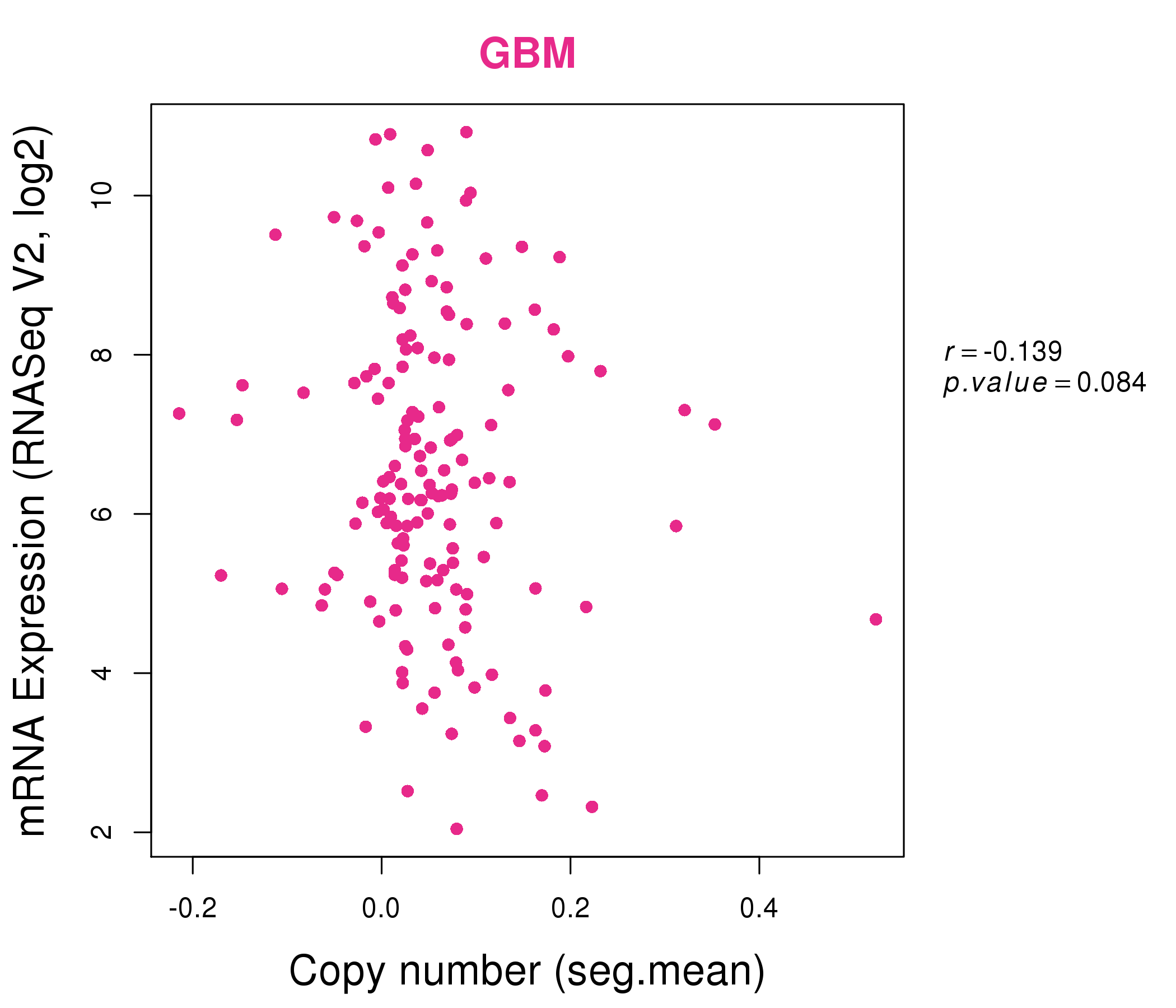
| * This plots show the correlation between CNV and gene expression. |
: Open all plots for all cancer types
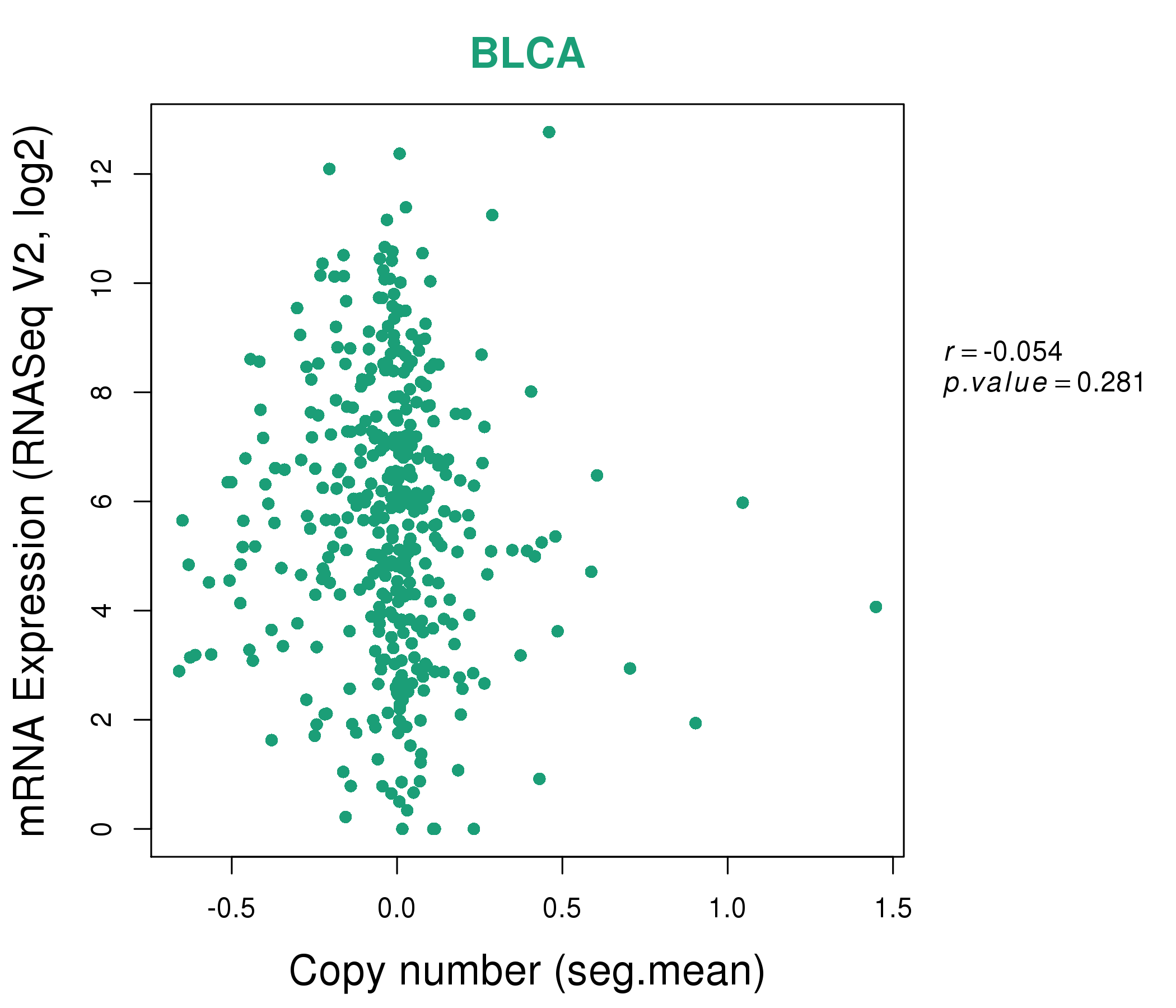 |
|
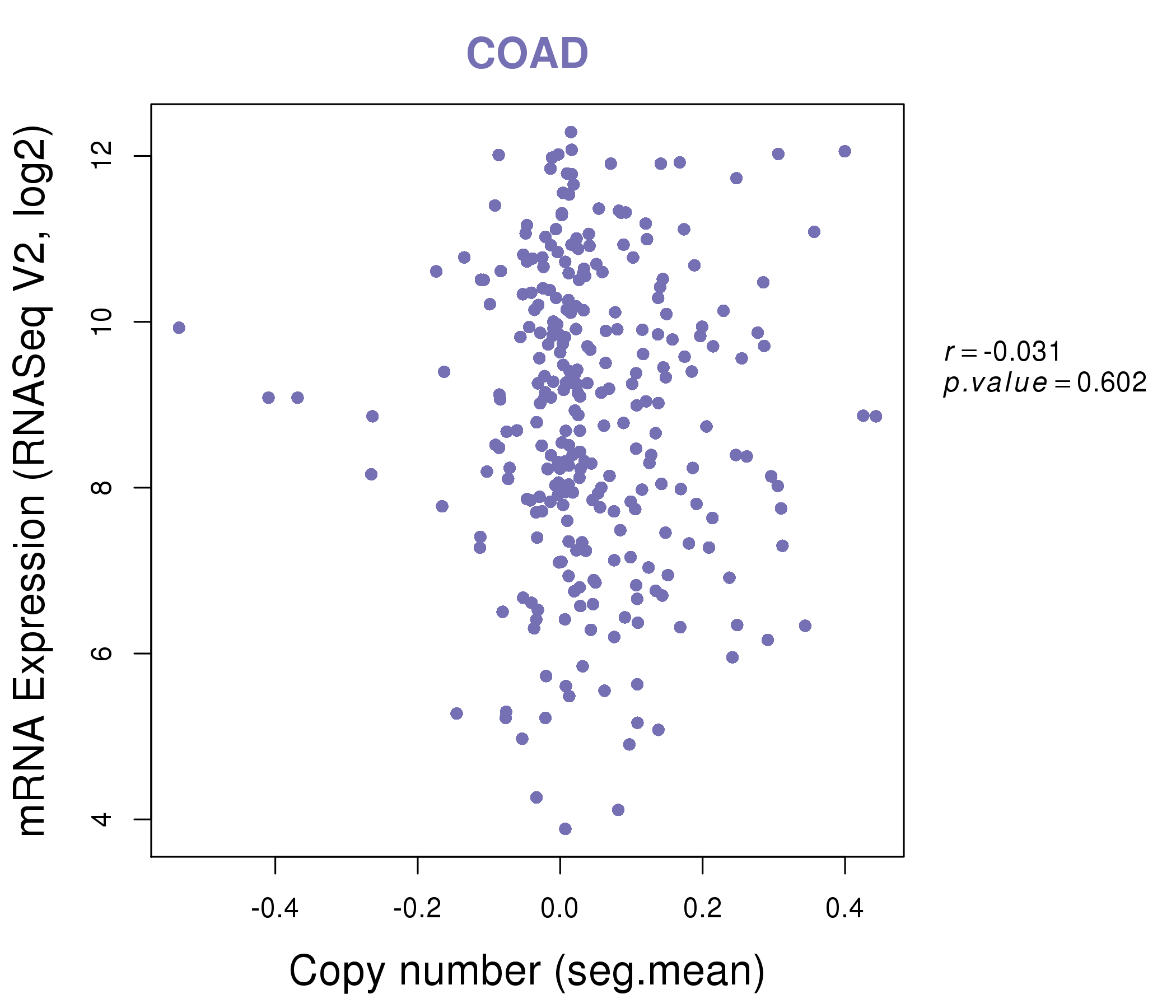 |
|
| Top |
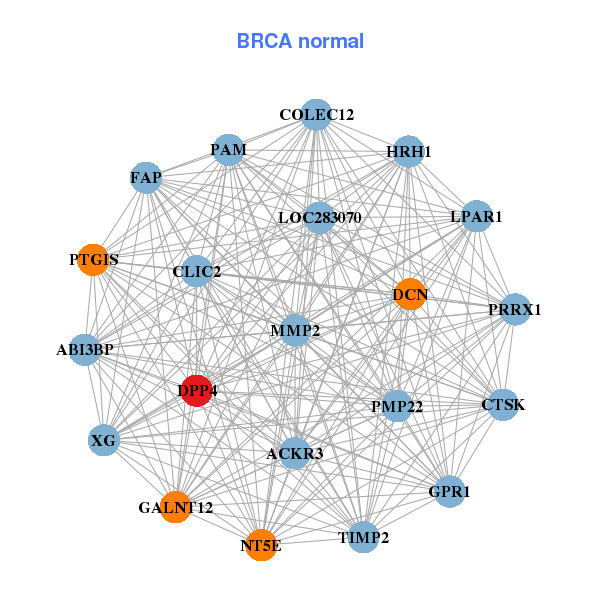
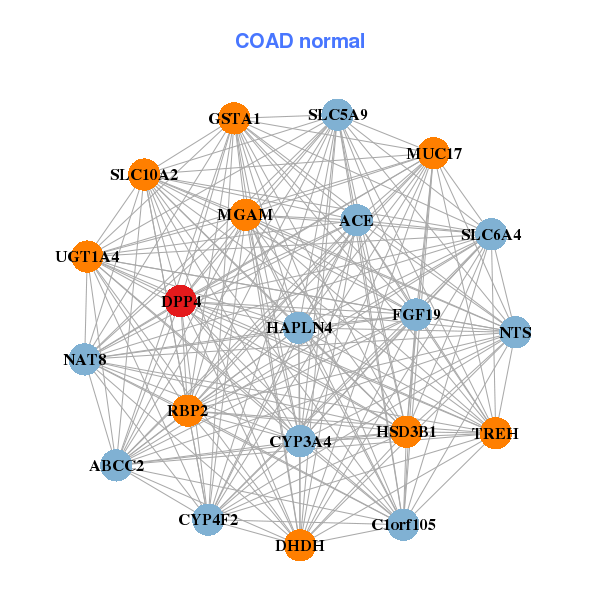
| Gene-Gene Network Information |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
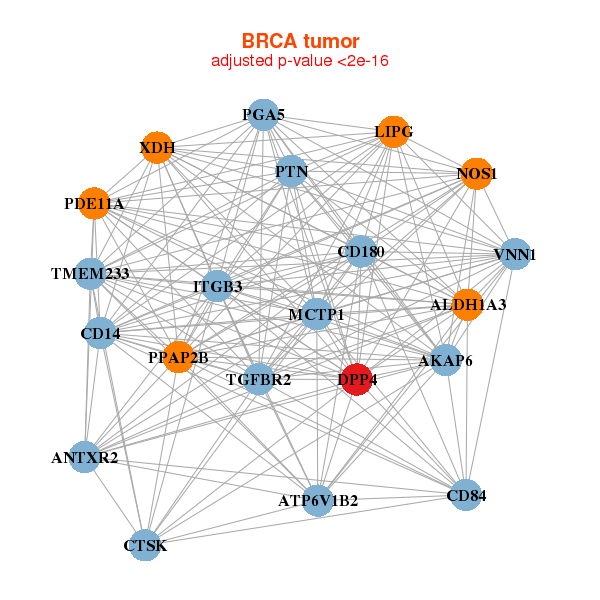 |
| ||||
| AKAP6,ALDH1A3,ANTXR2,ATP6V1B2,CD14,CD180,CD84, CTSK,DPP4,ITGB3,LIPG,MCTP1,NOS1,PDE11A, PGA5,PPAP2B,PTN,TGFBR2,TMEM233,VNN1,XDH | ABI3BP,CLIC2,COLEC12,CTSK,ACKR3,DCN,DPP4, FAP,GALNT12,GPR1,HRH1,LOC283070,LPAR1,MMP2, NT5E,PAM,PMP22,PRRX1,PTGIS,TIMP2,XG | ||||
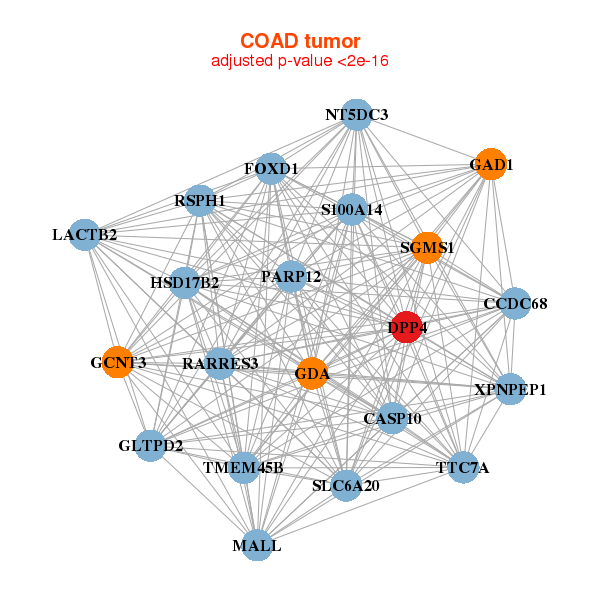 |
| ||||
| CASP10,CCDC68,DPP4,FOXD1,GAD1,GCNT3,GDA, GLTPD2,HSD17B2,LACTB2,MALL,NT5DC3,PARP12,RARRES3, RSPH1,S100A14,SGMS1,SLC6A20,TMEM45B,TTC7A,XPNPEP1 | ABCC2,ACE,C1orf105,CYP3A4,CYP4F2,DHDH,DPP4, FGF19,GSTA1,HAPLN4,HSD3B1,MGAM,MUC17,NAT8, NTS,RBP2,SLC10A2,SLC5A9,SLC6A4,TREH,UGT1A4 |
| * Co-Expression network figures were drawn using R package igraph. Only the top 20 genes with the highest correlations were shown. Red circle: input gene, orange circle: cell metabolism gene, sky circle: other gene |
: Open all plots for all cancer types
| Top |
: Open all interacting genes' information including KEGG pathway for all interacting genes from DAVID
| Top |
| Pharmacological Information for DPP4 |
| DB Category | DB Name | DB's ID and Url link |
| Chemistry | BindingDB | P27487; -. |
| Chemistry | ChEMBL | CHEMBL2111469; -. |
| Chemistry | GuidetoPHARMACOLOGY | 1612; -. |
| Organism-specific databases | PharmGKB | PA27467; -. |
| Organism-specific databases | CTD | 1803; -. |
| * Gene Centered Interaction Network. |
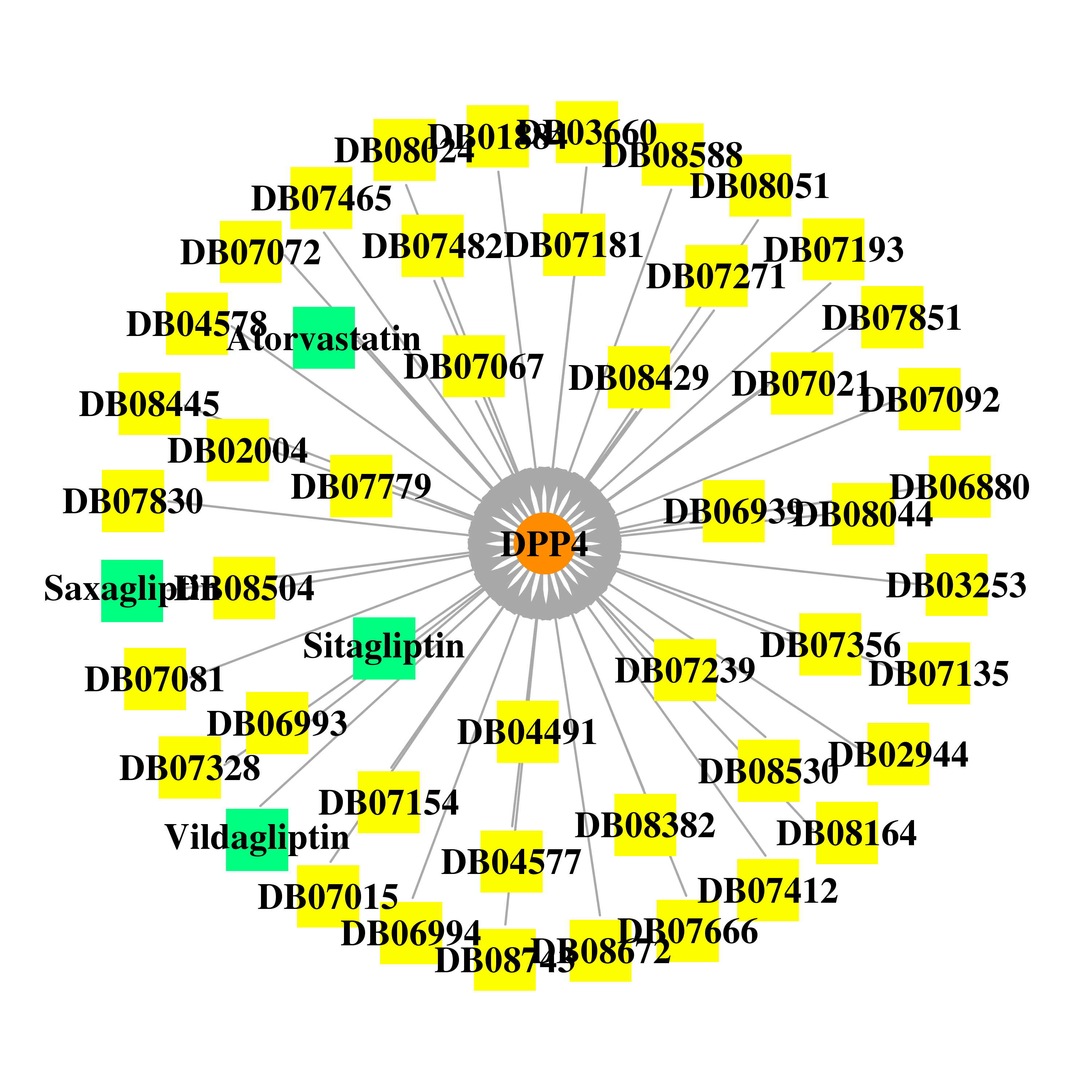 |
| * Drug Centered Interaction Network. |
| DrugBank ID | Target Name | Drug Groups | Generic Name | Drug Centered Network | Drug Structure |
| DB01076 | dipeptidyl-peptidase 4 | approved | Atorvastatin | 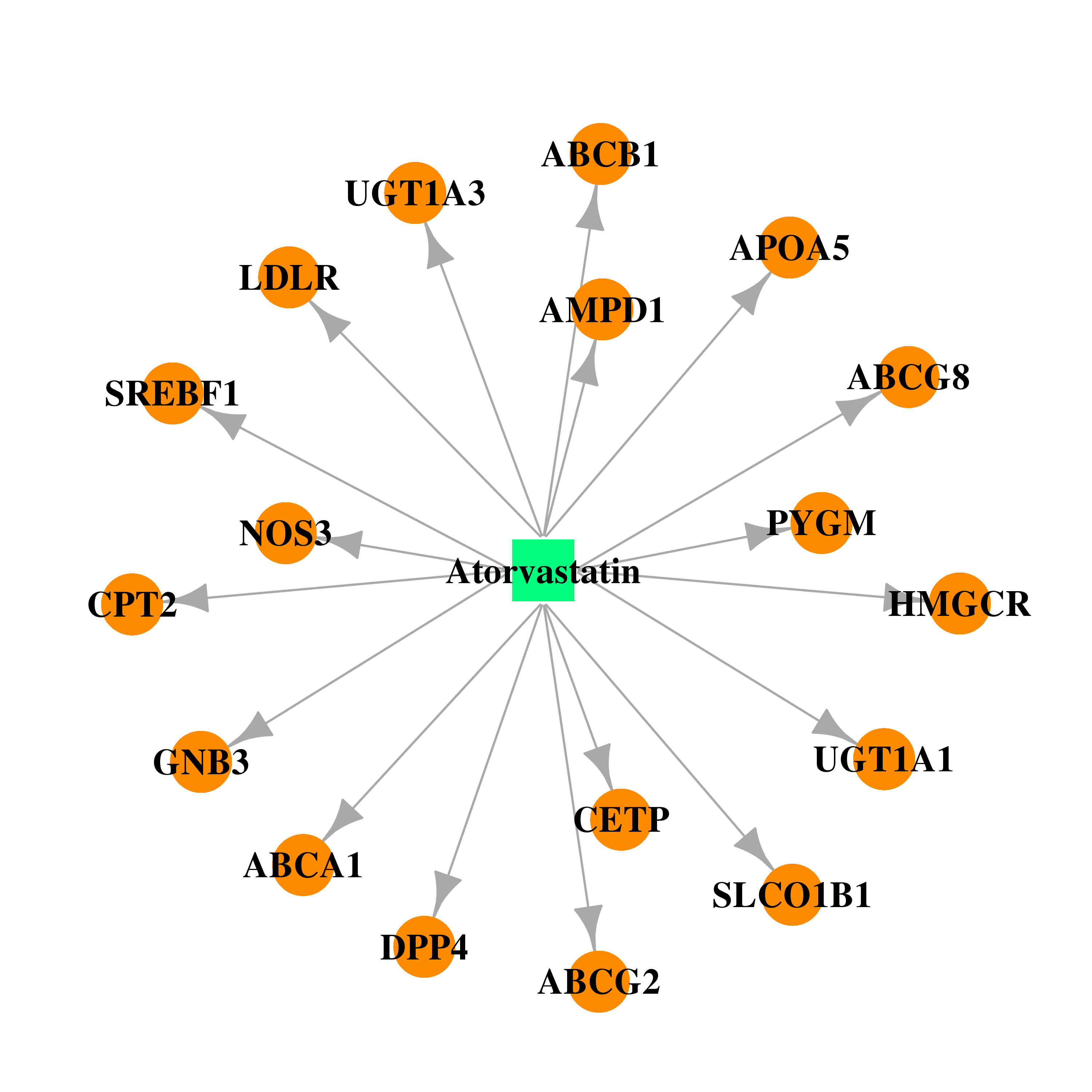 |  |
| DB01261 | dipeptidyl-peptidase 4 | approved; investigational | Sitagliptin |  |  |
| DB01884 | dipeptidyl-peptidase 4 | experimental | 2-Amino-3-Methyl-1-Pyrrolidin-1-Yl-Butan-1-One |  |  |
| DB02004 | dipeptidyl-peptidase 4 | experimental | 5-(Aminomethyl)-6-(2,4-Dichlorophenyl)-2-(3,5-Dimethoxyphenyl)Pyrimidin-4-Amine |  |  |
| DB02944 | dipeptidyl-peptidase 4 | experimental | Alpha-D-Mannose |  |  |
| DB03253 | dipeptidyl-peptidase 4 | experimental | (2s)-Pyrrolidin-2-Ylmethylamine |  |  |
| DB03660 | dipeptidyl-peptidase 4 | experimental | Iodo-Phenylalanine |  |  |
| DB04491 | dipeptidyl-peptidase 4 | experimental | Diisopropylphosphono Group |  |  |
| DB04577 | dipeptidyl-peptidase 4 | experimental | 1-(1-phenylcyclopentyl)methylamine |  |  |
| DB04578 | dipeptidyl-peptidase 4 | experimental | (S)-2-[(R)-3-amino-4-(2-fluorophenyl)butyryl]-1,2,3,4-tetrahydroisoquinoline-3-carboxamide |  |  |
| DB04876 | dipeptidyl-peptidase 4 | approved; investigational | Vildagliptin |  |  |
| DB06335 | dipeptidyl-peptidase 4 | approved; investigational | Saxagliptin |  | 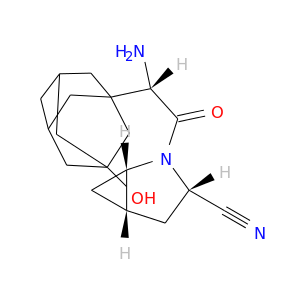 |
| DB06880 | dipeptidyl-peptidase 4 | experimental | (1S)-2-[(2S,5R)-2-(AMINOMETHYL)-5-PROP-1-YN-1-YLPYRROLIDIN-1-YL]-1-CYCLOPENTYL-2-OXOETHANAMINE |  | 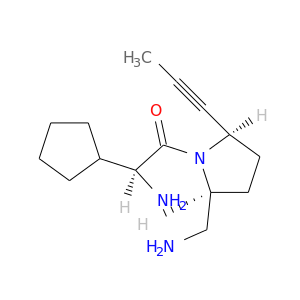 |
| DB06939 | dipeptidyl-peptidase 4 | experimental | N-(TRANS-4-{(1S,2S)-2-AMINO-3-[(3S)-3-FLUOROPYRROLIDIN-1-YL]-1-METHYL-3-OXOPROPYL}CYCLOHEXYL)-N-METHYLACETAMIDE |  |  |
| DB06993 | dipeptidyl-peptidase 4 | experimental | (2S,3S)-4-cyclopropyl-3-{(3R,5R)-3-[2-fluoro-4-(methylsulfonyl)phenyl]-1,2,4-oxadiazolidin-5-yl}-1-[(3S)-3-fluoropyrrolidin-1-yl]-1-oxobutan-2-amine |  |  |
| DB06994 | dipeptidyl-peptidase 4 | experimental | (2S,3S)-3-{3-[2-chloro-4-(methylsulfonyl)phenyl]-1,2,4-oxadiazol-5-yl}-1-cyclopentylidene-4-cyclopropyl-1-fluorobutan-2-amine |  |  |
| DB07015 | dipeptidyl-peptidase 4 | experimental | (3R,4R)-4-(pyrrolidin-1-ylcarbonyl)-1-(quinoxalin-2-ylcarbonyl)pyrrolidin-3-amine |  |  |
| DB07021 | dipeptidyl-peptidase 4 | experimental | (7R,8R)-8-(2,4,5-trifluorophenyl)-6,7,8,9-tetrahydroimidazo[1,2-a:4,5-c']dipyridin-7-amine |  |  |
| DB07067 | dipeptidyl-peptidase 4 | experimental | (2S,3S)-3-{3-[4-(METHYLSULFONYL)PHENYL]-1,2,4-OXADIAZOL-5-YL}-1-OXO-1-PYRROLIDIN-1-YLBUTAN-2-AMINE |  |  |
| DB07072 | dipeptidyl-peptidase 4 | experimental | (1S,2R,5S)-5-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-2-(2,4,5-TRIFLUOROPHENYL)CYCLOHEXANAMINE |  |  |
| DB07081 | dipeptidyl-peptidase 4 | experimental | (2R)-4-[(8R)-8-METHYL-2-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[1,5-A]PYRAZIN-7(8H)-YL]-4-OXO-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-AMINE |  |  |
| DB07092 | dipeptidyl-peptidase 4 | experimental | (2S,3S)-3-AMINO-4-(3,3-DIFLUOROPYRROLIDIN-1-YL)-N,N-DIMETHYL-4-OXO-2-(TRANS-4-[1,2,4]TRIAZOLO[1,5-A]PYRIDIN-6-YLCYCLOHEXYL)BUTANAMIDE |  | 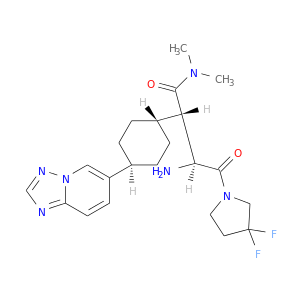 |
| DB07135 | dipeptidyl-peptidase 4 | experimental | (2S,3S)-3-AMINO-4-[(3S)-3-FLUOROPYRROLIDIN-1-YL]-N,N-DIMETHYL-4-OXO-2-(TRANS-4-[1,2,4]TRIAZOLO[1,5-A]PYRIDIN-5-YLCYCLOHEXYL)BUTANAMIDE |  | 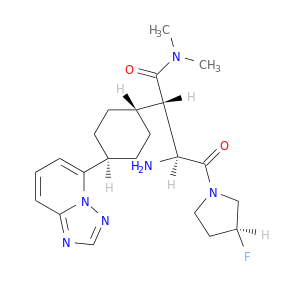 |
| DB07154 | dipeptidyl-peptidase 4 | experimental | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-METHYL-1,4-DIAZEPAN-2-ONE |  |  |
| DB07181 | dipeptidyl-peptidase 4 | experimental | 4'-[(1R)-1-amino-2-(2,5-difluorophenyl)ethyl]biphenyl-3-carboxamide |  |  |
| DB07193 | dipeptidyl-peptidase 4 | experimental | (2R,3R)-7-(methylsulfonyl)-3-(2,4,5-trifluorophenyl)-1,2,3,4-tetrahydropyrido[1,2-a]benzimidazol-2-amine |  |  |
| DB07239 | dipeptidyl-peptidase 4 | experimental | 7-(aminomethyl)-6-(2-chlorophenyl)-1-methyl-1H-benzimidazole-5-carbonitrile |  |  |
| DB07271 | dipeptidyl-peptidase 4 | experimental | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-(2,2,2-TRIFLUOROETHYL)-1,4-DIAZEPAN-2-ONE |  |  |
| DB07328 | dipeptidyl-peptidase 4 | experimental | METHYL 4-{[({[(2R,5S)-5-{[(2S)-2-(AMINOMETHYL)PYRROLIDIN-1-YL]CARBONYL}PYRROLIDIN-2-YL]METHYL}AMINO)CARBONYL]AMINO}BENZOATE |  |  |
| DB07356 | dipeptidyl-peptidase 4 | experimental | (1S)-2-[(2S,5R)-2-(AMINOMETHYL)-5-ETHYNYLPYRROLIDIN-1-YL]-1-CYCLOPENTYL-2-OXOETHANAMINE |  |  |
| DB07412 | dipeptidyl-peptidase 4 | experimental | 1-biphenyl-2-ylmethanamine |  |  |
| DB07465 | dipeptidyl-peptidase 4 | experimental | (1S,3S,5S)-2-{(2S)-2-amino-2-[(1R,3S,5R,7S)-3-hydroxytricyclo[3.3.1.1~3,7~]dec-1-yl]acetyl}-2-azabicyclo[3.1.0]hexane-3-carbonitrile |  |  |
| DB07482 | dipeptidyl-peptidase 4 | experimental | (2R)-N-[(2R)-2-(DIHYDROXYBORYL)-1-L-PROLYLPYRROLIDIN-2-YL]-N-[(5R)-5-(DIHYDROXYBORYL)-1-L-PROLYLPYRROLIDIN-2-YL]-L-PROLINAMIDE |  |  |
| DB07666 | dipeptidyl-peptidase 4 | experimental | (3R,4S)-1-{6-[3-(METHYLSULFONYL)PHENYL]PYRIMIDIN-4-YL}-4-(2,4,5-TRIFLUOROPHENYL)PYRROLIDIN-3-AMINE |  |  |
| DB07779 | dipeptidyl-peptidase 4 | experimental | N-({(2S)-1-[(3R)-3-AMINO-4-(2-FLUOROPHENYL)BUTANOYL]PYRROLIDIN-2-YL}METHYL)BENZAMIDE |  |  |
| DB07830 | dipeptidyl-peptidase 4 | experimental | (4R,5R)-5-AMINO-1-[2-(1,3-BENZODIOXOL-5-YL)ETHYL]-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-2-ONE |  |  |
| DB07851 | dipeptidyl-peptidase 4 | experimental | (3R,4S)-1-(3,4-DIMETHOXYPHENYL)-3-(3-METHYLPHENYL)PIPERIDIN-4-AMINE |  |  |
| DB08024 | dipeptidyl-peptidase 4 | experimental | 1-[2-(S)-AMINO-3-BIPHENYL-4-YL-PROPIONYL]-PYRROLIDINE-2-(S)-CARBONITRILE |  |  |
| DB08044 | dipeptidyl-peptidase 4 | experimental | (1S,6R)-3-{[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]CARBONYL}-6-(2,4,5-TRIFLUOROPHENYL)CYCLOHEX-3-EN-1-AMINE |  |  |
| DB08051 | dipeptidyl-peptidase 4 | experimental | (2R)-4-(2-BENZOYL-1,2-DIAZEPAN-1-YL)-4-OXO-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-AMINE |  |  |
| DB08164 | dipeptidyl-peptidase 4 | experimental | (3R,4R)-1-{6-[3-(METHYLSULFONYL)PHENYL]PYRIMIDIN-4-YL}-4-(2,4,5-TRIFLUOROPHENYL)PIPERIDIN-3-AMINE |  |  |
| DB08382 | dipeptidyl-peptidase 4 | experimental | 2-(4-{(3S,5S)-5-[(3,3-difluoropyrrolidin-1-yl)carbonyl]pyrrolidin-3-yl}piperazin-1-yl)pyrimidine |  |  |
| DB08429 | dipeptidyl-peptidase 4 | experimental | N-({(2S)-1-[(3R)-3-amino-4-(3-chlorophenyl)butanoyl]pyrrolidin-2-yl}methyl)-3-(methylsulfonyl)benzamide |  | 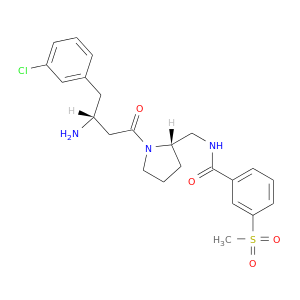 |
| DB08445 | dipeptidyl-peptidase 4 | experimental | (3R,4S)-1-[6-(6-METHOXYPYRIDIN-3-YL)PYRIMIDIN-4-YL]-4-(2,4,5-TRIFLUOROPHENYL)PYRROLIDIN-3-AMINE |  | 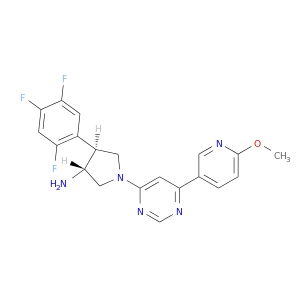 |
| DB08504 | dipeptidyl-peptidase 4 | experimental | 6-(4-{(1S,2S)-2-AMINO-1-[(DIMETHYLAMINO)CARBONYL]-3-[(3S)-3-FLUOROPYRROLIDIN-1-YL]-3-OXOPROPYL}PHENYL)-1H-[1,2,4]TRIAZOLO[1,5-A]PYRIDIN-4-IUM |  |  |
| DB08530 | dipeptidyl-peptidase 4 | experimental | 7-BENZYL-1,3-DIMETHYL-8-PIPERAZIN-1-YL-3,7-DIHYDRO-PURINE-2,6-DIONE |  |  |
| DB08588 | dipeptidyl-peptidase 4 | experimental | 2-({2-[(3R)-3-AMINOPIPERIDIN-1-YL]-4-OXOQUINAZOLIN-3(4H)-YL}METHYL)BENZONITRILE |  |  |
| DB08672 | dipeptidyl-peptidase 4 | experimental | 4-[(3R)-3-{[2-(4-FLUOROPHENYL)-2-OXOETHYL]AMINO}BUTYL]BENZAMIDE |  |  |
| DB08743 | dipeptidyl-peptidase 4 | experimental | 2-({8-[(3R)-3-AMINOPIPERIDIN-1-YL]-1,3-DIMETHYL-2,6-DIOXO-1,2,3,6-TETRAHYDRO-7H-PURIN-7-YL}METHYL)BENZONITRILE |  |  |
| Top |
| Cross referenced IDs for DPP4 |
| * We obtained these cross-references from Uniprot database. It covers 150 different DBs, 18 categories. http://www.uniprot.org/help/cross_references_section |
: Open all cross reference information
|
Copyright © 2016-Present - The Univsersity of Texas Health Science Center at Houston @ |